

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137838.10 - phase: 0
(500 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
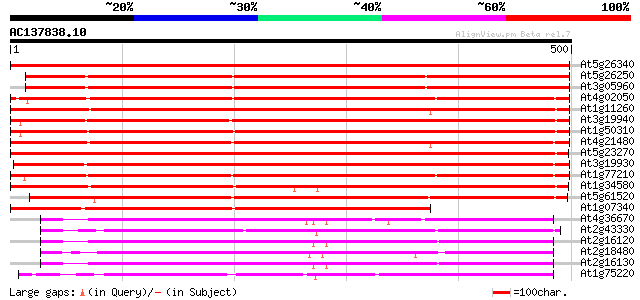
Score E
Sequences producing significant alignments: (bits) Value
At5g26340 hexose transporter - like protein 736 0.0
At5g26250 hexose transporter - like protein 587 e-168
At3g05960 putative hexose transporter 585 e-167
At4g02050 putative hexose transporter 583 e-167
At1g11260 glucose transporter 577 e-165
At3g19940 putative monosaccharide transport protein, STP4 552 e-157
At1g50310 hexose transporter, putative 546 e-155
At4g21480 glucose transporter 541 e-154
At5g23270 monosaccharide transporter 538 e-153
At3g19930 monosaccharide transport protein, STP4 528 e-150
At1g77210 unknown protein 515 e-146
At1g34580 monosaccharide transporter like protein 466 e-131
At5g61520 monosaccharide transporter STP3 460 e-130
At1g07340 hexose transporter like protein 408 e-114
At4g36670 sugar transporter like protein 201 1e-51
At2g43330 membrane transporter like protein 201 1e-51
At2g16120 putative sugar transporter 199 4e-51
At2g18480 putative sugar transporter 195 4e-50
At2g16130 putative sugar transporter 195 6e-50
At1g75220 integral membrane protein, putative 194 1e-49
>At5g26340 hexose transporter - like protein
Length = 526
Score = 736 bits (1899), Expect = 0.0
Identities = 358/499 (71%), Positives = 425/499 (84%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
M GGGF T ++ V FEA+IT V+ISCIMAATGGLMFGYDVG+SGGVTSMP FL+KFFP
Sbjct: 1 MTGGGFATSANGVEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMPDFLEKFFPV 60
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
VY++ +SNYCKYDNQ LQLFTSSLYLA L A+ AS TR LGR+ TML+AG+
Sbjct: 61 VYRKVVAGADKDSNYCKYDNQGLQLFTSSLYLAGLTATFFASYTTRTLGRRLTMLIAGVF 120
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNIT 180
FI+G L+A A L +LI GRILLGCGVGFANQAVP+FLSEIAPTRIRG LNI+FQLN+T
Sbjct: 121 FIIGVALNAGAQDLAMLIAGRILLGCGVGFANQAVPLFLSEIAPTRIRGGLNILFQLNVT 180
Query: 181 IGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGK 240
IGI ANLVN+ T+KIKGG+GWR+SL A IPA++LT+G+L+V +TPNSL+ERG ++GK
Sbjct: 181 IGILFANLVNYGTAKIKGGWGWRLSLGLAGIPALLLTVGALLVTETPNSLVERGRLDEGK 240
Query: 241 AVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGI 300
AVL +IRG +N+EPEF D+L AS++A EVK PF++L++ NRP L+IA+ +Q+FQQCTGI
Sbjct: 241 AVLRRIRGTDNVEPEFADLLEASRLAKEVKHPFRNLLQRRNRPQLVIAVALQIFQQCTGI 300
Query: 301 NAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMF 360
NAIMFYAPVLFSTLGF +DASLYS+V+TG VNVL TLVS+Y VDK GRRVLLLEA VQMF
Sbjct: 301 NAIMFYAPVLFSTLGFGSDASLYSAVVTGAVNVLSTLVSIYSVDKVGRRVLLLEAGVQMF 360
Query: 361 VSQVVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLET 420
SQVVI I+LG K+ D S +LSKG+A+LVVVM+CT+VA+FAWSWGPLGWLIPSETFPLET
Sbjct: 361 FSQVVIAIILGVKVTDTSTNLSKGFAILVVVMICTYVAAFAWSWGPLGWLIPSETFPLET 420
Query: 421 RSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIP 480
RSAGQSVTV N+LFTF+IAQAFLS+LC FKFGIF+FFSAWV +M VF +FL+PETKNIP
Sbjct: 421 RSAGQSVTVCVNLLFTFIIAQAFLSMLCHFKFGIFIFFSAWVLIMSVFVMFLLPETKNIP 480
Query: 481 IEDMAETVWKQHWFWRRFM 499
IE+M E VWK+HWFW RFM
Sbjct: 481 IEEMTERVWKKHWFWARFM 499
>At5g26250 hexose transporter - like protein
Length = 507
Score = 587 bits (1514), Expect = e-168
Identities = 290/485 (59%), Positives = 371/485 (75%), Gaps = 4/485 (0%)
Query: 15 FEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESN 74
F+A++T V I I+AA GGL+FGYD+GISGGVT+M FL++FFP VY+R + E+N
Sbjct: 14 FDAKMTVYVFICVIIAAVGGLIFGYDIGISGGVTAMDDFLKEFFPSVYERKKH--AHENN 71
Query: 75 YCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGKL 134
YCKYDNQ LQLFTSSLYLAALVAS AS KLGR+ TM LA I F++G L+A A +
Sbjct: 72 YCKYDNQFLQLFTSSLYLAALVASFFASATCSKLGRRPTMQLASIFFLIGVGLAAGAVNI 131
Query: 135 ILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTS 194
+LI GRILLG GVGF NQAVP+FLSEIAP R+RG LNI+FQL +TIGI IAN+VN+FTS
Sbjct: 132 YMLIIGRILLGFGVGFGNQAVPLFLSEIAPARLRGGLNIVFQLMVTIGILIANIVNYFTS 191
Query: 195 KIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGVENIEP 254
I YGWR++L GA IPA++L GSL++ +TP SLIER ++GK L KIRGVE+++
Sbjct: 192 SIHP-YGWRIALGGAGIPALILLFGSLLICETPTSLIERNKTKEGKETLKKIRGVEDVDE 250
Query: 255 EFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINAIMFYAPVLFSTL 314
E+E I+ A +A +VK P+ L+K +RPP +I + +Q FQQ TGINAIMFYAPVLF T+
Sbjct: 251 EYESIVHACDIARQVKDPYTKLMKPASRPPFVIGMLLQFFQQFTGINAIMFYAPVLFQTV 310
Query: 315 GFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGAKL 374
GF NDA+L S+V+TG +NVL T V ++ VDK GRR LLL++ V M + Q+VIGI+L AK
Sbjct: 311 GFGNDAALLSAVVTGTINVLSTFVGIFLVDKTGRRFLLLQSSVHMLICQLVIGIIL-AKD 369
Query: 375 QDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNML 434
D + +L++ A++VV+ VC +V FAWSWGPLGWLIPSETFPLETR+ G ++ V NM
Sbjct: 370 LDVTGTLARPQALVVVIFVCVYVMGFAWSWGPLGWLIPSETFPLETRTEGFALAVSCNMF 429
Query: 435 FTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDMAETVWKQHWF 494
FTF+IAQAFLS+LC K GIF FFS W+ VMG+F +F +PETK + I+DM ++VWK HW+
Sbjct: 430 FTFVIAQAFLSMLCAMKSGIFFFFSGWIVVMGLFALFFVPETKGVSIDDMRDSVWKLHWY 489
Query: 495 WRRFM 499
W+RFM
Sbjct: 490 WKRFM 494
>At3g05960 putative hexose transporter
Length = 507
Score = 585 bits (1509), Expect = e-167
Identities = 290/485 (59%), Positives = 372/485 (75%), Gaps = 4/485 (0%)
Query: 15 FEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESN 74
FEA++T V I ++AA GGL+FGYD+GISGGV++M FL++FFP V++R + V E+N
Sbjct: 13 FEAKMTVYVFICVMIAAVGGLIFGYDIGISGGVSAMDDFLKEFFPAVWERKKH--VHENN 70
Query: 75 YCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGKL 134
YCKYDNQ LQLFTSSLYLAALVAS +AS KLGR+ TM A I F++G L+A A L
Sbjct: 71 YCKYDNQFLQLFTSSLYLAALVASFVASATCSKLGRRPTMQFASIFFLIGVGLTAGAVNL 130
Query: 135 ILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTS 194
++LI GR+ LG GVGF NQAVP+FLSEIAP ++RG LNI+FQL +TIGI IAN+VN+FT+
Sbjct: 131 VMLIIGRLFLGFGVGFGNQAVPLFLSEIAPAQLRGGLNIVFQLMVTIGILIANIVNYFTA 190
Query: 195 KIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGVENIEP 254
+ YGWR++L GA IPAV+L GSL++ +TP SLIER E+GK L KIRGV++I
Sbjct: 191 TVHP-YGWRIALGGAGIPAVILLFGSLLIIETPTSLIERNKNEEGKEALRKIRGVDDIND 249
Query: 255 EFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINAIMFYAPVLFSTL 314
E+E I+ A +A++VK P++ L+K +RPP II + +Q+FQQ TGINAIMFYAPVLF T+
Sbjct: 250 EYESIVHACDIASQVKDPYRKLLKPASRPPFIIGMLLQLFQQFTGINAIMFYAPVLFQTV 309
Query: 315 GFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGAKL 374
GF +DA+L S+VITG +NVL T V +Y VD+ GRR LLL++ V M + Q++IGI+L AK
Sbjct: 310 GFGSDAALLSAVITGSINVLATFVGIYLVDRTGRRFLLLQSSVHMLICQLIIGIIL-AKD 368
Query: 375 QDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNML 434
+ +L + A++VV+ VC +V FAWSWGPLGWLIPSETFPLETRSAG +V V NM
Sbjct: 369 LGVTGTLGRPQALVVVIFVCVYVMGFAWSWGPLGWLIPSETFPLETRSAGFAVAVSCNMF 428
Query: 435 FTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDMAETVWKQHWF 494
FTF+IAQAFLS+LC + GIF FFS W+ VMG+F F IPETK I I+DM E+VWK HWF
Sbjct: 429 FTFVIAQAFLSMLCGMRSGIFFFFSGWIIVMGLFAFFFIPETKGIAIDDMRESVWKPHWF 488
Query: 495 WRRFM 499
W+R+M
Sbjct: 489 WKRYM 493
>At4g02050 putative hexose transporter
Length = 513
Score = 583 bits (1504), Expect = e-167
Identities = 279/506 (55%), Positives = 384/506 (75%), Gaps = 15/506 (2%)
Query: 1 MAGGGFTTGSSDVV------FEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFL 54
MAGG F G + V ++ ++T+ V+I+C++AA GG +FGYD+GISGGVTSM FL
Sbjct: 1 MAGGSF--GPTGVAKERAEQYQGKVTSYVIIACLVAAIGGSIFGYDIGISGGVTSMDEFL 58
Query: 55 QKFFPDVY-KRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQT 113
++FF VY K+ Q H ESNYCKYDNQ L FTSSLYLA LV++++ASP+TR GR+ +
Sbjct: 59 EEFFHTVYEKKKQAH---ESNYCKYDNQGLAAFTSSLYLAGLVSTLVASPITRNYGRRAS 115
Query: 114 MLLAGILFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNI 173
++ GI F++G+ L+A A L +L+ GRI+LG G+GF NQAVP++LSE+APT +RG LN+
Sbjct: 116 IVCGGISFLIGSGLNAGAVNLAMLLAGRIMLGVGIGFGNQAVPLYLSEVAPTHLRGGLNM 175
Query: 174 MFQLNITIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIER 233
MFQL TIGIF AN+VN+ T ++K +GWR+SL A PA+++T+G + +TPNSL+ER
Sbjct: 176 MFQLATTIGIFTANMVNYGTQQLKP-WGWRLSLGLAAFPALLMTLGGYFLPETPNSLVER 234
Query: 234 GFEEKGKAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQV 293
G E+G+ VL K+RG EN+ E +D++ AS++AN +K PF+++++ +RP L++AICM +
Sbjct: 235 GLTERGRRVLVKLRGTENVNAELQDMVDASELANSIKHPFRNILQKRHRPQLVMAICMPM 294
Query: 294 FQQCTGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLL 353
FQ TGIN+I+FYAPVLF T+GF +ASLYSS +TG V VL T +S+ VD+ GRR LL+
Sbjct: 295 FQILTGINSILFYAPVLFQTMGFGGNASLYSSALTGAVLVLSTFISIGLVDRLGRRALLI 354
Query: 354 EACVQMFVSQVVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPS 413
+QM + QV++ ++LG K D+ + LSKGY+++VV+ +C FV +F WSWGPLGW IPS
Sbjct: 355 TGGIQMIICQVIVAVILGVKFGDNQE-LSKGYSVIVVIFICLFVVAFGWSWGPLGWTIPS 413
Query: 414 ETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLI 473
E FPLETRSAGQS+TV N+LFTF+IAQAFL LLC FKFGIFLFF+ WV VM +F FL+
Sbjct: 414 EIFPLETRSAGQSITVAVNLLFTFIIAQAFLGLLCAFKFGIFLFFAGWVTVMTIFVYFLL 473
Query: 474 PETKNIPIEDMAETVWKQHWFWRRFM 499
PETK +PIE+M +W +HWFW++ +
Sbjct: 474 PETKGVPIEEMT-LLWSKHWFWKKVL 498
>At1g11260 glucose transporter
Length = 522
Score = 577 bits (1487), Expect = e-165
Identities = 276/501 (55%), Positives = 375/501 (74%), Gaps = 4/501 (0%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
M GGF G + ++T V+ +C++AA GGL+FGYD+GISGGVTSMPSFL++FFP
Sbjct: 1 MPAGGFVVGDGQKAYPGKLTPFVLFTCVVAAMGGLIFGYDIGISGGVTSMPSFLKRFFPS 60
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
VY++ QE + YC+YD+ L +FTSSLYLAAL++S++AS VTRK GR+ +ML GIL
Sbjct: 61 VYRKQQEDAST-NQYCQYDSPTLTMFTSSLYLAALISSLVASTVTRKFGRRLSMLFGGIL 119
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNIT 180
F G +++ A + +LI GRILLG G+GFANQAVP++LSE+AP + RGALNI FQL+IT
Sbjct: 120 FCAGALINGFAKHVWMLIVGRILLGFGIGFANQAVPLYLSEMAPYKYRGALNIGFQLSIT 179
Query: 181 IGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGK 240
IGI +A ++N+F +KIKGG+GWR+SL GA++PA+++T+GSL++ DTPNS+IERG E+ K
Sbjct: 180 IGILVAEVLNYFFAKIKGGWGWRLSLGGAVVPALIITIGSLVLPDTPNSMIERGQHEEAK 239
Query: 241 AVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGI 300
L +IRGV+++ EF+D++ ASK + ++ P+++L++ RP L +A+ + FQQ TGI
Sbjct: 240 TKLRRIRGVDDVSQEFDDLVAASKESQSIEHPWRNLLRRKYRPHLTMAVMIPFFQQLTGI 299
Query: 301 NAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMF 360
N IMFYAPVLF+T+GF DASL S+V+TG VNV TLVS+Y VD+ GRR L LE QM
Sbjct: 300 NVIMFYAPVLFNTIGFTTDASLMSAVVTGSVNVAATLVSIYGVDRWGRRFLFLEGGTQML 359
Query: 361 VSQVVIGIVLGAK--LQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPL 418
+ Q V+ +GAK + L K YA++VV +C +VA FAWSWGPLGWL+PSE FPL
Sbjct: 360 ICQAVVAACIGAKFGVDGTPGELPKWYAIVVVTFICIYVAGFAWSWGPLGWLVPSEIFPL 419
Query: 419 ETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKN 478
E RSA QS+TV NM+FTF+IAQ FL++LC KFG+FL F+ +V VM +F +PETK
Sbjct: 420 EIRSAAQSITVSVNMIFTFIIAQIFLTMLCHLKFGLFLVFAFFVVVMSIFVYIFLPETKG 479
Query: 479 IPIEDMAETVWKQHWFWRRFM 499
IPIE+M + VW+ HW+W RF+
Sbjct: 480 IPIEEMGQ-VWRSHWYWSRFV 499
>At3g19940 putative monosaccharide transport protein, STP4
Length = 514
Score = 552 bits (1422), Expect = e-157
Identities = 274/502 (54%), Positives = 373/502 (73%), Gaps = 6/502 (1%)
Query: 1 MAGGGFTT--GSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFF 58
MAGG F + G +E +TA V+++CI+AA GGL+FGYD+GISGGVTSM FL KFF
Sbjct: 1 MAGGAFVSEGGGGGRSYEGGVTAFVIMTCIVAAMGGLLFGYDLGISGGVTSMEEFLTKFF 60
Query: 59 PDVYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAG 118
P V + ++ ++ YCK+DNQ LQLFTSSLYLAALVAS +AS +TRK GRK +M + G
Sbjct: 61 PQVESQMKK-AKHDTAYCKFDNQMLQLFTSSLYLAALVASFMASVITRKHGRKVSMFIGG 119
Query: 119 ILFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLN 178
+ F++G + +A A + +LI GR+LLG GVGFANQ+ PV+LSE+AP +IRGALNI FQ+
Sbjct: 120 LAFLIGALFNAFAVNVSMLIIGRLLLGVGVGFANQSTPVYLSEMAPAKIRGALNIGFQMA 179
Query: 179 ITIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEK 238
ITIGI +ANL+N+ TSK+ +GWRVSL A +PAV++ +GS I+ DTPNS++ERG E+
Sbjct: 180 ITIGILVANLINYGTSKM-AQHGWRVSLGLAAVPAVVMVIGSFILPDTPNSMLERGKNEE 238
Query: 239 GKAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCT 298
K +L KIRG +N++ EF+D++ A + A +V++P+K++++S RP LI + FQQ T
Sbjct: 239 AKQMLKKIRGADNVDHEFQDLIDAVEAAKKVENPWKNIMESKYRPALIFCSAIPFFQQIT 298
Query: 299 GINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQ 358
GIN IMFYAPVLF TLGF +DA+L S+VITG VN+L T VS+Y VD+ GRR+L LE +Q
Sbjct: 299 GINVIMFYAPVLFKTLGFGDDAALMSAVITGVVNMLSTFVSIYAVDRYGRRLLFLEGGIQ 358
Query: 359 MFVSQVVIGIVLGAKL-QDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFP 417
MF+ Q+++G +GA+ + +L+ A ++ +C +VA FAWSWGPLGWL+PSE P
Sbjct: 359 MFICQLLVGSFIGARFGTSGTGTLTPATADWILAFICVYVAGFAWSWGPLGWLVPSEICP 418
Query: 418 LETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETK 477
LE R AGQ++ V NM FTFLI Q FL++LC KFG+F FF++ V +M VF FL+PETK
Sbjct: 419 LEIRPAGQAINVSVNMFFTFLIGQFFLTMLCHMKFGLFYFFASMVAIMTVFIYFLLPETK 478
Query: 478 NIPIEDMAETVWKQHWFWRRFM 499
+PIE+M VWKQHWFW++++
Sbjct: 479 GVPIEEMGR-VWKQHWFWKKYI 499
>At1g50310 hexose transporter, putative
Length = 517
Score = 546 bits (1407), Expect = e-155
Identities = 278/503 (55%), Positives = 367/503 (72%), Gaps = 7/503 (1%)
Query: 1 MAGGGFTT--GSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFF 58
MAGG F + G +E +T V+++CI+AA GGL+FGYD+GISGGVTSM FL KFF
Sbjct: 1 MAGGAFVSEGGGGGNSYEGGVTVFVIMTCIVAAMGGLLFGYDLGISGGVTSMEEFLSKFF 60
Query: 59 PDVYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAG 118
P+V K+ E E+ YCK+DNQ LQLFTSSLYLAAL +S +AS VTRK GRK +M + G
Sbjct: 61 PEVDKQMHEAR-RETAYCKFDNQLLQLFTSSLYLAALASSFVASAVTRKYGRKISMFVGG 119
Query: 119 ILFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLN 178
+ F++G++ +A A + +LI GR+LLG GVGFANQ+ PV+LSE+AP +IRGALNI FQ+
Sbjct: 120 VAFLIGSLFNAFATNVAMLIVGRLLLGVGVGFANQSTPVYLSEMAPAKIRGALNIGFQMA 179
Query: 179 ITIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEK 238
ITIGI IANL+N+ TS++ GWRVSL A +PAV++ +GS ++ DTPNS++ERG E+
Sbjct: 180 ITIGILIANLINYGTSQMAKN-GWRVSLGLAAVPAVIMVIGSFVLPDTPNSMLERGKYEQ 238
Query: 239 GKAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLV-KSHNRPPLIIAICMQVFQQC 297
+ +L KIRG +N++ EF+D+ A + A +V +P+K++ ++ RP L+ + FQQ
Sbjct: 239 AREMLQKIRGADNVDEEFQDLCDACEAAKKVDNPWKNIFQQAKYRPALVFCSAIPFFQQI 298
Query: 298 TGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACV 357
TGIN IMFYAPVLF TLGF +DASL S+VITG VNV+ TLVS+Y VD+ GRR+L LE +
Sbjct: 299 TGINVIMFYAPVLFKTLGFADDASLISAVITGAVNVVSTLVSIYAVDRYGRRILFLEGGI 358
Query: 358 QMFVSQVVIGIVLGAKL-QDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETF 416
QM VSQ+V+G ++G K S +L+ A ++ +C +VA FAWSWGPLGWL+PSE
Sbjct: 359 QMIVSQIVVGTLIGMKFGTTGSGTLTPATADWILAFICLYVAGFAWSWGPLGWLVPSEIC 418
Query: 417 PLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPET 476
PLE R AGQ++ V NM FTFLI Q FL++LC KFG+F FF V VM VF FL+PET
Sbjct: 419 PLEIRPAGQAINVSVNMFFTFLIGQFFLTMLCHMKFGLFYFFGGMVAVMTVFIYFLLPET 478
Query: 477 KNIPIEDMAETVWKQHWFWRRFM 499
K +PIE+M VWKQH FW+R+M
Sbjct: 479 KGVPIEEMGR-VWKQHPFWKRYM 500
>At4g21480 glucose transporter
Length = 508
Score = 541 bits (1394), Expect = e-154
Identities = 271/502 (53%), Positives = 369/502 (72%), Gaps = 8/502 (1%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
M G G + ++T V ++CI+AA GGL+FGYD+GISGGVT+M SF QKFFP
Sbjct: 1 MPSVGIVIGDGKKEYPGKLTLYVTVTCIVAAMGGLIFGYDIGISGGVTTMDSFQQKFFPS 60
Query: 61 VY-KRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGI 119
VY K+ ++H + YC++D+ L LFTSSLYLAAL +S++AS VTR+ GRK +MLL G+
Sbjct: 61 VYEKQKKDHD--SNQYCRFDSVSLTLFTSSLYLAALCSSLVASYVTRQFGRKISMLLGGV 118
Query: 120 LFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNI 179
LF G +L+ A + +LI GR+LLG G+GF NQ+VP++LSE+AP + RGALNI FQL+I
Sbjct: 119 LFCAGALLNGFATAVWMLIVGRLLLGFGIGFTNQSVPLYLSEMAPYKYRGALNIGFQLSI 178
Query: 180 TIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKG 239
TIGI +AN++N+F SKI +GWR+SL GA++PA+++T+GSLI+ DTPNS+IERG
Sbjct: 179 TIGILVANVLNFFFSKIS--WGWRLSLGGAVVPALIITVGSLILPDTPNSMIERGQFRLA 236
Query: 240 KAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTG 299
+A L KIRGV++I+ E D++ AS+ + V+ P+++L++ RP L +AI + FQQ TG
Sbjct: 237 EAKLRKIRGVDDIDDEINDLIIASEASKLVEHPWRNLLQRKYRPHLTMAILIPAFQQLTG 296
Query: 300 INAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQM 359
IN IMFYAPVLF T+GF +DA+L S+V+TG VNV T+VS+Y VDK GRR L LE QM
Sbjct: 297 INVIMFYAPVLFQTIGFGSDAALISAVVTGLVNVGATVVSIYGVDKWGRRFLFLEGGFQM 356
Query: 360 FVSQVVIGIVLGAK--LQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFP 417
+SQV + +GAK + L K YA++VV+ +C +VA+FAWSWGPLGWL+PSE FP
Sbjct: 357 LISQVAVAAAIGAKFGVDGTPGVLPKWYAIVVVLFICIYVAAFAWSWGPLGWLVPSEIFP 416
Query: 418 LETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETK 477
LE RSA QS+TV NM+FTFLIAQ FL +LC KFG+F+FF+ +V VM +F +PET+
Sbjct: 417 LEIRSAAQSITVSVNMIFTFLIAQVFLMMLCHLKFGLFIFFAFFVVVMSIFVYLFLPETR 476
Query: 478 NIPIEDMAETVWKQHWFWRRFM 499
+PIE+M VW+ HW+W +F+
Sbjct: 477 GVPIEEM-NRVWRSHWYWSKFV 497
>At5g23270 monosaccharide transporter
Length = 514
Score = 538 bits (1385), Expect = e-153
Identities = 271/500 (54%), Positives = 358/500 (71%), Gaps = 3/500 (0%)
Query: 1 MAGGGFTTGSSDVV-FEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFP 59
MAGG F S +E R+TA V+I+CI+AA GGL+FGYD+GISGGV SM FL KFFP
Sbjct: 1 MAGGAFIDESGHGGDYEGRVTAFVMITCIVAAMGGLLFGYDIGISGGVISMEDFLTKFFP 60
Query: 60 DVYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGI 119
DV ++ Q E+ YCKYDN+ L LFTSSLYLAAL AS +AS +TR GRK +M++ +
Sbjct: 61 DVLRQMQNKRGRETEYCKYDNELLTLFTSSLYLAALFASFLASTITRLFGRKVSMVIGSL 120
Query: 120 LFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNI 179
F+ G +L+ A L +LI GR+ LG GVGFANQ+VP++LSE+AP +IRGALNI FQL I
Sbjct: 121 AFLSGALLNGLAINLEMLIIGRLFLGVGVGFANQSVPLYLSEMAPAKIRGALNIGFQLAI 180
Query: 180 TIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKG 239
TIGI AN+VN+ T K++ G GWR+SL A +PAVM+ +G + DTPNS++ERG +EK
Sbjct: 181 TIGILAANIVNYVTPKLQNGIGWRLSLGLAGVPAVMMLVGCFFLPDTPNSILERGNKEKA 240
Query: 240 KAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTG 299
K +L KIRG +E EF ++ A + A +VK P+ +++++ RP L + FQQ TG
Sbjct: 241 KEMLQKIRGTMEVEHEFNELCNACEAAKKVKHPWTNIMQARYRPQLTFCTFIPFFQQLTG 300
Query: 300 INAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQM 359
IN IMFYAPVLF T+GF NDASL S+VITG VNVL T+VS+Y VDK GRR L L+ QM
Sbjct: 301 INVIMFYAPVLFKTIGFGNDASLISAVITGLVNVLSTIVSIYSVDKFGRRALFLQGGFQM 360
Query: 360 FVSQVVIGIVLGAKLQDHSD-SLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPL 418
V+Q+ +G ++G K + + +LS A +++ ++C +VA FAWSWGPLGWL+PSE PL
Sbjct: 361 IVTQIAVGSMIGWKFGFNGEGNLSGVDADIILALICLYVAGFAWSWGPLGWLVPSEICPL 420
Query: 419 ETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKN 478
E RSAGQS+ V NM FTF I Q FL++LC KFG+F FF+ V +M +F FL+PETK
Sbjct: 421 EIRSAGQSLNVSVNMFFTFFIGQFFLTMLCHMKFGLFYFFAGMVLIMTIFIYFLLPETKG 480
Query: 479 IPIEDMAETVWKQHWFWRRF 498
+PIE+M + VWK+H +W ++
Sbjct: 481 VPIEEMGK-VWKEHRYWGKY 499
>At3g19930 monosaccharide transport protein, STP4
Length = 514
Score = 528 bits (1361), Expect = e-150
Identities = 256/498 (51%), Positives = 359/498 (71%), Gaps = 5/498 (1%)
Query: 4 GGFTTGSSDVV-FEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVY 62
GGF + + V + ++T V ++C + A GGL+FGYD+GISGGVTSM FL++FFP VY
Sbjct: 3 GGFVSQTPGVRNYNYKLTPKVFVTCFIGAFGGLIFGYDLGISGGVTSMEPFLEEFFPYVY 62
Query: 63 KRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFI 122
K+ + + E+ YC++D+Q L LFTSSLY+AALV+S+ AS +TR GRK +M L G F
Sbjct: 63 KKMK--SAHENEYCRFDSQLLTLFTSSLYVAALVSSLFASTITRVFGRKWSMFLGGFTFF 120
Query: 123 VGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIG 182
+G+ + A + +L+ GRILLG GVGFANQ+VPV+LSE+AP +RGA N FQ+ I G
Sbjct: 121 IGSAFNGFAQNIAMLLIGRILLGFGVGFANQSVPVYLSEMAPPNLRGAFNNGFQVAIIFG 180
Query: 183 IFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAV 242
I +A ++N+FT+++KG GWR+SL A +PAVM+ +G+LI+ DTPNSLIERG+ E+ K +
Sbjct: 181 IVVATIINYFTAQMKGNIGWRISLGLACVPAVMIMIGALILPDTPNSLIERGYTEEAKEM 240
Query: 243 LTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINA 302
L IRG ++ EF+D++ AS+ + +VK P+K+++ RP LI+ + FQQ TGIN
Sbjct: 241 LQSIRGTNEVDEEFQDLIDASEESKQVKHPWKNIMLPRYRPQLIMTCFIPFFQQLTGINV 300
Query: 303 IMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVS 362
I FYAPVLF TLGF + ASL S+++TG + +LCT VSV+ VD+ GRR+L L+ +QM VS
Sbjct: 301 ITFYAPVLFQTLGFGSKASLLSAMVTGIIELLCTFVSVFTVDRFGRRILFLQGGIQMLVS 360
Query: 363 QVVIGIVLGAKL-QDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETR 421
Q+ IG ++G K + ++ K A L+V ++C +VA FAWSWGPLGWL+PSE PLE R
Sbjct: 361 QIAIGAMIGVKFGVAGTGNIGKSDANLIVALICIYVAGFAWSWGPLGWLVPSEISPLEIR 420
Query: 422 SAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPI 481
SA Q++ V NM FTFL+AQ FL++LC KFG+F FF+ +V +M +F ++PETKN+PI
Sbjct: 421 SAAQAINVSVNMFFTFLVAQLFLTMLCHMKFGLFFFFAFFVVIMTIFIYLMLPETKNVPI 480
Query: 482 EDMAETVWKQHWFWRRFM 499
E+M VWK HWFW +F+
Sbjct: 481 EEM-NRVWKAHWFWGKFI 497
>At1g77210 unknown protein
Length = 504
Score = 515 bits (1326), Expect = e-146
Identities = 257/503 (51%), Positives = 358/503 (71%), Gaps = 8/503 (1%)
Query: 1 MAGGGFTTGSS---DVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKF 57
MAGG T ++E RIT+ + +CI+ + GG +FGYD+G+SGGVTSM FL++F
Sbjct: 1 MAGGALTDEGGLKRAHLYEHRITSYFIFACIVGSMGGSLFGYDLGVSGGVTSMDDFLKEF 60
Query: 58 FPDVYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLA 117
FP +YKR Q H + E++YCKYDNQ L LFTSSLY A L+++ AS VTR GR+ ++L+
Sbjct: 61 FPGIYKRKQMH-LNETDYCKYDNQILTLFTSSLYFAGLISTFGASYVTRIYGRRGSILVG 119
Query: 118 GILFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQL 177
+ F +G V++A+A +++LI GRI LG G+GF NQAVP++LSE+AP +IRG +N +FQL
Sbjct: 120 SVSFFLGGVINAAAKNILMLILGRIFLGIGIGFGNQAVPLYLSEMAPAKIRGTVNQLFQL 179
Query: 178 NITIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEE 237
IGI +ANL+N+ T +I +GWR+SL A +PA+++ +G L++ +TPNSL+E+G E
Sbjct: 180 TTCIGILVANLINYKTEQIH-PWGWRLSLGLATVPAILMFLGGLVLPETPNSLVEQGKLE 238
Query: 238 KGKAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLII-AICMQVFQQ 296
K KAVL K+RG NIE EF+D++ AS A VK+PF++L+ NRP L+I AI + FQQ
Sbjct: 239 KAKAVLIKVRGTNNIEAEFQDLVEASDAARAVKNPFRNLLARRNRPQLVIGAIGLPAFQQ 298
Query: 297 CTGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEAC 356
TG+N+I+FYAPV+F +LGF ASL SS IT V+ ++S+Y DK GRR LLLEA
Sbjct: 299 LTGMNSILFYAPVMFQSLGFGGSASLISSTITNAALVVAAIMSMYSADKFGRRFLLLEAS 358
Query: 357 VQMFVSQVVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETF 416
V+MF VV+G+ L K + + L K +++VV++C FV ++ SWGP+GWL+PSE F
Sbjct: 359 VEMFCYMVVVGVTLALKFGEGKE-LPKSLGLILVVLICLFVLAYGRSWGPMGWLVPSELF 417
Query: 417 PLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPET 476
PLETRSAGQSV V N+ FT LIAQ FL LC K+GIFL F+ + MG F FL+PET
Sbjct: 418 PLETRSAGQSVVVCVNLFFTALIAQCFLVSLCHLKYGIFLLFAGLILGMGSFVYFLLPET 477
Query: 477 KNIPIEDMAETVWKQHWFWRRFM 499
K +PIE++ +W+QHW W++++
Sbjct: 478 KQVPIEEV-YLLWRQHWLWKKYV 499
>At1g34580 monosaccharide transporter like protein
Length = 506
Score = 466 bits (1200), Expect = e-131
Identities = 237/505 (46%), Positives = 338/505 (66%), Gaps = 12/505 (2%)
Query: 1 MAGGGFTTG-SSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFP 59
MAGGG SS +A+ITAAVV+SCI+AA+ GL+FGYD+GISGGVT+M FL+KFFP
Sbjct: 1 MAGGGLALDVSSAGNIDAKITAAVVMSCIVAASCGLIFGYDIGISGGVTTMKPFLEKFFP 60
Query: 60 DVYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGI 119
V K+ E + YC YD+Q L FTSSLY+A LVAS++AS +T GR+ TM+L G
Sbjct: 61 SVLKKASEAKT--NVYCVYDSQLLTAFTSSLYVAGLVASLVASRLTAAYGRRTTMILGGF 118
Query: 120 LFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNI 179
F+ G +++ A + +LI GRILLG GVGF NQA PV+LSE+AP R RGA NI F I
Sbjct: 119 TFLFGALINGLAANIAMLISGRILLGFGVGFTNQAAPVYLSEVAPPRWRGAFNIGFSCFI 178
Query: 180 TIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKG 239
++G+ ANL+N+ T + G WR+SL A +PA ++T+G L + DTP+SL+ RG ++
Sbjct: 179 SMGVVAANLINYGTDSHRNG--WRISLGLAAVPAAIMTVGCLFISDTPSSLLARGKHDEA 236
Query: 240 KAVLTKIRGVENI---EPEFEDILRASKVANEVKSPF--KDLVKSHNRPPLIIAICMQVF 294
L K+RGVENI E E +++R+S++A E ++ K +++ RP L++A+ + F
Sbjct: 237 HTSLLKLRGVENIADVETELAELVRSSQLAIEARAELFMKTILQRRYRPHLVVAVVIPCF 296
Query: 295 QQCTGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLE 354
QQ TGI FYAPVLF ++GF + +L ++ I G VN+ L+S +D+ GRR L +
Sbjct: 297 QQLTGITVNAFYAPVLFRSVGFGSGPALIATFILGFVNLGSLLLSTMVIDRFGRRFLFIA 356
Query: 355 ACVQMFVSQVVIGIVLGAKLQDHSDS-LSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPS 413
+ M + Q+ + ++L + D + KGYA+ VVV++C + A F WSWGPL WL+PS
Sbjct: 357 GGILMLLCQIAVAVLLAVTVGATGDGEMKKGYAVTVVVLLCIYAAGFGWSWGPLSWLVPS 416
Query: 414 ETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLI 473
E FPL+ R AGQS++V N TF ++Q FL+ LC FK+G FLF+ W+F M +F + +
Sbjct: 417 EIFPLKIRPAGQSLSVAVNFAATFALSQTFLATLCDFKYGAFLFYGGWIFTMTIFVIMFL 476
Query: 474 PETKNIPIEDMAETVWKQHWFWRRF 498
PETK IP++ M + VW++HW+W+RF
Sbjct: 477 PETKGIPVDSMYQ-VWEKHWYWQRF 500
>At5g61520 monosaccharide transporter STP3
Length = 514
Score = 460 bits (1183), Expect = e-130
Identities = 233/485 (48%), Positives = 327/485 (67%), Gaps = 9/485 (1%)
Query: 18 RITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESN--- 74
+IT VV SC+MAA GG++FGYD+G+SGGV SM FL++FFP VYK +E N
Sbjct: 19 KITYFVVASCVMAAMGGVIFGYDIGVSGGVMSMGPFLKRFFPKVYKLQEEDRRRRGNSNN 78
Query: 75 -YCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGK 133
YC +++Q L FTSSLY++ L+A+++AS VTR GRK ++ L G+ F+ G L SA
Sbjct: 79 HYCLFNSQLLTSFTSSLYVSGLIATLLASSVTRSWGRKPSIFLGGVSFLAGAALGGSAQN 138
Query: 134 LILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFT 193
+ +LI R+LLG GVGFANQ+VP++LSE+AP + RGA++ FQL I IG AN++N+ T
Sbjct: 139 VAMLIIARLLLGVGVGFANQSVPLYLSEMAPAKYRGAISNGFQLCIGIGFLSANVINYET 198
Query: 194 SKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIER-GFEEKGKAVLTKIRGVENI 252
IK +GWR+SLA A IPA +LT+GSL + +TPNS+I+ G K + +L ++RG ++
Sbjct: 199 QNIK--HGWRISLATAAIPASILTLGSLFLPETPNSIIQTTGDVHKTELMLRRVRGTNDV 256
Query: 253 EPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINAIMFYAPVLFS 312
+ E D++ AS ++ + F L++ RP L++A+ + FQQ TGIN + FYAPVL+
Sbjct: 257 QDELTDLVEASSGSDTDSNAFLKLLQRKYRPELVMALVIPFFQQVTGINVVAFYAPVLYR 316
Query: 313 TLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGA 372
T+GF SL S+++TG V TL+S+ VD+ GR+ L L +QM VSQV IG+++
Sbjct: 317 TVGFGESGSLMSTLVTGIVGTSSTLLSMLVVDRIGRKTLFLIGGLQMLVSQVTIGVIVMV 376
Query: 373 KLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTN 432
H + +GY VVV+VC +VA F WSWGPLGWL+PSE FPLE RS QSVTV +
Sbjct: 377 A-DVHDGVIKEGYGYAVVVLVCVYVAGFGWSWGPLGWLVPSEIFPLEIRSVAQSVTVAVS 435
Query: 433 MLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDMAETVWKQH 492
+FTF +AQ+ +LC F+ GIF F+ W+ VM V +PETKN+PIE + +W++H
Sbjct: 436 FVFTFAVAQSAPPMLCKFRAGIFFFYGGWLVVMTVAVQLFLPETKNVPIEKVV-GLWEKH 494
Query: 493 WFWRR 497
WFWRR
Sbjct: 495 WFWRR 499
>At1g07340 hexose transporter like protein
Length = 383
Score = 408 bits (1048), Expect = e-114
Identities = 213/376 (56%), Positives = 275/376 (72%), Gaps = 5/376 (1%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
MA G F A++T V + C++AA GGLMFGYD+GISGGVTSM +FL FFP
Sbjct: 1 MAVGSMNVEEGTKAFPAKLTGQVFLCCVIAAVGGLMFGYDIGISGGVTSMDTFLLDFFPH 60
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
VY++ +H V E+NYCK+D+Q LQLFTSSLYLA + AS I+S V+R GRK T++LA I
Sbjct: 61 VYEK--KHRVHENNYCKFDDQLLQLFTSSLYLAGIFASFISSYVSRAFGRKPTIMLASIF 118
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNIT 180
F+VG +L+ SA +L +LI GRILLG G+GF NQ VP+F+SEIAP R RG LN+MFQ IT
Sbjct: 119 FLVGAILNLSAQELGMLIGGRILLGFGIGFGNQTVPLFISEIAPARYRGGLNVMFQFLIT 178
Query: 181 IGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGK 240
IGI A+ VN+ TS +K GWR SL GA +PA++L +GS + +TP SLIERG +EKGK
Sbjct: 179 IGILAASYVNYLTSTLKN--GWRYSLGGAAVPALILLIGSFFIHETPASLIERGKDEKGK 236
Query: 241 AVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDL-VKSHNRPPLIIAICMQVFQQCTG 299
VL KIRG+E+IE EF +I A++VA +VKSPFK+L KS NRPPL+ +Q FQQ TG
Sbjct: 237 QVLRKIRGIEDIELEFNEIKYATEVATKVKSPFKELFTKSENRPPLVCGTLLQFFQQFTG 296
Query: 300 INAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQM 359
IN +MFYAPVLF T+G ++ASL S+V+T GVN + T++S+ VD AGRR LL+E +QM
Sbjct: 297 INVVMFYAPVLFQTMGSGDNASLISTVVTNGVNAIATVISLLVVDFAGRRCLLMEGALQM 356
Query: 360 FVSQVVIGIVLGAKLQ 375
+Q+ IG +L A L+
Sbjct: 357 TATQMTIGGILLAHLK 372
>At4g36670 sugar transporter like protein
Length = 493
Score = 201 bits (510), Expect = 1e-51
Identities = 144/479 (30%), Positives = 238/479 (49%), Gaps = 47/479 (9%)
Query: 28 IMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESNYCKYDNQKLQLFT 87
I+A+ ++FGYD G+ G V K ++ ++++ T
Sbjct: 22 IVASIVSIIFGYDTGVMSGAM---------------------VFIEEDLKTNDVQIEVLT 60
Query: 88 SSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGKLILLIFGRILLGCG 147
L L ALV S++A + +GR+ T++LA ILF++G++L +L+ GR G G
Sbjct: 61 GILNLCALVGSLLAGRTSDIIGRRYTIVLASILFMLGSILMGWGPNYPVLLSGRCTAGLG 120
Query: 148 VGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTSKIKGGYGWRVSLA 207
VGFA PV+ +EIA RG L + L I+IGI + +VN+F SK+ GWR+ L
Sbjct: 121 VGFALMVAPVYSAEIATASHRGLLASLPHLCISIGILLGYIVNYFFSKLPMHIGWRLMLG 180
Query: 208 GAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKI-RGVENIEPEFEDILRAS--- 263
A +P+++L G L + ++P LI +G ++GK +L + E E F+DI A+
Sbjct: 181 IAAVPSLVLAFGILKMPESPRWLIMQGRLKEGKEILELVSNSPEEAELRFQDIKAAAGID 240
Query: 264 -KVANEV----------KSPFKDLVKSHN---RPPLIIAICMQVFQQCTGINAIMFYAPV 309
K ++V + +K+L+ R L+ A+ + FQ +GI A++ Y P
Sbjct: 241 PKCVDDVVKMEGKKTHGEGVWKELILRPTPAVRRVLLTALGIHFFQHASGIEAVLLYGPR 300
Query: 310 LFSTLGFHNDASLYSSVITGGVNVLCT---LVSVYFVDKAGRRVLLLEACVQMFVSQVVI 366
+F G L+ ++T GV ++ T + +DK GRR LLL + M ++ ++
Sbjct: 301 IFKKAGITTKDKLF--LVTIGVGIMKTTFIFTATLLLDKVGRRKLLLTSVGGMVIALTML 358
Query: 367 GIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQS 426
G G + ++ +L +V +FVA F+ GP+ W+ SE FPL+ R+ G S
Sbjct: 359 G--FGLTMAQNAGGKLAWALVLSIVAAYSFVAFFSIGLGPITWVYSSEVFPLKLRAQGAS 416
Query: 427 VTVFTNMLFTFLIAQAFLSLL-CLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDM 484
+ V N + ++ +FLSL + G F F+ V F FL+PETK +E++
Sbjct: 417 LGVAVNRVMNATVSMSFLSLTSAITTGGAFFMFAGVAAVAWNFFFFLLPETKGKSLEEI 475
>At2g43330 membrane transporter like protein
Length = 509
Score = 201 bits (510), Expect = 1e-51
Identities = 126/469 (26%), Positives = 234/469 (49%), Gaps = 28/469 (5%)
Query: 28 IMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESNYCKYDNQKLQLFT 87
+ A GGL+FGYD G+ G +Y + V +S++ + +
Sbjct: 36 VTAGIGGLLFGYDTGVISGAL------------LYIKDDFEVVKQSSFLQ------ETIV 77
Query: 88 SSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGKLILLIFGRILLGCG 147
S + A++ + + GRK+ L A ++F G ++ A+A +LI GR+L+G G
Sbjct: 78 SMALVGAMIGAAAGGWINDYYGRKKATLFADVVFAAGAIVMAAAPDPYVLISGRLLVGLG 137
Query: 148 VGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTSKIKGGYGWRVSLA 207
VG A+ PV+++E +P+ +RG L L IT G F++ LVN +++ G + W + ++
Sbjct: 138 VGVASVTAPVYIAEASPSEVRGGLVSTNVLMITGGQFLSYLVNSAFTQVPGTWRWMLGVS 197
Query: 208 GAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGVENIEPEFEDILRASKVAN 267
G +PAV+ + L + ++P L + + + VL + + +E E + + A +
Sbjct: 198 G--VPAVIQFILMLFMPESPRWLFMKNRKAEAIQVLARTYDISRLEDEIDHLSAAEEEEK 255
Query: 268 EVKSP--FKDLVKSHN-RPPLIIAICMQVFQQCTGINAIMFYAPVLFSTLGFH-NDASLY 323
+ K + D+ +S R + +Q FQQ TGIN +M+Y+P + GFH N +L+
Sbjct: 256 QRKRTVGYLDVFRSKELRLAFLAGAGLQAFQQFTGINTVMYYSPTIVQMAGFHSNQLALF 315
Query: 324 SSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGAKLQDHSDSLSK 383
S+I +N T+V +YF+D GR+ L L + + +S +++ + + + SD
Sbjct: 316 LSLIVAAMNAAGTVVGIYFIDHCGRKKLALSSLFGVIISLLILSVSFFKQSETSSD--GG 373
Query: 384 GYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNMLFTFLIAQAF 443
Y L V+ + ++ FA GP+ W + SE +P + R ++ N + ++AQ F
Sbjct: 374 LYGWLAVLGLALYIVFFAPGMGPVPWTVNSEIYPQQYRGICGGMSATVNWISNLIVAQTF 433
Query: 444 LSLLCLFKFGIFLFFSAWVFVMGV-FTVFLIPETKNIPIEDMAETVWKQ 491
L++ G+ A + V+ V F + +PET+ + ++ E +WK+
Sbjct: 434 LTIAEAAGTGMTFLILAGIAVLAVIFVIVFVPETQGLTFSEV-EQIWKE 481
>At2g16120 putative sugar transporter
Length = 511
Score = 199 bits (505), Expect = 4e-51
Identities = 146/477 (30%), Positives = 228/477 (47%), Gaps = 42/477 (8%)
Query: 28 IMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESNYCKYDNQKLQLFT 87
I+A+ ++ GYD+G+ G + + + K + +L++
Sbjct: 31 ILASMTSIILGYDIGVMSGAS---------------------IFIKDDLKLSDVQLEILM 69
Query: 88 SSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGKLILLIFGRILLGCG 147
L + +LV S A + LGR+ T++LAG F G +L A ++ GR + G G
Sbjct: 70 GILNIYSLVGSGAAGRTSDWLGRRYTIVLAGAFFFCGALLMGFATNYPFIMVGRFVAGIG 129
Query: 148 VGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTSKIKGGYGWRVSLA 207
VG+A PV+ +E+AP RG L ++ I IGI + + N+F SK+ GWR L
Sbjct: 130 VGYAMMIAPVYTAEVAPASSRGFLTSFPEIFINIGILLGYVSNYFFSKLPEHLGWRFMLG 189
Query: 208 GAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGV-ENIEPEFEDILRASKVA 266
+P+V L +G L + ++P L+ +G VL K E +DI RA +
Sbjct: 190 VGAVPSVFLAIGVLAMPESPRWLVLQGRLGDAFKVLDKTSNTKEEAISRLDDIKRAVGIP 249
Query: 267 NEV--------------KSPFKDLVKSHN---RPPLIIAICMQVFQQCTGINAIMFYAPV 309
+++ K +KDL+ R LI + + QQ +GI+A++ Y+P
Sbjct: 250 DDMTDDVIVVPNKKSAGKGVWKDLLVRPTPSVRHILIACLGIHFAQQASGIDAVVLYSPT 309
Query: 310 LFSTLGFHN-DASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGI 368
+FS G + + L ++V G V L +V VD+ GRR LLL + MF+S +G
Sbjct: 310 IFSKAGLKSKNDQLLATVAVGVVKTLFIVVGTCVVDRFGRRALLLTSMGGMFLSLTALGT 369
Query: 369 VLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVT 428
L ++ +L K L V V TFVA+F+ GP+ W+ SE FP+ R+ G S+
Sbjct: 370 SLTVINRNPGQTL-KWAIGLAVTTVMTFVATFSIGAGPVTWVYCSEIFPVRLRAQGASLG 428
Query: 429 VFTNMLFTFLIAQAFLSL-LCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDM 484
V N L + +I FLSL L G FL F+ VF +PET+ IP+E+M
Sbjct: 429 VMLNRLMSGIIGMTFLSLSKGLTIGGAFLLFAGVAAAAWVFFFTFLPETRGIPLEEM 485
>At2g18480 putative sugar transporter
Length = 508
Score = 195 bits (496), Expect = 4e-50
Identities = 138/479 (28%), Positives = 235/479 (48%), Gaps = 48/479 (10%)
Query: 28 IMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESNYCKYDNQKLQLFT 87
I+A+ ++FGYD G+ G Q F D K ++ ++++
Sbjct: 27 IVASIISIIFGYDTGVMSGA-------QIFIRDDLK--------------INDTQIEVLA 65
Query: 88 SSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGKLILLIFGRILLGCG 147
L L ALV S+ A + +GR+ T+ L+ ++F+VG+VL +L+ GR + G G
Sbjct: 66 GILNLCALVGSLTAGKTSDVIGRRYTIALSAVIFLVGSVLMGYGPNYPVLMVGRCIAGVG 125
Query: 148 VGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTSKIKGGYGWRVSLA 207
VGFA PV+ +EI+ RG L + +L I++GI + + N+ K+ GWR+ L
Sbjct: 126 VGFALMIAPVYSAEISSASHRGFLTSLPELCISLGILLGYVSNYCFGKLTLKLGWRLMLG 185
Query: 208 GAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGV-ENIEPEFEDILRASKV- 265
A P+++L G + ++P L+ +G E+ K ++ + E E F DIL A++V
Sbjct: 186 IAAFPSLILAFGITRMPESPRWLVMQGRLEEAKKIMVLVSNTEEEAEERFRDILTAAEVD 245
Query: 266 -------------ANEVKSPFKDLV---KSHNRPPLIIAICMQVFQQCTGINAIMFYAPV 309
N KS +++LV + R LI A+ + F+ TGI A++ Y+P
Sbjct: 246 VTEIKEVGGGVKKKNHGKSVWRELVIKPRPAVRLILIAAVGIHFFEHATGIEAVVLYSPR 305
Query: 310 LFSTLG-FHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMF--VSQVVI 366
+F G D L ++V G +++ + +DK GRR LLL + M ++ + +
Sbjct: 306 IFKKAGVVSKDKLLLATVGVGLTKAFFIIIATFLLDKVGRRKLLLTSTGGMVFALTSLAV 365
Query: 367 GIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQS 426
+ + + + +LS L +V FVA F+ GP+ W+ SE FPL R+ G S
Sbjct: 366 SLTMVQRFGRLAWALS-----LSIVSTYAFVAFFSIGLGPITWVYSSEIFPLRLRAQGAS 420
Query: 427 VTVFTNMLFTFLIAQAFLSLL-CLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDM 484
+ V N + ++ +FLS+ + G+F F+ F F++PETK +P+E+M
Sbjct: 421 IGVAVNRIMNATVSMSFLSMTKAITTGGVFFVFAGIAVAAWWFFFFMLPETKGLPLEEM 479
>At2g16130 putative sugar transporter
Length = 511
Score = 195 bits (495), Expect = 6e-50
Identities = 141/477 (29%), Positives = 226/477 (46%), Gaps = 42/477 (8%)
Query: 28 IMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESNYCKYDNQKLQLFT 87
I+A+ ++ GYD+G+ G + + K + +L++
Sbjct: 31 ILASMTSIILGYDIGVMSGAA---------------------IFIKDDLKLSDVQLEILM 69
Query: 88 SSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGKLILLIFGRILLGCG 147
L + +L+ S A + +GR+ T++LAG F G +L A ++ GR + G G
Sbjct: 70 GILNIYSLIGSGAAGRTSDWIGRRYTIVLAGFFFFCGALLMGFATNYPFIMVGRFVAGIG 129
Query: 148 VGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTSKIKGGYGWRVSLA 207
VG+A PV+ +E+AP RG L+ ++ I IGI + + N+F +K+ GWR L
Sbjct: 130 VGYAMMIAPVYTTEVAPASSRGFLSSFPEIFINIGILLGYVSNYFFAKLPEHIGWRFMLG 189
Query: 208 GAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGV-ENIEPEFEDILRASKVA 266
+P+V L +G L + ++P L+ +G VL K E DI RA +
Sbjct: 190 IGAVPSVFLAIGVLAMPESPRWLVMQGRLGDAFKVLDKTSNTKEEAISRLNDIKRAVGIP 249
Query: 267 NEV--------------KSPFKDLVKSHN---RPPLIIAICMQVFQQCTGINAIMFYAPV 309
+++ K +KDL+ R LI + + QQ +GI+A++ Y+P
Sbjct: 250 DDMTDDVIVVPNKKSAGKGVWKDLLVRPTPSVRHILIACLGIHFSQQASGIDAVVLYSPT 309
Query: 310 LFSTLGFHN-DASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGI 368
+FS G + + L ++V G V L +V VD+ GRR LLL + MF S +G
Sbjct: 310 IFSRAGLKSKNDQLLATVAVGVVKTLFIVVGTCLVDRFGRRALLLTSMGGMFFSLTALGT 369
Query: 369 VLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVT 428
L ++ +L K L V V TFVA+F+ GP+ W+ SE FP+ R+ G S+
Sbjct: 370 SLTVIDRNPGQTL-KWAIGLAVTTVMTFVATFSLGAGPVTWVYASEIFPVRLRAQGASLG 428
Query: 429 VFTNMLFTFLIAQAFLSL-LCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDM 484
V N L + +I FLSL L G FL F+ VF +PET+ +P+E++
Sbjct: 429 VMLNRLMSGIIGMTFLSLSKGLTIGGAFLLFAGVAVAAWVFFFTFLPETRGVPLEEI 485
>At1g75220 integral membrane protein, putative
Length = 487
Score = 194 bits (492), Expect = 1e-49
Identities = 143/482 (29%), Positives = 232/482 (47%), Gaps = 41/482 (8%)
Query: 9 GSSDVVFEARITAAVVISCIM-AATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQE 67
GSS V+ ++ I+ V++C++ A G + FG+ G S P T++
Sbjct: 36 GSSQVIRDSSIS---VLACVLIVALGPIQFGFTCGYSS-------------PTQAAITKD 79
Query: 68 HTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVL 127
+ S Y +F S + A+V ++ + + +GRK ++++A I I+G +
Sbjct: 80 LGLTVSEY--------SVFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIAAIPNIIGWLC 131
Query: 128 SASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIAN 187
+ A L GR+L G GVG + VPV+++EIAP +RG L + QL++TIGI +A
Sbjct: 132 ISFAKDTSFLYMGRLLEGFGVGIISYTVPVYIAEIAPQNMRGGLGSVNQLSVTIGIMLAY 191
Query: 188 LVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIR 247
L+ F WR+ I+P +L G + ++P L + G ++ + L +R
Sbjct: 192 LLGLFVP-------WRILAVLGILPCTLLIPGLFFIPESPRWLAKMGMTDEFETSLQVLR 244
Query: 248 GVE-NIEPEFEDILRASKVANEVKS---PFKDLVKSHNRPPLIIAICMQVFQQCTGINAI 303
G E +I E +I R+ VA+ K F DL + PL++ I + V QQ GIN +
Sbjct: 245 GFETDITVEVNEIKRS--VASSTKRNTVRFVDLKRRRYYFPLMVGIGLLVLQQLGGINGV 302
Query: 304 MFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQ 363
+FY+ +F + G + + V G + V+ T +S + VDKAGRR+LL + V M +S
Sbjct: 303 LFYSSTIFESAGVTSSNAATFGV--GAIQVVATAISTWLVDKAGRRLLLTISSVGMTISL 360
Query: 364 VVIGIVLGAKLQDHSDS-LSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRS 422
V++ K DS + ++L VV V V F+ GP+ WLI SE P+ +
Sbjct: 361 VIVAAAFYLKEFVSPDSDMYSWLSILSVVGVVAMVVFFSLGMGPIPWLIMSEILPVNIKG 420
Query: 423 AGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIE 482
S+ N F++LI LL G F + VF +PETK +E
Sbjct: 421 LAGSIATLANWFFSWLITMTANLLLAWSSGGTFTLYGLVCAFTVVFVTLWVPETKGKTLE 480
Query: 483 DM 484
++
Sbjct: 481 EL 482
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.141 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,301,612
Number of Sequences: 26719
Number of extensions: 409621
Number of successful extensions: 1611
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 1300
Number of HSP's gapped (non-prelim): 103
length of query: 500
length of database: 11,318,596
effective HSP length: 103
effective length of query: 397
effective length of database: 8,566,539
effective search space: 3400915983
effective search space used: 3400915983
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC137838.10


