

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137831.4 + phase: 0 /partial
(420 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
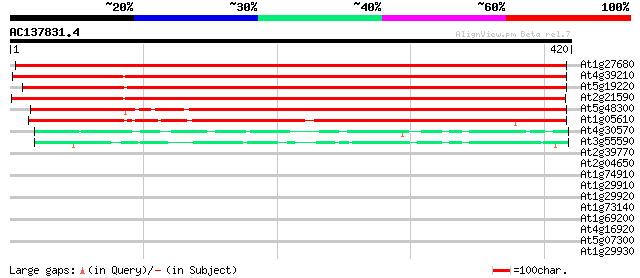
Score E
Sequences producing significant alignments: (bits) Value
At1g27680 ADP-glucose pyrophosphorylase, putative 621 e-178
At4g39210 glucose-1-phosphate adenylyltransferase (APL3) 568 e-162
At5g19220 Glucose-1-phosphate adenylyltransferase (ApL1/adg2) 567 e-162
At2g21590 putative ADP-glucose pyrophosphorylase large subunit 566 e-162
At5g48300 ADPG pyrophosphorylase small subunit (gb|AAC39441.1) 464 e-131
At1g05610 putative ADP-glucose pyrophosphorylase, small subunit ... 290 1e-78
At4g30570 GDP-mannose pyrophosphorylase like protein 53 4e-07
At3g55590 mannose-1-phosphate guanylyltransferase-like protein 53 4e-07
At2g39770 GDP-mannose pyrophosphorylase 41 0.001
At2g04650 putative GDP-mannose pyrophosphorylase 32 0.86
At1g74910 putative GDP-mannose pyrophosphorylase 31 1.5
At1g29910 chlorophyll a/b-binding protein 30 1.9
At1g29920 chlorophyll a/b-binding protein 30 1.9
At1g73140 hypothetical protein 30 3.3
At1g69200 putative fructokinase 29 5.5
At4g16920 disease resistance RPP5 like protein 28 7.2
At5g07300 copine-like protein 28 9.5
At1g29930 unknown protein 28 9.5
>At1g27680 ADP-glucose pyrophosphorylase, putative
Length = 518
Score = 621 bits (1602), Expect = e-178
Identities = 294/413 (71%), Positives = 351/413 (84%)
Query: 5 PIFAGQEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEIN 64
P+ Q A+PK VASIILGGGAGTRLFPLT KRAKPAVP GGCYRL+DIPMSNCINS I
Sbjct: 73 PLLRTQNADPKNVASIILGGGAGTRLFPLTSKRAKPAVPIGGCYRLIDIPMSNCINSGIR 132
Query: 65 KIYVLTQFNSQSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRR 124
KI++LTQFNS SLNRH++RTYN G GVN G F+EVLAATQT G++G KWFQGTADAVR+
Sbjct: 133 KIFILTQFNSFSLNRHLSRTYNFGNGVNFGDGFVEVLAATQTSGDAGKKWFQGTADAVRQ 192
Query: 125 FLWLFEDAEHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGL 184
F+W+FEDA+ +N+E++L+L GD LYRMDYM VQKHI S ADI+VSCLP+D SRASDFGL
Sbjct: 193 FIWVFEDAKTKNVEHVLILSGDHLYRMDYMNFVQKHIESNADITVSCLPMDESRASDFGL 252
Query: 185 VKVDERGRIHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKL 244
+K+D+ G+I QF EKPKGD L++M VDTS+ GL +EA + PYIASMG+YVF+ +VL KL
Sbjct: 253 LKIDQSGKIIQFSEKPKGDDLKAMQVDTSILGLPPKEAAESPYIASMGVYVFRKEVLLKL 312
Query: 245 LRSCYPNANDFGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSFFDANLALMDKPPKFQLY 304
LRS YP +NDFGSE+IP+A + VQA LFN YWEDIGTI SFFDANLAL ++PPKFQ Y
Sbjct: 313 LRSSYPTSNDFGSEIIPLAVGEHNVQAFLFNDYWEDIGTIGSFFDANLALTEQPPKFQFY 372
Query: 305 DQSKPIFTCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTM 364
DQ P FT PRFLPPTK++KC++++S+VS GCFLREC V+HSIVGIRSRL SGV+L+DTM
Sbjct: 373 DQKTPFFTSPRFLPPTKVDKCRILDSIVSHGCFLRECSVQHSIVGIRSRLESGVELQDTM 432
Query: 365 MMGADYYETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
MMGAD+Y+TEAEIASLL+ G VP+G+G+NTKI CIIDKNA+IG NV IAN +
Sbjct: 433 MMGADFYQTEAEIASLLAEGKVPVGVGQNTKIKNCIIDKNAKIGKNVVIANAD 485
>At4g39210 glucose-1-phosphate adenylyltransferase (APL3)
Length = 521
Score = 568 bits (1463), Expect = e-162
Identities = 270/415 (65%), Positives = 339/415 (81%), Gaps = 1/415 (0%)
Query: 3 QSPIFAGQEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSE 62
Q +F + A+PK VA+IILGGG G +LFPLT++ A PAVP GGCYR++DIPMSNCINS
Sbjct: 75 QPSMFERRRADPKNVAAIILGGGDGAKLFPLTKRAATPAVPVGGCYRMIDIPMSNCINSC 134
Query: 63 INKIYVLTQFNSQSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAV 122
INKI+VLTQFNS SLNRH+ARTY G G+N G F+EVLAATQT GE+G KWFQGTADAV
Sbjct: 135 INKIFVLTQFNSASLNRHLARTY-FGNGINFGDGFVEVLAATQTPGEAGKKWFQGTADAV 193
Query: 123 RRFLWLFEDAEHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDF 182
R+FLW+FEDA++RNIENI++L GD LYRM+YM+ VQ H++S ADI++SC PVD SRAS++
Sbjct: 194 RKFLWVFEDAKNRNIENIIILSGDHLYRMNYMDFVQHHVDSKADITLSCAPVDESRASEY 253
Query: 183 GLVKVDERGRIHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLR 242
GLV +D GR+ F EKP G L+SM DT++ GLS QEA K PYIASMG+Y FK + L
Sbjct: 254 GLVNIDRSGRVVHFSEKPTGIDLKSMQTDTTMHGLSHQEAAKSPYIASMGVYCFKTEALL 313
Query: 243 KLLRSCYPNANDFGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSFFDANLALMDKPPKFQ 302
KLL YP++NDFGSE+IP A KD VQ ++ YWEDIGTIKSF++AN+AL+++ PKF+
Sbjct: 314 KLLTWRYPSSNDFGSEIIPAAIKDHNVQGYIYRDYWEDIGTIKSFYEANIALVEEHPKFE 373
Query: 303 LYDQSKPIFTCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKD 362
YDQ+ P +T PRFLPPTK EKC++VNS++S GCFL EC ++ SI+G RSRL+ GV+L+D
Sbjct: 374 FYDQNTPFYTSPRFLPPTKTEKCRIVNSVISHGCFLGECSIQRSIIGERSRLDYGVELQD 433
Query: 363 TMMMGADYYETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
T+M+GAD Y+TE+EIASLL+ G+VPIGIG++TKI KCIIDKNA+IG NV I NK+
Sbjct: 434 TLMLGADSYQTESEIASLLAEGNVPIGIGRDTKIRKCIIDKNAKIGKNVVIMNKD 488
>At5g19220 Glucose-1-phosphate adenylyltransferase (ApL1/adg2)
Length = 522
Score = 567 bits (1462), Expect = e-162
Identities = 268/408 (65%), Positives = 337/408 (81%), Gaps = 1/408 (0%)
Query: 10 QEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVL 69
++ +P+TVASIILGGGAGTRLFPLT++RAKPAVP GG YRL+D+PMSNCINS INK+Y+L
Sbjct: 83 EKRDPRTVASIILGGGAGTRLFPLTKRRAKPAVPIGGAYRLIDVPMSNCINSGINKVYIL 142
Query: 70 TQFNSQSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLF 129
TQ+NS SLNRH+AR YN G+ G ++EVLAATQT GESG +WFQGTADAVR+F WLF
Sbjct: 143 TQYNSASLNRHLARAYN-SNGLGFGDGYVEVLAATQTPGESGKRWFQGTADAVRQFHWLF 201
Query: 130 EDAEHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDE 189
EDA ++IE++L+L GD LYRMDYM+ +Q H S ADIS+SC+P+D RASDFGL+K+D+
Sbjct: 202 EDARSKDIEDVLILSGDHLYRMDYMDFIQDHRQSGADISISCIPIDDRRASDFGLMKIDD 261
Query: 190 RGRIHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCY 249
+GR+ F EKPKGD L++M VDT++ GLS +EA K PYIASMG+YVFK ++L LLR +
Sbjct: 262 KGRVISFSEKPKGDDLKAMAVDTTILGLSKEEAEKKPYIASMGVYVFKKEILLNLLRWRF 321
Query: 250 PNANDFGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSFFDANLALMDKPPKFQLYDQSKP 309
P ANDFGSE+IP +AK+F V A LFN YWEDIGTI+SFF+ANLAL + P F YD +KP
Sbjct: 322 PTANDFGSEIIPFSAKEFYVNAYLFNDYWEDIGTIRSFFEANLALTEHPGAFSFYDAAKP 381
Query: 310 IFTCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTMMMGAD 369
I+T R LPP+K++ ++++S++S G FL C +EHSIVGIRSR+ S VQLKDT+M+GAD
Sbjct: 382 IYTSRRNLPPSKIDNSKLIDSIISHGSFLTNCLIEHSIVGIRSRVGSNVQLKDTVMLGAD 441
Query: 370 YYETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
YYETEAE+A+LL+ G+VPIGIG+NTKI +CIIDKNAR+G NV IAN E
Sbjct: 442 YYETEAEVAALLAEGNVPIGIGENTKIQECIIDKNARVGKNVIIANSE 489
>At2g21590 putative ADP-glucose pyrophosphorylase large subunit
Length = 523
Score = 566 bits (1459), Expect = e-162
Identities = 272/415 (65%), Positives = 337/415 (80%), Gaps = 1/415 (0%)
Query: 2 LQSPIFAGQEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINS 61
+++ +F ++ +P+ VA+IILGGG G +LFPLT + A PAVP GGCYRL+DIPMSNCINS
Sbjct: 76 VKTSMFERRKVDPQNVAAIILGGGNGAKLFPLTMRAATPAVPVGGCYRLIDIPMSNCINS 135
Query: 62 EINKIYVLTQFNSQSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADA 121
INKI+VLTQFNS SLNRH+ARTY G G+N GG F+EVLAATQT GE+G KWFQGTADA
Sbjct: 136 CINKIFVLTQFNSASLNRHLARTY-FGNGINFGGGFVEVLAATQTPGEAGKKWFQGTADA 194
Query: 122 VRRFLWLFEDAEHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASD 181
VR+FLW+FEDA++RNIENIL+L GD LYRM+YM+ VQ H++S ADI++SC PV SRAS+
Sbjct: 195 VRKFLWVFEDAKNRNIENILILSGDHLYRMNYMDFVQSHVDSNADITLSCAPVSESRASN 254
Query: 182 FGLVKVDERGRIHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVL 241
FGLVK+D GR+ F EKP G L+SM DT++ GLS QEA PYIASMG+Y FK + L
Sbjct: 255 FGLVKIDRGGRVIHFSEKPTGVDLKSMQTDTTMLGLSHQEATDSPYIASMGVYCFKTEAL 314
Query: 242 RKLLRSCYPNANDFGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSFFDANLALMDKPPKF 301
LL YP++NDFGSEVIP A +D VQ +F YWEDIGTIK+F++ANLAL+++ PKF
Sbjct: 315 LNLLTRQYPSSNDFGSEVIPAAIRDHDVQGYIFRDYWEDIGTIKTFYEANLALVEERPKF 374
Query: 302 QLYDQSKPIFTCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLK 361
+ YD P +T PRFLPPTK EKC++V+S++S GCFLREC V+ SI+G RSRL+ GV+L+
Sbjct: 375 EFYDPETPFYTSPRFLPPTKAEKCRMVDSIISHGCFLRECSVQRSIIGERSRLDYGVELQ 434
Query: 362 DTMMMGADYYETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANK 416
DT+M+GADYY+TE+EIASLL+ G VPIGIGK+TKI KCIIDKNA+IG NV I NK
Sbjct: 435 DTLMLGADYYQTESEIASLLAEGKVPIGIGKDTKIRKCIIDKNAKIGKNVIIMNK 489
>At5g48300 ADPG pyrophosphorylase small subunit (gb|AAC39441.1)
Length = 520
Score = 464 bits (1193), Expect = e-131
Identities = 232/406 (57%), Positives = 305/406 (74%), Gaps = 11/406 (2%)
Query: 16 TVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVLTQFNSQ 75
+V IILGGGAGTRL+PLT+KRAKPAVP G YRL+DIP+SNC+NS I+KIYVLTQFNS
Sbjct: 89 SVLGIILGGGAGTRLYPLTKKRAKPAVPLGANYRLIDIPVSNCLNSNISKIYVLTQFNSA 148
Query: 76 SLNRHIARTY--NLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFEDAE 133
SLNRH++R Y N+GG N G F+EVLAA Q+ WFQGTADAVR++LWLFE+
Sbjct: 149 SLNRHLSRAYASNMGGYKNEG--FVEVLAAQQS--PENPNWFQGTADAVRQYLWLFEE-- 202
Query: 134 HRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDERGRI 193
N+ L+L GD LYRMDY + +Q H + ADI+V+ LP+D RA+ FGL+K+DE GRI
Sbjct: 203 -HNVLEYLILAGDHLYRMDYEKFIQAHRETDADITVAALPMDEQRATAFGLMKIDEEGRI 261
Query: 194 HQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNAN 253
+F EKPKG+ L++M VDT++ GL Q A++ P+IASMGIYV DV+ LLR+ +P AN
Sbjct: 262 IEFAEKPKGEHLKAMKVDTTILGLDDQRAKEMPFIASMGIYVVSRDVMLDLLRNQFPGAN 321
Query: 254 DFGSEVIPMAAK-DFKVQACLFNGYWEDIGTIKSFFDANLALMDKP-PKFQLYDQSKPIF 311
DFGSEVIP A +VQA L++GYWEDIGTI++F++ANL + KP P F YD+S PI+
Sbjct: 322 DFGSEVIPGATSLGLRVQAYLYDGYWEDIGTIEAFYNANLGITKKPVPDFSFYDRSAPIY 381
Query: 312 TCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTMMMGADYY 371
T PR+LPP+K+ V +S++ +GC ++ CK+ HS+VG+RS ++ G ++D+++MGADYY
Sbjct: 382 TQPRYLPPSKMLDADVTDSVIGEGCVIKNCKIHHSVVGLRSCISEGAIIEDSLLMGADYY 441
Query: 372 ETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
ET E + L + G VPIGIGKN+ I + IIDKNARIG+NV I N +
Sbjct: 442 ETATEKSLLSAKGSVPIGIGKNSHIKRAIIDKNARIGDNVKIINSD 487
>At1g05610 putative ADP-glucose pyrophosphorylase, small subunit
precursor
Length = 480
Score = 290 bits (742), Expect = 1e-78
Identities = 162/408 (39%), Positives = 252/408 (61%), Gaps = 18/408 (4%)
Query: 15 KTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVLTQFNS 74
++VA+I+ GGG+ + L+PLT+ R+K A+P YRL+D +SNCINS I KIY +TQFNS
Sbjct: 53 QSVAAIVFGGGSDSELYPLTKTRSKGAIPIAANYRLIDAVISNCINSGITKIYAITQFNS 112
Query: 75 QSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFEDAEH 134
SLN H+++ Y+ G G+ F+EV+AA Q+L + G WFQGTADA+RR LW+FE+
Sbjct: 113 TSLNSHLSKAYS-GFGLG-KDRFVEVIAAYQSLEDQG--WFQGTADAIRRCLWVFEEFP- 167
Query: 135 RNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDERGRIH 194
+ LVL G LY+MDY L++ H S ADI++ L FG ++VD +
Sbjct: 168 --VTEFLVLPGHHLYKMDYKMLIEDHRRSRADITIVGLSSVTDHDFGFGFMEVDSTNAVT 225
Query: 195 QFMEKPKGDLLR-SMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNAN 253
+F K + DL+ + T G S+ + S GIYV + + KLLR C +
Sbjct: 226 RFTIKGQQDLISVANRTATRSDGTSSCS------VPSAGIYVIGREQMVKLLRECLIKSK 279
Query: 254 DFGSEVIPMA-AKDFKVQACLFNGYWEDIGTIKSFFDANLALMDK-PPKFQLYDQSKPIF 311
D SE+IP A ++ KV+A +F+GYWED+ +I +++ AN+ + + + YD+ P++
Sbjct: 280 DLASEIIPGAISEGMKVKAHMFDGYWEDVRSIGAYYRANMESIKRCRLDLKFYDRQCPLY 339
Query: 312 TCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTMMMGADYY 371
T PR LPP+ + + NS++ DGC L +C + S+VG+R+R+ V ++D++++G+D Y
Sbjct: 340 TMPRCLPPSSMSVAVITNSIIGDGCILDKCVIRGSVVGMRTRIADEVIVEDSIIVGSDIY 399
Query: 372 ETEAEI--ASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
E E ++ ++ IGIG+ ++I + I+DKNARIG NV I N++
Sbjct: 400 EMEEDVRRKGKEKKIEIRIGIGEKSRIRRAIVDKNARIGKNVMIINRD 447
>At4g30570 GDP-mannose pyrophosphorylase like protein
Length = 351
Score = 52.8 bits (125), Expect = 4e-07
Identities = 84/406 (20%), Positives = 162/406 (39%), Gaps = 75/406 (18%)
Query: 19 SIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVLTQFNSQSLN 78
++IL GG GTRL PLT KP V FG ++ + + + ++ + +
Sbjct: 3 ALILVGGFGTRLRPLTLSMPKPLVDFGNKPMILH-QIEALKGAGVTEVVLAINHQQPEVM 61
Query: 79 RHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFEDAEHRNIE 138
+ + Y + I T+ LG +G A+ R + E + +
Sbjct: 62 LNFVKEYEKKLEIK-----ITFSQETEPLGTAGPL-------ALARDKLVDESGQPFFVL 109
Query: 139 NILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDE-RGRIHQFM 197
N V+C L +E+++ H + A+ S+ VD S +G+V +E R+ F+
Sbjct: 110 NSDVICEYPL-----LEMIEFHKTNRAEASIMVTEVDDP--SKYGVVVTEEGTARVESFV 162
Query: 198 EKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKL-LRSCYPNANDFG 256
EKPK + ++ GIY+ VL ++ LR
Sbjct: 163 EKPKHFVGNKINA---------------------GIYLLSPSVLDRIELRR-----TSIE 196
Query: 257 SEVIPMAAKDFKVQACLFNGYWEDIGTIKSFFDANL----ALMDKPPKFQLYDQSKPIFT 312
E+ P A + K+ A + G+W DIG K + +L +K P + + T
Sbjct: 197 KEIFPKIASEKKLYAMVLPGFWMDIGQPKDYITGQRMYLNSLREKTP--------QELAT 248
Query: 313 CPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTMMMGADYYE 372
+ + + ++++ +GC + ++G ++SGV+L +M + +
Sbjct: 249 GDNIIGNVLVHE----SAVIGEGCLIG----PDVVIGPGCVIDSGVRLFGCTVMRGVWIK 300
Query: 373 TEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKEV 418
A I++ + D +G+ ++ + +G +V +A+ EV
Sbjct: 301 EHACISNSIVGWDST--VGRWARVFNITV-----LGKDVNVADAEV 339
>At3g55590 mannose-1-phosphate guanylyltransferase-like protein
Length = 364
Score = 52.8 bits (125), Expect = 4e-07
Identities = 90/417 (21%), Positives = 158/417 (37%), Gaps = 83/417 (19%)
Query: 19 SIILGGGAGTRLFPLTQKRAKPAVPFGG---------CYRLVDIP-MSNCINSEINKIYV 68
++IL GG GTRL PLT KP V F + + + + IN E ++ V
Sbjct: 3 ALILVGGFGTRLRPLTLSLPKPLVDFANKPMILHQIEALKAIGVDEVVLAINYEPEQLLV 62
Query: 69 LTQFNSQSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWL 128
+++F++ LG + C E L L + +K G+
Sbjct: 63 MSKFSNDV-------EATLGIKITCSQE-TEPLGTAGPLALARDKLVDGSG--------- 105
Query: 129 FEDAEHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVD 188
+ VL D + E++ H + S+ VD S +G+V ++
Sbjct: 106 ---------QPFFVLNSDVISDYPLEEMIAFHNAHGGEASIMVTKVD--EPSKYGVVVME 154
Query: 189 ER-GRIHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRS 247
E GR+ +F+EKPK + V + + GIY+ VL ++
Sbjct: 155 EATGRVERFVEKPK------LFVGNKI---------------NAGIYLLNPSVLDRI--E 191
Query: 248 CYPNANDFGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSFFDANLALMDKPPKFQLYDQS 307
P + + E+ P A+ K+ A L G+W DIG + + +D K
Sbjct: 192 LRPTSIE--KEIFPQIAEAEKLYAMLLPGFWMDIGQPRDYITGLRLYLDSLRK----KSP 245
Query: 308 KPIFTCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTMMMG 367
+ T P L +++ + + +GC + + +G + SGV+L +M
Sbjct: 246 SKLATGPHILGNVLVDE----TAEIGEGCLIG----PNVAIGPGCVVESGVRLSHCTVMR 297
Query: 368 ADYYETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARI------GNNVTIANKEV 418
+ + A I+S + +G + M I+ KN + V + NKE+
Sbjct: 298 GVHVKRYACISSSIIGWHSTVGQWARVENMS-ILGKNVYVCDEIYCNGGVVLHNKEI 353
>At2g39770 GDP-mannose pyrophosphorylase
Length = 361
Score = 41.2 bits (95), Expect = 0.001
Identities = 61/273 (22%), Positives = 104/273 (37%), Gaps = 53/273 (19%)
Query: 19 SIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLV-DIPMSNCINSEINKIYVLTQFNSQSL 77
++IL GG GTRL PLT KP V F ++ I + ++++ + + + +
Sbjct: 3 ALILVGGFGTRLRPLTLSFPKPLVDFANKPMILHQIEALKAVG--VDEVVLAINYQPEVM 60
Query: 78 NRHIAR-TYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFEDAEHRN 136
+ L + C E L L + +K G+
Sbjct: 61 LNFLKDFETKLEIKITCSQE-TEPLGTAGPLALARDKLLDGSG----------------- 102
Query: 137 IENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDER-GRIHQ 195
E VL D + E+++ H + + S+ VD S +G+V ++E GR+ +
Sbjct: 103 -EPFFVLNSDVISEYPLKEMLEFHKSHGGEASIMVTKVD--EPSKYGVVVMEESTGRVEK 159
Query: 196 FMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKL-LRSCYPNAND 254
F+EKPK ++V + + GIY+ VL K+ LR
Sbjct: 160 FVEKPK------LYVGNKI---------------NAGIYLLNPSVLDKIELR-----PTS 193
Query: 255 FGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSF 287
E P A + A + G+W DIG + +
Sbjct: 194 IEKETFPKIAAAQGLYAMVLPGFWMDIGQPRDY 226
>At2g04650 putative GDP-mannose pyrophosphorylase
Length = 406
Score = 31.6 bits (70), Expect = 0.86
Identities = 21/74 (28%), Positives = 38/74 (50%), Gaps = 3/74 (4%)
Query: 15 KTVASIILGGGA-GTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCIN-SEINKIYVLTQF 72
K VA I++GG GTR PL+ KP +P G ++ P+S C S + +I+++ +
Sbjct: 5 KVVAVIMVGGPTKGTRFRPLSFNTPKPLIPLAG-QPMIHHPISACKKISNLAQIFLIGFY 63
Query: 73 NSQSLNRHIARTYN 86
+ +++ N
Sbjct: 64 EEREFALYVSSISN 77
>At1g74910 putative GDP-mannose pyrophosphorylase
Length = 415
Score = 30.8 bits (68), Expect = 1.5
Identities = 23/80 (28%), Positives = 37/80 (45%), Gaps = 3/80 (3%)
Query: 9 GQEANPKTVASIILGGGA-GTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCIN-SEINKI 66
G K VA I++GG GTR PL+ KP P G +V P+S C + +I
Sbjct: 2 GSSMEEKVVAVIMVGGPTKGTRFRPLSLNIPKPLFPIAG-QPMVHHPISACKRIPNLAQI 60
Query: 67 YVLTQFNSQSLNRHIARTYN 86
Y++ + + +++ N
Sbjct: 61 YLVGFYEEREFALYVSAISN 80
>At1g29910 chlorophyll a/b-binding protein
Length = 267
Score = 30.4 bits (67), Expect = 1.9
Identities = 19/45 (42%), Positives = 23/45 (50%), Gaps = 4/45 (8%)
Query: 1 ALQSPIFAGQEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFG 45
AL SP FAG+ N AS +LG G T + + AKP P G
Sbjct: 7 ALSSPAFAGKAVNLSPAASEVLGSGRVT----MRKTVAKPKGPSG 47
>At1g29920 chlorophyll a/b-binding protein
Length = 267
Score = 30.4 bits (67), Expect = 1.9
Identities = 19/45 (42%), Positives = 23/45 (50%), Gaps = 4/45 (8%)
Query: 1 ALQSPIFAGQEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFG 45
AL SP FAG+ N AS +LG G T + + AKP P G
Sbjct: 7 ALSSPAFAGKAVNLSPAASEVLGSGRVT----MRKTVAKPKGPSG 47
>At1g73140 hypothetical protein
Length = 403
Score = 29.6 bits (65), Expect = 3.3
Identities = 13/27 (48%), Positives = 17/27 (62%)
Query: 389 GIGKNTKIMKCIIDKNARIGNNVTIAN 415
G G N +IMK + D +R+G NVT N
Sbjct: 311 GTGSNQEIMKIVGDVLSRVGENVTFLN 337
>At1g69200 putative fructokinase
Length = 614
Score = 28.9 bits (63), Expect = 5.5
Identities = 19/75 (25%), Positives = 35/75 (46%), Gaps = 6/75 (8%)
Query: 152 DYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDERGRIHQFMEKP-KGDLLRSMHV 210
DY + + ++N C + + +DG R + +K+ +RGR+ KP D L +
Sbjct: 278 DYGQAMLYYLNVCK-VQTRSVKIDGKRVTACSTMKISKRGRLKSTCIKPCAEDSLSKSEI 336
Query: 211 DTSVFGLSAQEARKF 225
+ V +EA+ F
Sbjct: 337 NVDVL----KEAKMF 347
>At4g16920 disease resistance RPP5 like protein
Length = 1304
Score = 28.5 bits (62), Expect = 7.2
Identities = 24/93 (25%), Positives = 40/93 (42%), Gaps = 10/93 (10%)
Query: 203 DLLRSMHVDTSVFGLSAQEARKFPYIASMG-IYVFKLDVLRKLLRSCYPNANDFGSEVIP 261
+L S+ + SV G S + K ++ M + D + + LR CY N E+
Sbjct: 370 ELAGSLPLGLSVLGSSLKGRDKDEWVKMMPRLRNDSDDKIEETLRVCYDRLNKKNREL-- 427
Query: 262 MAAKDFKVQACLFNGYWEDIGTIKSFFDANLAL 294
FK AC FNG+ + +K + ++ L
Sbjct: 428 -----FKCIACFFNGF--KVSNVKELLEDDVGL 453
>At5g07300 copine-like protein
Length = 586
Score = 28.1 bits (61), Expect = 9.5
Identities = 22/79 (27%), Positives = 35/79 (43%), Gaps = 5/79 (6%)
Query: 40 PAVPFGGCYRLVDIPMSNCINSEINKIYVLTQFNSQSLNRHIARTYNLG-GGVNCGGSFI 98
PA FG R +DIP+S+C N + Y +N + +N+ G G I
Sbjct: 397 PAWGFGA--RPIDIPVSHCFNLNGSSTYCEVDGIQGIMNAYNGALFNVSFAGPTLFGPVI 454
Query: 99 EVLA--ATQTLGESGNKWF 115
A A+ +L +S K++
Sbjct: 455 NAAATIASDSLAQSAKKYY 473
>At1g29930 unknown protein
Length = 267
Score = 28.1 bits (61), Expect = 9.5
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 4/45 (8%)
Query: 1 ALQSPIFAGQEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFG 45
AL SP FAG+ AS +LG G T + + AKP P G
Sbjct: 7 ALSSPAFAGKAVKLSPAASEVLGSGRVT----MRKTVAKPKGPSG 47
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,335,548
Number of Sequences: 26719
Number of extensions: 397229
Number of successful extensions: 823
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 793
Number of HSP's gapped (non-prelim): 21
length of query: 420
length of database: 11,318,596
effective HSP length: 102
effective length of query: 318
effective length of database: 8,593,258
effective search space: 2732656044
effective search space used: 2732656044
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137831.4


