

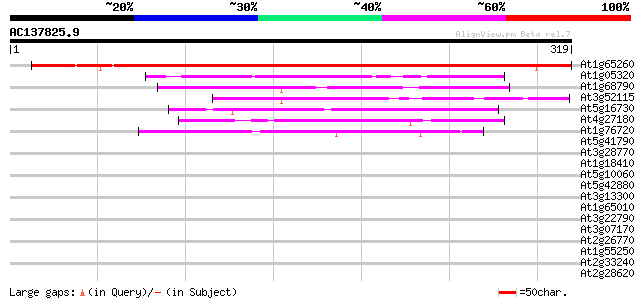
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137825.9 - phase: 0
(319 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g65260 unknown protein 411 e-115
At1g05320 unknown protein 49 3e-06
At1g68790 putative nuclear matrix constituent protein 1 (NMCP1) 45 7e-05
At3g52115 gamma response I protein 44 9e-05
At5g16730 putative protein 43 3e-04
At4g27180 heavy chain polypeptide of kinesin-like protein (katB) 43 3e-04
At1g76720 putative translation initiation factor IF-2 42 4e-04
At5g41790 myosin heavy chain-like protein 39 0.003
At3g28770 hypothetical protein 39 0.003
At1g18410 kinesin-related protein, putative 39 0.004
At5g10060 unknown protein 39 0.005
At5g42880 putative protein 37 0.011
At3g13300 unknown protein 37 0.019
At1g65010 hypothetical protein 37 0.019
At3g22790 unknown protein 36 0.024
At3g07170 unknown protein 36 0.024
At2g26770 unknown protein 36 0.032
At1g55250 unknown protein 36 0.032
At2g33240 putative myosin heavy chain 35 0.041
At2g28620 putative kinesin-like spindle protein 35 0.041
>At1g65260 unknown protein
Length = 330
Score = 411 bits (1057), Expect = e-115
Identities = 221/312 (70%), Positives = 266/312 (84%), Gaps = 7/312 (2%)
Query: 13 SAPLQSSSSSSSLR-KPLATSFFGTRPANTVKFRGMRIA--KPVRGGGAIGAGMNLFERF 69
S+P SS+ SLR PL TSFFG + ++ +R+A +R G GA MNLFERF
Sbjct: 21 SSPSTSSNRPCSLRILPLRTSFFGNS-SGALRVNVLRLACDNRLRCNGH-GATMNLFERF 78
Query: 70 TRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAATQA 129
+RVVKSYANA++SSFEDPEKILEQ V+EMN DLTKMRQATAQVLASQK+++NKYKAA Q+
Sbjct: 79 SRVVKSYANALISSFEDPEKILEQTVIEMNSDLTKMRQATAQVLASQKQLQNKYKAAQQS 138
Query: 130 SEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLES 189
S++WY++AQLAL KG+EDLAREALKRRKSFADNA+ALK QLDQQKGVVD+LVSNTRLLES
Sbjct: 139 SDDWYKRAQLALAKGDEDLAREALKRRKSFADNATALKTQLDQQKGVVDNLVSNTRLLES 198
Query: 190 KIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQ 249
KIQEA++KKDTL ARA++AKT+TKV EM+G VNTSGALSAFEKMEEKVM MES+A+AL Q
Sbjct: 199 KIQEAKAKKDTLLARARTAKTATKVQEMIGTVNTSGALSAFEKMEEKVMAMESEADALTQ 258
Query: 250 LTSDDLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGRSSTRTGT--PFRDADIET 307
+ +D+LEGKF MLE+SSVDDDLA+LKKELSGSSKKGELPPGRS+ T PF+D++IE
Sbjct: 259 IGTDELEGKFQMLETSSVDDDLADLKKELSGSSKKGELPPGRSTVAASTRYPFKDSEIEN 318
Query: 308 ELEQLRQRSKEF 319
EL +LR+++ +F
Sbjct: 319 ELNELRRKANDF 330
>At1g05320 unknown protein
Length = 841
Score = 49.3 bits (116), Expect = 3e-06
Identities = 52/205 (25%), Positives = 95/205 (45%), Gaps = 21/205 (10%)
Query: 78 NAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAATQASE-EWYRK 136
N + S+ E+ EK N DL ++ Q LA+Q + ++A E E Y++
Sbjct: 484 NTLESTIEELEK--------ENGDLAEVNIKLNQKLANQGSETDDFQAKLSVLEAEKYQQ 535
Query: 137 AQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEARS 196
A+ LQ EDL ++ R+ S+L+ + +Q + S + L++++Q +S
Sbjct: 536 AK-ELQITIEDLTKQLTSERERLRSQISSLEEEKNQVNEIYQSTKNELVKLQAQLQVDKS 594
Query: 197 KKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSDDLE 256
K D + ++ + K S V+E S S FE++E + ++ + E + +LTS E
Sbjct: 595 KSDDMVSQIE--KLSALVAEK------SVLESKFEQVE---IHLKEEVEKVAELTSKLQE 643
Query: 257 GKFAMLESSSVDDDLANLKKELSGS 281
K + +++ L KEL S
Sbjct: 644 HKHKASDRDVLEEKAIQLHKELQAS 668
Score = 36.6 bits (83), Expect = 0.019
Identities = 44/206 (21%), Positives = 90/206 (43%), Gaps = 20/206 (9%)
Query: 78 NAIVSSFEDPEKILEQAVLEMNDDLTKMRQATA----QVLASQKRMENKYKAATQASEEW 133
NA++ + E+ LE+ E+++ T+ + A L QK ME+ T+A
Sbjct: 305 NAVMEKLKSSEERLEKQAREIDEATTRSIELEALHKHSELKVQKTMEDFSSRDTEA---- 360
Query: 134 YRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQE 193
K+ K E+ R + + +L+ +LDQ + L L+ KIQE
Sbjct: 361 --KSLTEKSKDLEEKIRVYEGKLAEACGQSLSLQEELDQSSAENELLADTNNQLKIKIQE 418
Query: 194 ARSKKDTLKARA--QSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLT 251
D+ K A + + T+ +++ + + + E+ + +V+ EA G
Sbjct: 419 LEGYLDSEKETAIEKLNQKDTEAKDLITKLKSHE--NVIEEHKRQVL------EASGVAD 470
Query: 252 SDDLEGKFAMLESSSVDDDLANLKKE 277
+ +E + A+L+ ++++ + L+KE
Sbjct: 471 TRKVEVEEALLKLNTLESTIEELEKE 496
Score = 35.4 bits (80), Expect = 0.041
Identities = 42/213 (19%), Positives = 91/213 (42%), Gaps = 26/213 (12%)
Query: 95 VLEMNDDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRKAQLALQKG--EEDLAREA 152
++E+ D + ++ QK+ ++ + + S+E Y K L + + + +
Sbjct: 112 MIELEDRIRISALEAEKLEELQKQSASELEEKLKISDERYSKTDALLSQALSQNSVLEQK 171
Query: 153 LKRRKSFADNASALKAQL--DQQKGVVDSLV-----SNTRLLESKIQEARSKKDTLKARA 205
LK + ++ S LK+ L +++G S+ LES + ++ ++ L+
Sbjct: 172 LKSLEELSEKVSELKSALIVAEEEGKKSSIQMQEYQEKVSKLESSLNQSSARNSELEEDL 231
Query: 206 QSAKTSTKVSEMLGNVNTSGALS----------AFEKMEEKVMTMESQAEALGQLTSDDL 255
+ A E +GNV+T ++ EK EEK+ +E+ Q+ + L
Sbjct: 232 RIALQKGAEHEDIGNVSTKRSVELQGLFQTSQLKLEKAEEKLKDLEAI-----QVKNSSL 286
Query: 256 EG--KFAMLESSSVDDDLANLKKELSGSSKKGE 286
E AM + + ++L + ++L S ++ E
Sbjct: 287 EATLSVAMEKERDLSENLNAVMEKLKSSEERLE 319
>At1g68790 putative nuclear matrix constituent protein 1 (NMCP1)
Length = 1085
Score = 44.7 bits (104), Expect = 7e-05
Identities = 44/203 (21%), Positives = 85/203 (41%), Gaps = 17/203 (8%)
Query: 85 EDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRKAQLALQKG 144
+D + +L+ E +L +MR++ + L +K + + EE K + AL+K
Sbjct: 354 DDQKAVLDSRRREFEMELEQMRRSLDEELEGKKAEIEQLQVEISHKEEKLAKREAALEKK 413
Query: 145 EEDLAREAL---KRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKKDTL 201
EE + ++ R K+ + ALKA+ + + N RLLE K + K +
Sbjct: 414 EEGVKKKEKDLDARLKTVKEKEKALKAEEKKLH------MENERLLEDKECLRKLKDEIE 467
Query: 202 KARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSDDLEGKFAM 261
+ ++ K +++ E ++ + EE+V + Q+E Q+ E + +
Sbjct: 468 EIGTETTKQESRIREEHESLRIT--------KEERVEFLRLQSELKQQIDKVKQEEELLL 519
Query: 262 LESSSVDDDLANLKKELSGSSKK 284
E + D +KE KK
Sbjct: 520 KEREELKQDKERFEKEWEALDKK 542
>At3g52115 gamma response I protein
Length = 588
Score = 44.3 bits (103), Expect = 9e-05
Identities = 42/207 (20%), Positives = 92/207 (44%), Gaps = 23/207 (11%)
Query: 116 QKRMENKYKAATQASEEWYRKAQLALQKGEEDLAREAL----KRRKSFADNASALKAQLD 171
+K+M+++ K A + +YR +L KG ++L+ + + KS L+ +L
Sbjct: 122 KKKMKSRSKMVGDARDLYYRLVELLQVKGLDELSEDGINMIVSEVKSLKMKTEFLQEELS 181
Query: 172 QQKGVVDSLVSNTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFE 231
++ V ++L+ L ++ + K +++ Q KT +V E G L
Sbjct: 182 KKTLVTENLLKKLEYLSTEAADGERKLSSVEEEKQRLKTRLQVFE-----ENVGRL---- 232
Query: 232 KMEEKVMTMESQAEALGQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGR 291
E+++ ++ G+ + L+GK + E ++ N K++++ K+ + G+
Sbjct: 233 ---EEILRQKTDEVEEGKTALEVLQGKLKLTER-----EMLNCKQKIADHEKEKTVVMGK 284
Query: 292 SSTRTGTPFRDADIETELEQLRQRSKE 318
+ + R +LE LR +S+E
Sbjct: 285 A--KDDMQGRHGSYLADLEALRCQSEE 309
>At5g16730 putative protein
Length = 853
Score = 42.7 bits (99), Expect = 3e-04
Identities = 45/195 (23%), Positives = 85/195 (43%), Gaps = 13/195 (6%)
Query: 91 LEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKA-----ATQASEEWYRKAQLALQKGE 145
LE E N L K + AT++V Q+ E K K +++ EE +KA +L
Sbjct: 417 LETVKEEKNRALKKEQDATSRV---QRLSEEKSKLLSDLESSKEEEEKSKKAMESLASAL 473
Query: 146 EDLAREALK-RRKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKKDTLKAR 204
+++ E + + K + + Q+D K V+ + E+ + EAR + D L +
Sbjct: 474 HEVSSEGRELKEKLLSQGDHEYETQIDDLKLVIKA---TNEKYENMLDEARHEIDVLVSA 530
Query: 205 AQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTS-DDLEGKFAMLE 263
+ K + S+ + + ++ +KMEE V +M + L L + E A +
Sbjct: 531 VEQTKKHFESSKKDWEMKEANLVNYVKKMEEDVASMGKEMNRLDNLLKRTEEEADAAWKK 590
Query: 264 SSSVDDDLANLKKEL 278
+ D L +++E+
Sbjct: 591 EAQTKDSLKEVEEEI 605
Score = 35.4 bits (80), Expect = 0.041
Identities = 55/248 (22%), Positives = 97/248 (38%), Gaps = 39/248 (15%)
Query: 91 LEQAVLEMNDDLTKMRQATAQVLASQKRMEN---KYKAATQA-------SEEWYRKAQLA 140
LE ++ + DL R A+V + +E +AA A S EW KA
Sbjct: 270 LEDEIVVLKRDLESARGFEAEVKEKEMIVEKLNVDLEAAKMAESNAHSLSNEWQSKA--- 326
Query: 141 LQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRL--LESKIQEARSKK 198
K E+ EA K +S + + ++ QL+ SN +L E++I + + +
Sbjct: 327 --KELEEQLEEANKLERSASVSLESVMKQLEG---------SNDKLHDTETEITDLKERI 375
Query: 199 DTLKARAQSAKTSTKVSEM-LGNVNTS------------GALSAFEKMEEKVMTMESQAE 245
TL+ K +VSE LG+V L ++ + + + E A
Sbjct: 376 VTLETTVAKQKEDLEVSEQRLGSVEEEVSKNEKEVEKLKSELETVKEEKNRALKKEQDAT 435
Query: 246 ALGQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGRSSTRTGTPFRDADI 305
+ Q S++ + LESS +++ + E S+ GR D +
Sbjct: 436 SRVQRLSEEKSKLLSDLESSKEEEEKSKKAMESLASALHEVSSEGRELKEKLLSQGDHEY 495
Query: 306 ETELEQLR 313
ET+++ L+
Sbjct: 496 ETQIDDLK 503
>At4g27180 heavy chain polypeptide of kinesin-like protein (katB)
Length = 745
Score = 42.7 bits (99), Expect = 3e-04
Identities = 40/192 (20%), Positives = 79/192 (40%), Gaps = 23/192 (11%)
Query: 97 EMNDDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRKAQLALQKGEEDLAREALKRR 156
E ++++ + + ++ S ++ ++KA QLA K +D + +K++
Sbjct: 212 EAHENIKRGEKERTGIVESIGNLKGQFKALQD---------QLAASKVSQD---DVMKQK 259
Query: 157 KSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKKDTLKARAQSAKTSTKVSE 216
+ +LK ++ Q K D ++ L+++ + KDT+ K E
Sbjct: 260 DELVNEIVSLKVEIQQVKDDRDRHITEIETLQAEATKQNDFKDTINELESKCSVQNKEIE 319
Query: 217 MLGNVNTSGA-------LSAFEKMEEKVMTMESQAEALGQLTSDDLEGKFAMLESSSVDD 269
L + + LS FEKM E E Q E++ +L E + ++E +
Sbjct: 320 ELQDQLVASERKLQVADLSTFEKMNE----FEEQKESIMELKGRLEEAELKLIEGEKLRK 375
Query: 270 DLANLKKELSGS 281
L N +EL G+
Sbjct: 376 KLHNTIQELKGN 387
Score = 28.9 bits (63), Expect = 3.9
Identities = 34/157 (21%), Positives = 63/157 (39%), Gaps = 9/157 (5%)
Query: 120 ENKYKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDS 179
+ K K A + +E+ ++ L+ EE+L + RK+FA Q+ K +
Sbjct: 87 QEKLKNAMEMNEKHCADLEVNLKVKEEELNMVIDELRKNFA------SVQVQLAKEQTEK 140
Query: 180 LVSNTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMT 239
L +N L K +EAR ++L+A T+ N ++ ++E +
Sbjct: 141 LAANESL--GKEREARIAVESLQAAITEELAKTQGELQTANQRIQAVNDMYKLLQEYNSS 198
Query: 240 MESQAEAL-GQLTSDDLEGKFAMLESSSVDDDLANLK 275
++ L G L K E + + + + NLK
Sbjct: 199 LQLYNSKLQGDLDEAHENIKRGEKERTGIVESIGNLK 235
>At1g76720 putative translation initiation factor IF-2
Length = 1224
Score = 42.0 bits (97), Expect = 4e-04
Identities = 50/213 (23%), Positives = 92/213 (42%), Gaps = 22/213 (10%)
Query: 74 KSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAATQASEEW 133
K A A SS E E+ E++V E K + A K + +A + E
Sbjct: 344 KKAAAAATSSVEAKEEKQEESVTEPLQPKKKDAKGKAAEKKIPKHVREMQEALARRQEAE 403
Query: 134 YRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNT---RLLESK 190
RK + + EE L +E +RR+ A A +A+ +++ + L+ +LL +K
Sbjct: 404 ERKKK----EEEEKLRKEEEERRRQEELEAQAEEAKRKRKEKEKEKLLRKKLEGKLLTAK 459
Query: 191 IQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEK--------------MEEK 236
+ K++ K + +A V++ +G+ +S K +E++
Sbjct: 460 QKTEAQKREAFKNQLLAAGRGLPVADDVGDATSSKRPIYANKKKPSRQKGNDTSVQVEDE 519
Query: 237 VMTMESQAEALGQLTSDDLEGKFAMLESSSVDD 269
V E+ A LG++ S+D E K +LES++ +
Sbjct: 520 VEPQENHAATLGEVGSEDTE-KVDLLESANTGE 551
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 39.3 bits (90), Expect = 0.003
Identities = 58/270 (21%), Positives = 112/270 (41%), Gaps = 39/270 (14%)
Query: 78 NAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQ--------KRMENKYKAATQA 129
+A +++ E+ +K L +LE+ D+L K Q+ Q L ++ + EN+ + +
Sbjct: 505 SASLNAAEEEKKSLSSMILEITDEL-KQAQSKVQELVTELAESKDTLTQKENELSSFVEV 563
Query: 130 SEEWYR--KAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLL 187
E R +Q+ + + A E +K +++ K L QQ + + +
Sbjct: 564 HEAHKRDSSSQVKELEARVESAEEQVKELNQNLNSSEEEKKILSQQ---ISEMSIKIKRA 620
Query: 188 ESKIQEARSKKDTLKA-------RAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTM 240
ES IQE S+ + LK S + + + + G + E E +V+ +
Sbjct: 621 ESTIQELSSESERLKGSHAEKDNELFSLRDIHETHQRELSTQLRGLEAQLESSEHRVLEL 680
Query: 241 ESQAEALGQ----------LTSDDLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPG 290
+A + TSD+LE M++ + D + LK++L+ K L
Sbjct: 681 SESLKAAEEESRTMSTKISETSDELERTQIMVQELTADS--SKLKEQLAEKESKLFLLTE 738
Query: 291 RSSTRTGTPFRD-----ADIETELEQLRQR 315
+ S ++ ++ A +E ELE +R R
Sbjct: 739 KDS-KSQVQIKELEATVATLELELESVRAR 767
Score = 34.7 bits (78), Expect = 0.071
Identities = 49/264 (18%), Positives = 113/264 (42%), Gaps = 28/264 (10%)
Query: 78 NAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQK--------RMENKYKAATQA 129
N ++S E+ +KIL Q + EM+ + K ++T Q L+S+ +N+ +
Sbjct: 593 NQNLNSSEEEKKILSQQISEMSIKI-KRAESTIQELSSESERLKGSHAEKDNELFSLRDI 651
Query: 130 SEEWYRKAQLALQKGEEDL-------------AREALKRRKSFADNASALKAQLDQQKGV 176
E R+ L+ E L + A + ++ + S +L++ + +
Sbjct: 652 HETHQRELSTQLRGLEAQLESSEHRVLELSESLKAAEEESRTMSTKISETSDELERTQIM 711
Query: 177 VDSLVSNTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEK 236
V L +++ L+ ++ E SK L + +K+ ++ E+ V T E + +
Sbjct: 712 VQELTADSSKLKEQLAEKESKLFLLTEK--DSKSQVQIKELEATVATLEL--ELESVRAR 767
Query: 237 VMTMESQAEALGQLTSD-DLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGRSSTR 295
++ +E++ + + + + + + S ++ + ELS ++K E +SS+
Sbjct: 768 IIDLETEIASKTTVVEQLEAQNREMVARISELEKTMEERGTELSALTQKLEDNDKQSSSS 827
Query: 296 TGTPFRDAD-IETELEQLRQRSKE 318
T + D + EL+ + + +E
Sbjct: 828 IETLTAEIDGLRAELDSMSVQKEE 851
Score = 33.5 bits (75), Expect = 0.16
Identities = 43/208 (20%), Positives = 92/208 (43%), Gaps = 17/208 (8%)
Query: 90 ILEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRKAQLALQKGEEDLA 149
++E D +++ QV +S+K + + A EE + L + +L+
Sbjct: 229 LVEVHETHQRDSSIHVKELEEQVESSKKLVAELNQTLNNAEEE-----KKVLSQKIAELS 283
Query: 150 REALKRRKSFADNASALKAQLDQQKGVVD-SLVSNTRLLESKIQEARSKKDTLKARAQSA 208
E + + + + S QL + V D L S + E+ +E+ ++ L+A+ +S+
Sbjct: 284 NEIKEAQNTIQELVSE-SGQLKESHSVKDRDLFSLRDIHETHQRESSTRVSELEAQLESS 342
Query: 209 K-------TSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSDDLEGKFAM 261
+ K +E +S L +K+E+ T++ + LG+L D + K +
Sbjct: 343 EQRISDLTVDLKDAEEENKAISSKNLEIMDKLEQAQNTIKELMDELGEL-KDRHKEKESE 401
Query: 262 LES--SSVDDDLANLKKELSGSSKKGEL 287
L S S D +A++K+ L + ++ ++
Sbjct: 402 LSSLVKSADQQVADMKQSLDNAEEEKKM 429
>At3g28770 hypothetical protein
Length = 2081
Score = 39.3 bits (90), Expect = 0.003
Identities = 38/176 (21%), Positives = 68/176 (38%), Gaps = 9/176 (5%)
Query: 97 EMNDDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRKAQLALQKGEEDLAREALKRR 156
E N + T Q ++ ME++ K A E +K+Q Q ++ E L +
Sbjct: 1230 EKNKPKDDKKNTTKQSGGKKESMESESKEA-----ENQQKSQATTQADSDESKNEILMQA 1284
Query: 157 KSFADNASALKAQLDQQKGVV----DSLVSNTRLLESKIQEARSKKDTLKARAQSAKTST 212
S AD+ S +A D+ K + DS + R E ++ S + K + + +
Sbjct: 1285 DSQADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNK 1344
Query: 213 KVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSDDLEGKFAMLESSSVD 268
+ SG + E K + +++A Q SD+ + + M S D
Sbjct: 1345 PKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQAD 1400
Score = 38.1 bits (87), Expect = 0.006
Identities = 38/176 (21%), Positives = 68/176 (38%), Gaps = 9/176 (5%)
Query: 97 EMNDDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRKAQLALQKGEEDLAREALKRR 156
E N + T Q ++ ME++ K A E +K+Q Q ++ E L +
Sbjct: 1341 EKNKPKDDKKNTTKQSGGKKESMESESKEA-----ENQQKSQATTQADSDESKNEILMQA 1395
Query: 157 KSFADNASALKAQLDQQKGVV----DSLVSNTRLLESKIQEARSKKDTLKARAQSAKTST 212
S AD+ S +A D+ K + DS + R E ++ S + K + + +
Sbjct: 1396 DSQADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNK 1455
Query: 213 KVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSDDLEGKFAMLESSSVD 268
+ SG + E K + +++A Q SD+ + + M S D
Sbjct: 1456 PKDDKKNTTEQSGGKKESMESESKEAENQQKSQATTQGESDESKNEILMQADSQAD 1511
>At1g18410 kinesin-related protein, putative
Length = 1081
Score = 38.9 bits (89), Expect = 0.004
Identities = 47/198 (23%), Positives = 85/198 (42%), Gaps = 12/198 (6%)
Query: 85 EDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRKAQLALQKG 144
E +K EQ L+M +K + ATA + K +E K A+ A RKA +
Sbjct: 370 ETTKKAYEQQCLQME---SKTKGATAGIEDRVKELEQMRKDASVA-----RKALEERVRE 421
Query: 145 EEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKKDTLKAR 204
E + +EA + + + L+ D+ V S+ R LE QE + +L+A+
Sbjct: 422 LEKMGKEADAVKMNLEEKVKELQKYKDETITVTTSIEGKNRELEQFKQETMTVTTSLEAQ 481
Query: 205 AQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSDDL---EGKFAM 261
+ + + K M N + E+ +++ MT+ + +A + +L + K
Sbjct: 482 NRELEQAIK-ETMTVNTSLEAKNRELEQSKKETMTVNTSLKAKNRELEQNLVHWKSKAKE 540
Query: 262 LESSSVDDDLANLKKELS 279
+E S + + +KELS
Sbjct: 541 MEEKSELKNRSWSQKELS 558
Score = 29.3 bits (64), Expect = 3.0
Identities = 30/131 (22%), Positives = 58/131 (43%), Gaps = 14/131 (10%)
Query: 185 RLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQA 244
R + ++ + R++ KAR + ++ KV E L + + + K+EEK E
Sbjct: 289 RRISTQSEHLRTQNSVFKAREEKYQSRIKVLETLASGTSEENETEKSKLEEKKKDKE--- 345
Query: 245 EALGQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGRSSTRT-GTPFRDA 303
+D+ G E+ + +++ L++EL + K E + ++T G
Sbjct: 346 --------EDMVG--IEKENGHYNLEISTLRRELETTKKAYEQQCLQMESKTKGATAGIE 395
Query: 304 DIETELEQLRQ 314
D ELEQ+R+
Sbjct: 396 DRVKELEQMRK 406
>At5g10060 unknown protein
Length = 469
Score = 38.5 bits (88), Expect = 0.005
Identities = 35/149 (23%), Positives = 72/149 (47%), Gaps = 7/149 (4%)
Query: 71 RVVKSYANAIVSSFEDPEKILEQ---AVLEMNDDLTKMRQATAQVLASQKRMENKYKAAT 127
R KS + SS EKI V E +++ +M + + V +K ++ +A +
Sbjct: 153 RESKSSRTKLASSGGVAEKIASAYHLVVAENSNEEAEMNKCKSAVKRIRKMEKDVEEACS 212
Query: 128 QASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLD----QQKGVVDSLVSN 183
A + RK+ + EE L R+ +++ KS + S+L QL +Q+ +D+L +
Sbjct: 213 TAKDNPKRKSLAKELEEEEYLLRQCIEKLKSVQGSRSSLVNQLKDALREQESELDNLKAQ 272
Query: 184 TRLLESKIQEARSKKDTLKARAQSAKTST 212
++ + + +EA++ + L ++K +T
Sbjct: 273 IQVAKEQTEEAQNMQKRLNDEDYTSKQTT 301
>At5g42880 putative protein
Length = 751
Score = 37.4 bits (85), Expect = 0.011
Identities = 47/229 (20%), Positives = 97/229 (41%), Gaps = 42/229 (18%)
Query: 94 AVLEMNDDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRKA----------QLALQK 143
AVLE + M + +++A+++ +E+ + A +A E+ + A + L+
Sbjct: 301 AVLEAKEIERTMDGLSIELIATKELLESVHTAHLEAEEKRFSVAMARDQDVYNWEKELKM 360
Query: 144 GEEDLAR-----EALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLE-----SKIQE 193
E D+ R A K+ + ASAL+ L + + S LLE + ++
Sbjct: 361 VENDIERLNQEVRAADDVKAKLETASALQHDLKTELAAFTDISSGNLLLEKNDIHAAVES 420
Query: 194 ARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSD 253
AR + + +KA + A + K +++ +G+L + E + + Q E+ G ++
Sbjct: 421 ARRELEEVKANIEKAASEVKKLKII-----AGSLQSELGRERQDLEETKQKESTGLARTN 475
Query: 254 DLEGKFAMLES-----------------SSVDDDLANLKKELSGSSKKG 285
D + ++E+ ++ D + KELS +K+G
Sbjct: 476 DKDAGEELVETAKKLEQATKEAEDAKALATASRDELRMAKELSEQAKRG 524
Score = 35.0 bits (79), Expect = 0.054
Identities = 55/214 (25%), Positives = 88/214 (40%), Gaps = 16/214 (7%)
Query: 111 QVLASQKRMENKYKAATQ---ASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALK 167
Q L K+ E+ A T A EE A+ Q +E A +A + D K
Sbjct: 458 QDLEETKQKESTGLARTNDKDAGEELVETAKKLEQATKE--AEDAKALATASRDELRMAK 515
Query: 168 AQLDQQKGVVDSLVSNTRLLESKIQ-EARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGA 226
+Q K + ++ S RL+E+K + EA + L A A T+ S+ +N S
Sbjct: 516 ELSEQAKRGMSTIES--RLVEAKKEMEAARASEKLALAAIKALQETESSQRFEEINNSPR 573
Query: 227 ---LSAFEKMEEKVMTMESQAEALGQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSSK 283
+S E E +ES+ EA +L+ + + A E S + + L + +E+ S +
Sbjct: 574 SIIISVEEYYELSKQALESEEEANTRLSEIVSQIEVAKEEESRILEKLEEVNREM--SVR 631
Query: 284 KGELPPGRSSTRTGTPFRDADIETELEQLRQRSK 317
K EL RD + E E + RS+
Sbjct: 632 KAELKEANGKAEKA---RDGKLGMEQELRKWRSE 662
Score = 30.8 bits (68), Expect = 1.0
Identities = 34/186 (18%), Positives = 71/186 (37%), Gaps = 28/186 (15%)
Query: 102 LTKMRQATAQVLASQKRMENKYKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFAD 161
+T + Q + +K ++ + + +A E+ R+A+LA E+ +AL+
Sbjct: 153 ITDWKAHKIQTIERRKMVDEELEKIQEAMPEYKREAELA-----EEAKYDALE------- 200
Query: 162 NASALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKKDTLKAR--------AQSAKTSTK 213
+L+ KG+++ L E + Q+A+ + + R A A + K
Sbjct: 201 -------ELENTKGLIEELKLELEKAEKEEQQAKQDSELAQMRVEEMEKGVANEASVAVK 253
Query: 214 VSEMLGNVNTSGALSAFEKMEEKV-MTMESQAEALGQLTSDDLEGKFAMLESSSVDDDLA 272
+ A S + E++ M + L + A+LE+ ++ +
Sbjct: 254 TQLEVAKARQVSATSELRSVREEIEMVSNEYKDMLREKELAAERADIAVLEAKEIERTMD 313
Query: 273 NLKKEL 278
L EL
Sbjct: 314 GLSIEL 319
>At3g13300 unknown protein
Length = 1326
Score = 36.6 bits (83), Expect = 0.019
Identities = 39/180 (21%), Positives = 76/180 (41%), Gaps = 19/180 (10%)
Query: 80 IVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENKY-KAATQASEEWYRKAQ 138
+ SF + + L + ++ L M++ QV+ASQK M+ + AAT + ++ +
Sbjct: 900 LADSFNEQSQSLSHPMTDLLPQLLAMQETMNQVMASQKEMQRQLSNAATGPIGKESKRLE 959
Query: 139 LALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKK 198
+AL + E KS NA AL A++ ++ V N + L Q+ +
Sbjct: 960 VALGRMIE----------KSSKSNADALWARIQEE------TVKNEKALRDHAQQIVNAT 1003
Query: 199 DTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMT--MESQAEALGQLTSDDLE 256
++ +A + + L + + A S +E+ V + ES +G + L+
Sbjct: 1004 TNFMSKELNAMFEKTIKKELAAIGPALARSVVPVIEKTVSSAITESFQRGIGDKAVNQLD 1063
>At1g65010 hypothetical protein
Length = 1318
Score = 36.6 bits (83), Expect = 0.019
Identities = 45/223 (20%), Positives = 90/223 (40%), Gaps = 20/223 (8%)
Query: 67 ERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAA 126
E+ + K A AI ++ EK++E+A + + LA+QKR E ++
Sbjct: 72 EQIELLKKDKAKAI-DDLKESEKLVEEA-----------NEKLKEALAAQKRAEESFEVE 119
Query: 127 TQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRL 186
+ E + A+QK + E R A + SAL + ++ + V L
Sbjct: 120 KFRAVELEQAGLEAVQKKDVTSKNELESIRSQHALDISALLSTTEELQRVKHELSMTADA 179
Query: 187 LESKIQEARSKKDTLKARAQSAKTST----KVSEMLGNVNTSGALSAFE---KMEEKVMT 239
+ A + A+ A+ ++ +LG+ A+ E K++ ++
Sbjct: 180 KNKALSHAEEATKIAEIHAEKAEILASELGRLKALLGSKEEKEAIEGNEIVSKLKSEIEL 239
Query: 240 MESQAEALGQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSS 282
+ + E + L S L+ + ++E VD + A + + + SS
Sbjct: 240 LRGELEKVSILES-SLKEQEGLVEQLKVDLEAAKMAESCTNSS 281
Score = 32.7 bits (73), Expect = 0.27
Identities = 28/132 (21%), Positives = 56/132 (42%), Gaps = 8/132 (6%)
Query: 154 KRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKKDTLKARAQSAKTSTK 213
+ K + + L + ++ + +SLV L++ +QE ++ A + + +K
Sbjct: 831 EENKELRERETTLLKKAEELSELNESLVDKASKLQTVVQENEELRERETAYLKKIEELSK 890
Query: 214 VSEMLGNVNTSGALSAFEK--MEEKVMTMESQAEALGQLTSD------DLEGKFAMLESS 265
+ E+L + T +S EK ++E+ + E L ++ D +L G +E
Sbjct: 891 LHEILSDQETKLQISNHEKEELKERETAYLKKIEELSKVQEDLLNKENELHGMVVEIEDL 950
Query: 266 SVDDDLANLKKE 277
D LA K E
Sbjct: 951 RSKDSLAQKKIE 962
Score = 31.6 bits (70), Expect = 0.60
Identities = 44/225 (19%), Positives = 92/225 (40%), Gaps = 19/225 (8%)
Query: 73 VKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQK----RMENKYKAATQ 128
++ Y + + E+ K LE V + +L ++ + L ++K ++N T+
Sbjct: 352 LEEYGRQVCIAKEEASK-LENLVESIKSELEISQEEKTRALDNEKAATSNIQNLLDQRTE 410
Query: 129 ASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASA------LKAQLDQQKGVVDSLV- 181
S E R ++ +K ++D+ L +++ +++ A + +L + VDSL
Sbjct: 411 LSIELER-CKVEEEKSKKDMESLTLALQEASTESSEAKATLLVCQEELKNCESQVDSLKL 469
Query: 182 ---SNTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVM 238
E +++AR++ D+LK+ S + + S+ + +K EE+
Sbjct: 470 ASKETNEKYEKMLEDARNEIDSLKSTVDSIQNEFENSKAGWEQKELHLMGCVKKSEEENS 529
Query: 239 TMESQAEALGQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSSK 283
+ + + L L + E A E + L N K G K
Sbjct: 530 SSQEEVSRLVNLLKESEEDACARKEEEA---SLKNNLKVAEGEVK 571
>At3g22790 unknown protein
Length = 1694
Score = 36.2 bits (82), Expect = 0.024
Identities = 43/176 (24%), Positives = 74/176 (41%), Gaps = 18/176 (10%)
Query: 151 EALKRRK-SFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKKDTLKARAQSAK 209
E+L+ K F + L +QL + L+ LLE+ + A + Q K
Sbjct: 649 ESLRGEKYEFIAERANLLSQLQIMTENMQKLLEKNSLLETSLSGANIE-------LQCVK 701
Query: 210 TSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSD--DLEGKFAMLESSSV 267
+K E + + ++ E + + + E LG L +LEGK+A L+
Sbjct: 702 EKSKCFEEFFQLLKNDKAELIKERESLISQLNAVKEKLGVLEKKFTELEGKYADLQREKQ 761
Query: 268 DDDLANLKKELSGSSKKGELPPGRSSTRTGTPFRDADIETEL----EQLRQRSKEF 319
+L + +S +++K E R+S T R AD++ + E+ R R KEF
Sbjct: 762 FKNLQVEELRVSLATEKQE----RASYERSTDTRLADLQNNVSFLREECRSRKKEF 813
>At3g07170 unknown protein
Length = 203
Score = 36.2 bits (82), Expect = 0.024
Identities = 40/166 (24%), Positives = 77/166 (46%), Gaps = 17/166 (10%)
Query: 100 DDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRKAQLALQKGEEDLAREALKRR--- 156
D T++RQ T + ++R ++K++ +S+ K QL+ ++ + R L++R
Sbjct: 26 DISTRVRQVTGK----RQRQDDKWEHDLFSSD----KPQLSNRRVDPRDLRLKLQKRHHG 77
Query: 157 -KSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKKDTLKARAQSAKTSTKVS 215
+S + S ++ DQ G ++ N+ +SK + AR ++ ++ KTS++ +
Sbjct: 78 SQSGREAGSGVRDLRDQLSGTMNQQPKNSDPPKSKAEAARPSMKSVATETETRKTSSQAT 137
Query: 216 EMLGNVNTSGALSAFEKME-EKVMTM----ESQAEALGQLTSDDLE 256
S S E + EK T E +AL +T DDL+
Sbjct: 138 RKKSQQADSSVDSFLESLGLEKYSTAFQVEEVDMDALMHMTDDDLK 183
>At2g26770 unknown protein
Length = 496
Score = 35.8 bits (81), Expect = 0.032
Identities = 52/209 (24%), Positives = 80/209 (37%), Gaps = 46/209 (22%)
Query: 109 TAQVLASQKRM---------ENKYKAATQASEEWYRKAQLALQK---------GEEDLAR 150
TAQ++ QKR+ E AA + SEE K +L+K E L
Sbjct: 62 TAQLMDQQKRLSVRDLAHKFEKGLAAAAKLSEEAKLKEATSLEKHVLLKKLRDALESLRG 121
Query: 151 EALKRRKSFADNA----SALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKK--DTLKAR 204
R K + A AL QL Q++G E I++A KK LK
Sbjct: 122 RVAGRNKDDVEEAIAMVEALAVQLTQREG------------ELFIEKAEVKKLASFLKQA 169
Query: 205 AQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSDDL------EGK 258
++ AK A +A +++EE + E + A G+ +DL +
Sbjct: 170 SEDAKKLVDEERAFARAEIESARAAVQRVEEALREHEQMSRASGKQDMEDLMKEVQEARR 229
Query: 259 FAMLESSS----VDDDLANLKKELSGSSK 283
ML S ++ +L L+ +L+ SK
Sbjct: 230 IKMLHQPSRVMDMEYELRALRNQLAEKSK 258
>At1g55250 unknown protein
Length = 899
Score = 35.8 bits (81), Expect = 0.032
Identities = 52/224 (23%), Positives = 94/224 (41%), Gaps = 23/224 (10%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
+NL + S E EK L+ ++E N + T + + +R + K
Sbjct: 418 LNLRAESLEAANHKTTTVGSRIEVLEKKLQSCIIEKNG----LELETEEAIQDSERQDIK 473
Query: 123 YK----AATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVD 178
+ A+T + E +AQL K +D A++AL R+ +L + D+QKG+ D
Sbjct: 474 SEFIAMASTLSKEMEMMEAQL---KRWKDTAQDALYLREQAQSLRVSLSNKADEQKGLED 530
Query: 179 SLVSNTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVM 238
++ E K +A +K LK + Q ++ + N L+ + + K
Sbjct: 531 KCAK--QMAEIKSLKALIEK-LLKEKLQLQNLASICTR---ECNDDRGLAEIKDSQRKA- 583
Query: 239 TMESQAEALGQLTSD---DLEGKFAMLESSSVDDDLANLKKELS 279
++QAE L + + +L K A S+ + LA K E++
Sbjct: 584 --QAQAEELKNVLDEHFLELRVKAAHETESACQERLATAKAEIA 625
>At2g33240 putative myosin heavy chain
Length = 1611
Score = 35.4 bits (80), Expect = 0.041
Identities = 45/191 (23%), Positives = 84/191 (43%), Gaps = 29/191 (15%)
Query: 82 SSFEDPEKILEQAV--LEMNDDLTKMRQATAQVLASQKRM----ENKYKAATQASEEWYR 135
S+ +D + E+ LEM +DL + +++S +R ++KY+ ++ SEE +
Sbjct: 982 SALQDMQLEFEELAKELEMTNDLAAENEQLKDLVSSLQRKIDESDSKYEETSKLSEERVK 1041
Query: 136 KAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEAR 195
+ + +G ++ K+ S L+ ++D D LV LLE KI E
Sbjct: 1042 QEVPVIDQGVIIKLEAENQKLKALV---STLEKKIDSLDRKHDDLVD---LLERKIDETE 1095
Query: 196 SK--------KDTLKARAQSAKTSTKVSEMLGN-----VNTSGAL----SAFEKMEEKVM 238
K ++ LK + K + S + V+T L ++ +++EEKV
Sbjct: 1096 KKYEEASKLCEERLKQVVDTEKKYEEASRLCEERLKQVVDTETKLIELKTSMQRLEEKVS 1155
Query: 239 TMESQAEALGQ 249
ME++ + L Q
Sbjct: 1156 DMEAEDKILRQ 1166
>At2g28620 putative kinesin-like spindle protein
Length = 1076
Score = 35.4 bits (80), Expect = 0.041
Identities = 43/183 (23%), Positives = 69/183 (37%), Gaps = 18/183 (9%)
Query: 78 NAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRKA 137
N++ S EK L+ M ++ +AT + S +++ KY ++ ++
Sbjct: 587 NSVAGSVSQQEKQLQDMENVMVSFVSAKTKATETLRGSLAQLKEKYNTGIKSLDDIAGNL 646
Query: 138 QLALQKGEEDLAREALKRR-------KSFADNA----SALKAQLDQQKGVVDSLVSNTRL 186
Q DL E K K F A L+ L Q+ + + R
Sbjct: 647 DKDSQSTLNDLNSEVTKHSCALEDMFKGFTSEAYTLLEGLQGSLHNQEEKLSAFTQQQRD 706
Query: 187 LESKIQEARSKKDTL-----KARAQSAKTSTKVSEMLGNVNTSGALSAF-EKMEEKVMTM 240
L S+ ++ T+ K A TK++E NVN LSAF +K EE +
Sbjct: 707 LHSRSMDSAKSVSTVMLDFFKTLDTHANKLTKLAEDAQNVNEQ-KLSAFTKKFEESIANE 765
Query: 241 ESQ 243
E Q
Sbjct: 766 EKQ 768
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.123 0.316
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,837,012
Number of Sequences: 26719
Number of extensions: 228685
Number of successful extensions: 1194
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 132
Number of HSP's that attempted gapping in prelim test: 1129
Number of HSP's gapped (non-prelim): 192
length of query: 319
length of database: 11,318,596
effective HSP length: 99
effective length of query: 220
effective length of database: 8,673,415
effective search space: 1908151300
effective search space used: 1908151300
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137825.9


