

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137825.14 + phase: 0
(552 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
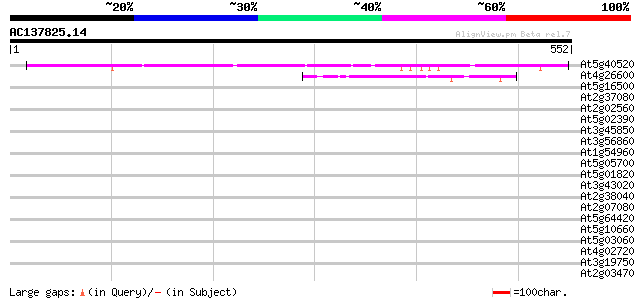
Score E
Sequences producing significant alignments: (bits) Value
At5g40520 putative protein 308 4e-84
At4g26600 unknown protein 45 1e-04
At5g16500 protein kinase-like protein 40 0.003
At2g37080 putative myosin heavy chain 39 0.008
At2g02560 unknown protein 37 0.037
At5g02390 putative protein 36 0.049
At3g45850 kinesin-related protein - like 36 0.064
At3g56860 unknown protein 35 0.083
At1g54960 NPK1-related protein kinase 2 (ANP2) 35 0.14
At5g05700 arginine-tRNA-protein transferase 1 homolog 34 0.18
At5g01820 unknown protein 34 0.18
At3g43020 putative protein 34 0.18
At2g38040 carboxyltransferase alpha subunit (CAC3) 34 0.18
At2g07080 putative gag-protease polyprotein 34 0.18
At5g64420 putative protein 34 0.24
At5g10660 vacuolar calcium binding protein - like 34 0.24
At5g03060 putative protein 34 0.24
At4g02720 unknown protein 34 0.24
At3g19750 hypothetical protein 33 0.32
At2g03470 putative MYB family transcription factor 33 0.32
>At5g40520 putative protein
Length = 693
Score = 308 bits (790), Expect = 4e-84
Identities = 214/591 (36%), Positives = 319/591 (53%), Gaps = 70/591 (11%)
Query: 17 LDNLTTKGLYLLAMIVGGDTVKYERTRSNLKKIIKSSLSSVLSSKRNKDQQLETRKQLFQ 76
LD+LT KGLYL+AMI+ G + +++TR +K+II+ S+ + + E QL Q
Sbjct: 107 LDSLTNKGLYLIAMILTGGSTSFDKTRLKMKEIIRDSVRRDFGKNKEGIGKEEIINQLHQ 166
Query: 77 LLNNPQNYRQRCELLPGSESQFY--HAAVVKVLCGLEKLPSQTLLAMRRKLKGIKAPMPQ 134
+L++P N+R+ C + G + A +KVL L+ L +QTL AM+RKLKG + +PQ
Sbjct: 167 VLSDPANFREDCRMNLGRTPTLHSHRDAAMKVLNELDSLSTQTLRAMKRKLKGSRM-IPQ 225
Query: 135 LQPFKNGWGRNHLIRQVNKISRKMLLELDGGNKLQEPLANAMSVADFSLKLIIGFGSTFL 194
L+ + G R+ LI QV + S KML EL G+KLQE LA A+SV D SLKL G+ +
Sbjct: 226 LKNSRRGQRRSDLINQVRQASEKMLSELSAGDKLQEQLAKALSVVDLSLKLSPGYKTAAA 285
Query: 195 EECYQFSPEVKSLQSDIMQAILSVEKKEVVPLPVLRELQLLIEPKATVVNKSLRKAFVNL 254
+ ++FSPE K+LQ++I++A+ + K V L+ L L ++P+A V N SLR A
Sbjct: 286 TDFFRFSPETKNLQNEIVKAVWLLRK---VRFRELKRLHLCLDPEAEVSNDSLRSAVRKT 342
Query: 255 LTEFLFECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQI 314
L E+LFECSD+++ PKSL++ L ++N + H V +E IEEE + IL+VSAQ QI
Sbjct: 343 LIEYLFECSDLDTIPKSLMEALSLVNSRTGNVEHKVC-PREAIEEETECILNVSAQVKQI 401
Query: 315 IQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDK 374
+P+YE DQ F DAYME+ EDSD+ D D+DD EN C++ +E L + +
Sbjct: 402 FCQCIPNYELDQDFGDAYMEDL-EDSDDNDDDDDDDDDDDEN---CNLFDNEKLGNKEES 457
Query: 375 NCDTINLESK--------------------------MRNTPYNNQ--------------C 394
C I LE K +T NQ
Sbjct: 458 RCRNIKLEVKDTQDDAMESSDSDHEESGAECLVLDPTDSTHETNQHDFSSSVNRVVVRDL 517
Query: 395 QEEITEQFS-----TPMSRKNC---DSSAVSLDK-----EPDENIVKRHEFHESYTEAAP 441
E IT + TP S K+ D +++ K E D + ++F +
Sbjct: 518 PESITRVYPRSLYVTPTSNKSIVISDRHDIAMSKTRVKVERDIEMEVDNQFSSRSLFSVE 577
Query: 442 RDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDKTSMLAYNLIGRMLEEFAIAEDLNLDLS 501
KS+ E+KP +KNQYLA Q+ D+TS++A+NLIGR+LE+FA + LNL+
Sbjct: 578 NIKSDDHGEQKP----RRRNKNQYLAIQEISDETSLVAHNLIGRLLEKFADRQRLNLETD 633
Query: 502 KRSYLNCDKQSEDIKETEE--QSSSRKRKGGPPIVRVIEELIPSFPDRYLL 550
+RSYL + + ++ E E Q+SS+ + I+ V++E I S + L+
Sbjct: 634 ERSYLGGESRLQEKVEVSEKKQASSQAKSDEAIILSVVKEQITSLEESVLM 684
>At4g26600 unknown protein
Length = 671
Score = 45.1 bits (105), Expect = 1e-04
Identities = 52/215 (24%), Positives = 91/215 (42%), Gaps = 17/215 (7%)
Query: 289 DVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDE 348
+V+ +E EEE + S + ++ DL D + ++G D+ ++ +D D++ D DE
Sbjct: 56 EVSTDEEEEEEENEQ----SDEGSESGSDLFSDGD-EEGNNDS--DDDDDDDDDDDDDDE 108
Query: 349 DDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQ-EEITEQFSTPMS 407
D +E+ L S + SD D + LE K R + + ++ ++F +
Sbjct: 109 DAEPLAEDFLDGSDNEEVTMGSDLDSDSGGSKLERKSRAIDRKRKKEVQDADDEFKMNIK 168
Query: 408 RKNCDSSAVSLDKEPDENIVKRHEFH--ESYTEAAPRDKSNFCEEKKPIPTKYSGHKNQY 465
K D + KE +E + + + R SNF K + K H+
Sbjct: 169 EKP-DEFQLPTQKELEEEARRPPDLPSLQMRIREIVRILSNF----KDLKPKGDKHERND 223
Query: 466 LAAQDACDKTSMLAYN--LIGRMLEEFAIAEDLNL 498
Q D +S YN LIG ++E F + E + L
Sbjct: 224 YVGQLKADLSSYYGYNEFLIGTLIEMFPVVELMEL 258
>At5g16500 protein kinase-like protein
Length = 636
Score = 40.4 bits (93), Expect = 0.003
Identities = 56/225 (24%), Positives = 96/225 (41%), Gaps = 40/225 (17%)
Query: 197 CYQFSPEVKSLQSDIMQAI--LSVEKKEVVPLPVLRELQLLIEPKATVVNKSLRKAFVNL 254
C Q P + L SD+M A+ LS+ ++ +P ATV +S R +++
Sbjct: 331 CLQEEPTARPLISDVMVALSFLSMSTEDGIP--------------ATVPMESFRDKSMSI 376
Query: 255 LTEFLFECSDMNSTPKSLLQILDVINKCSNKS-THDVTLQKEHIEEEVDSILSVSAQTNQ 313
CS TP + + DV NK S+ S + D +KE E+ + S Q
Sbjct: 377 ALSRHGSCS---VTPFCISR-KDVGNKSSSSSDSEDEEEEKEQKAEKEEESTSKKRQ--- 429
Query: 314 IIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSD 373
E ++ TD+ ++SD+ S+KD+++ Q S + S S+
Sbjct: 430 ---------EQEETATDS-----DDESDSNSEKDQEEEQSQLEKARESSSSSSDSGSER- 474
Query: 374 KNCDTINLESKMRNTPYNN-QCQEEITEQFSTPMSRKNCDSSAVS 417
++ D N ++ Y+N +EE E+ S+ S K+ + S S
Sbjct: 475 RSIDETNATAQSLKISYSNYSSEEEDNEKLSSKSSCKSNEESTFS 519
>At2g37080 putative myosin heavy chain
Length = 583
Score = 38.9 bits (89), Expect = 0.008
Identities = 80/416 (19%), Positives = 174/416 (41%), Gaps = 53/416 (12%)
Query: 43 RSNLKKIIKSSLSSVLSSKRNKDQQLETRKQLFQLLNNPQNYRQRCELLPGSESQFYHAA 102
+ LKK + +S K +DQ ET++QL ++ + + L + + +
Sbjct: 87 QEELKKAKEQLSASEALKKEAQDQAEETKQQLMEINASEDSRIDELRKLSQERDKAWQSE 146
Query: 103 VVKVLCGLEKLPSQTLLAMRRKLKGIKAPMPQLQPFKN-GWGRNHLIRQVNKISRKMLLE 161
++ + + S L + +++ +KA + + + +N N + V K+ ++
Sbjct: 147 -LEAMQRQHAMDSAALSSTMNEVQKLKAQLSESENVENLRMELNETLSLVEKLRGELFDA 205
Query: 162 LDGGNKLQEPLANA---MSVADFSLKLIIGFGSTFLEECYQFSPEVKSLQSDIMQAILSV 218
+G + E ++ + +A+ +L+++ G E C + E++ +S++ S+
Sbjct: 206 KEGEAQAHEIVSGTEKQLEIANLTLEMLRSDGMKMSEACNSLTTELEQSKSEVR----SL 261
Query: 219 EKKEVVPLPVLRELQLLIEPKATVVNKSLRKAFVNLLTEFLFECSDMNSTPKSLLQILDV 278
E+ ++R+L+ E + N + + V L E ++N + + Q+
Sbjct: 262 EQ-------LVRQLEEEDEARG---NANGDSSSVEELKE------EINVARQEISQL--- 302
Query: 279 INKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFT--DAYMEEQ 336
KS +VT ++ H EE + S L + Q+ E G+ +A + E+
Sbjct: 303 ------KSAVEVTERRYH-EEYIQSTLQIRTAYEQV-------DEVKSGYAQREAELGEE 348
Query: 337 SEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSD-SDKNCDTINLESKMRNTPYNNQCQ 395
+ + E D + E L V +E LNS +K +NLE+ + NQ +
Sbjct: 349 LKKTKAERDSLHERLMDKEAKLRILVDENEILNSKIKEKEEVYLNLENSL------NQNE 402
Query: 396 EEITEQFSTPMSRKNCDSSAVSLDKEPD-ENIVKRHEFHESYTEAAPRDKSNFCEE 450
E T + + + A +DKE + ++++ ++E S E +K+ +E
Sbjct: 403 PEDTGELK-KLESDVMELRANLMDKEMELQSVMSQYESLRSEMETMQSEKNKAIDE 457
>At2g02560 unknown protein
Length = 1217
Score = 36.6 bits (83), Expect = 0.037
Identities = 24/80 (30%), Positives = 43/80 (53%), Gaps = 10/80 (12%)
Query: 278 VINKCSNKSTHDVTLQKEHIEEEVDSIL-----SVSAQTNQIIQDLLPDYEFDQGFTDAY 332
+IN C++ S +D L +E+ + ++S L +S ++I+ L +D FTD
Sbjct: 255 LINYCTSASENDEEL-REYSLQALESFLLRCPRDISPYCDEILNLTLEYISYDPNFTD-- 311
Query: 333 MEEQSEDSDNESDKDEDDSQ 352
ED+DNE+ +DE+D +
Sbjct: 312 --NMEEDTDNETLEDEEDDE 329
>At5g02390 putative protein
Length = 835
Score = 36.2 bits (82), Expect = 0.049
Identities = 27/106 (25%), Positives = 49/106 (45%), Gaps = 11/106 (10%)
Query: 333 MEEQSEDSDNESDKDEDDSQCSEN-----SLHCSVTPSECLNSDSDKNCDTINLESKMRN 387
+EE+ + DN S+ ED SQ SE+ ++ S ++ D+ T+++E + R+
Sbjct: 550 IEEEQDGLDNISEISEDHSQSSEHETLDQTMDASEDSPVDAETEQDREISTLDVEHETRS 609
Query: 388 TPY------NNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIV 427
NN +I E S ++ D+ VS K+ DE ++
Sbjct: 610 LKESSEESPNNVSTVDIDENASVFDISRDLDTETVSTSKQLDEEVL 655
>At3g45850 kinesin-related protein - like
Length = 1058
Score = 35.8 bits (81), Expect = 0.064
Identities = 37/184 (20%), Positives = 77/184 (41%), Gaps = 40/184 (21%)
Query: 180 DFSLKLIIGFGSTFLEECYQFSPEVKSLQSDIMQAILSVEKKEVVPLPVLRELQLLIEPK 239
+F + ++ + +E +Q E++S SD+ +E+K+ K
Sbjct: 517 EFVISNLLKSEKSLVERAFQLRTELESASSDVSNLFSKIERKD----------------K 560
Query: 240 ATVVNKSLRKAFVNLLTEFLFECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEE 299
N+ L + F + LT Q L++++K S +Q +H+EE
Sbjct: 561 IEDGNRFLIQKFQSQLT-----------------QQLELLHKTVASSVTQQEVQLKHMEE 603
Query: 300 EVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLH 359
+++S +S ++ + ++D L + G E DN + K + +SQ + +SL+
Sbjct: 604 DMESFVSTKSEATEELRDRLSKLKRVYG-------SGIEALDNIAVKLDGNSQSTFSSLN 656
Query: 360 CSVT 363
V+
Sbjct: 657 SEVS 660
>At3g56860 unknown protein
Length = 478
Score = 35.4 bits (80), Expect = 0.083
Identities = 24/91 (26%), Positives = 41/91 (44%), Gaps = 7/91 (7%)
Query: 283 SNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDN 342
SN++ K+ EEE ++ NQ Q + + E+++ + EE+ ED D+
Sbjct: 12 SNEAEEPSQKLKQTPEEEQQLVIK-----NQDNQGDVEEVEYEE--VEEEQEEEVEDDDD 64
Query: 343 ESDKDEDDSQCSENSLHCSVTPSECLNSDSD 373
E D DE++ Q N + + T D D
Sbjct: 65 EDDGDENEDQTDGNRIEAAATSGSGNQEDDD 95
>At1g54960 NPK1-related protein kinase 2 (ANP2)
Length = 651
Score = 34.7 bits (78), Expect = 0.14
Identities = 39/171 (22%), Positives = 74/171 (42%), Gaps = 22/171 (12%)
Query: 257 EFLFEC----SDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEH----IEEEVDSILSVS 308
+FL +C ++ T LL+ V K ++ D+T ++ + E+ +I S
Sbjct: 305 DFLLKCLQQEPNLRPTASELLKHPFVTGKQKESASKDLTSFMDNSCSPLPSELTNITSYQ 364
Query: 309 AQTNQIIQDL--LPDYEFDQGFTDAYMEEQSE--DSDNESDKDEDDSQC---SENSL--H 359
T+ + D+ L F + ++ S S+N D D+D+ C EN L +
Sbjct: 365 TSTSDDVGDICNLGSLTCTLAFPEKSIQNNSLCLKSNNGYDDDDDNDMCLIDDENFLTYN 424
Query: 360 CSVTPSECLNSDSDKNCDTIN-----LESKMRNTPYNNQCQEEITEQFSTP 405
PS N+D+ K+CDT++ L+ K N + + +++ + P
Sbjct: 425 GETGPSLDNNTDAKKSCDTMSEISDILKCKFDENSGNGETETKVSMEVDHP 475
>At5g05700 arginine-tRNA-protein transferase 1 homolog
Length = 632
Score = 34.3 bits (77), Expect = 0.18
Identities = 34/124 (27%), Positives = 60/124 (47%), Gaps = 17/124 (13%)
Query: 303 SILSVSAQTNQIIQDLLPDYE-FDQGFT-DAYM----EEQSEDSDNESDKDEDDSQCSEN 356
S+L+ +++T +++ ++E +QG T D +M E++ ED D++ D D+DD + E
Sbjct: 505 SLLAGASET--LVEPAASEHEDMEQGETNDNFMGCSDEDEDEDEDDD-DDDDDDEEMYET 561
Query: 357 SLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAV 416
S S+ + D+D N I L Y +Q + + Q TP+ RK +
Sbjct: 562 ESEDSHIESDPGSKDNDINNILIGL--------YGSQYRYKEMRQIITPVGRKQLEPMLQ 613
Query: 417 SLDK 420
S K
Sbjct: 614 SYRK 617
>At5g01820 unknown protein
Length = 442
Score = 34.3 bits (77), Expect = 0.18
Identities = 17/49 (34%), Positives = 27/49 (54%)
Query: 304 ILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQ 352
+L + QT I++++ D F QG+ D + EDSD + DE DS+
Sbjct: 256 LLDTNPQTRITIEEIIHDPWFKQGYDDRMSKFHLEDSDMKLPADETDSE 304
>At3g43020 putative protein
Length = 366
Score = 34.3 bits (77), Expect = 0.18
Identities = 39/167 (23%), Positives = 71/167 (42%), Gaps = 20/167 (11%)
Query: 298 EEEVDSILSVSAQTNQIIQDLLP---DYEFDQ-----GFTDAYMEEQSEDSDNESDKDED 349
EE+VDS + V +NQ ++ LLP D+ D+ D E SE+ + +++ +
Sbjct: 181 EEDVDSKMQVDPSSNQSVEKLLPLNQDHIIDRVPAIHSGLDLPKEHNSEEQQSNANEADV 240
Query: 350 DSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEEITEQFSTPMSRK 409
DS+ S+ S+ +SD D + + E + + T + +++
Sbjct: 241 DSKMSD-----SLDRDPASHSDLDLAIEQSSKEEQAKVTLES----VAVSQSLLQTQPEV 291
Query: 410 NCDSSAVSLDKEPDENIVKRHEFHESYTE--AAPRDKSNFCEEKKPI 454
N D + D+ D + H+ ++ E A D S E +KPI
Sbjct: 292 NFDGDPMK-DQITDSRDIGGHDLDDNQEEGYVAVSDSSPAREREKPI 337
>At2g38040 carboxyltransferase alpha subunit (CAC3)
Length = 769
Score = 34.3 bits (77), Expect = 0.18
Identities = 58/279 (20%), Positives = 117/279 (41%), Gaps = 21/279 (7%)
Query: 94 SESQFYHAAVVKVLCGLEKLPSQ--TLLAMRRKLKGIKAPMPQLQPFKNGWGRNHLIRQV 151
SE H +++ + EKL + T L + +K+ + L+ F +
Sbjct: 503 SEEHLMHPVLIEKI---EKLKEEFNTRLTDAPNYESLKSKLNMLRDFSRAKAASEATSLK 559
Query: 152 NKISRKMLLELDGGNKLQEPLANAMSVADFSLKLIIGFGSTFLEECYQFSPEVKSLQSDI 211
+I+++ +D ++ A VA S+F E +V + ++
Sbjct: 560 KEINKRFQEAVDRPEIREKVEAIKAEVASSG-------ASSFDELPDALKEKVLKTKGEV 612
Query: 212 MQAILSVEKKEVVPLPVLRELQLLIEPKATVVNKSLRKAFVNLLTEFLFECSDMNSTP-- 269
+ V K + L +++ Q + N++L++ L E + ++ TP
Sbjct: 613 EAEMAGVLKSMGLELDAVKQNQKDTAEQIYAANENLQEKLEKLNQEITSKIEEVVRTPEI 672
Query: 270 KSLLQILDVINKCSNKSTHDVT--LQK-EHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQ 326
KS++++L V ++K T VT QK E +E+++ ++ + T+ + Q+ + E +
Sbjct: 673 KSMVELLKVETAKASK-TPGVTEAYQKIEALEQQIKQKIAEALNTSGL-QEKQDELEKEL 730
Query: 327 GFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPS 365
E+S+ S E D D++DS SE+ V PS
Sbjct: 731 AAARELAAEESDGSVKEDDDDDEDS--SESGKSEMVNPS 767
>At2g07080 putative gag-protease polyprotein
Length = 627
Score = 34.3 bits (77), Expect = 0.18
Identities = 33/125 (26%), Positives = 54/125 (42%), Gaps = 9/125 (7%)
Query: 418 LDKEPDENIVKRHEFHESYTEAAPRDKSNFCEEK------KPIPTKYSGHKNQYLAAQDA 471
L EPDE IVK + A + ++K + +P K++ HK A +
Sbjct: 116 LKMEPDEKIVKFSSKISALANEAEVMGKTYKDQKLVKKLLRCLPPKFAAHKAVMRVAGNT 175
Query: 472 CDKTSMLAYNLIGRMLEEFAIAEDLNLDLSKRSYLNCDKQSEDIKETEEQSSSRKRKGGP 531
DK S + +L+G + E A+ + SK N D+ SE +E ++ + R G
Sbjct: 176 -DKISFV--DLVGMLKLEEMKADQDKVKPSKNIAFNADQGSEQFQEIKDGMALLARNFGK 232
Query: 532 PIVRV 536
+ RV
Sbjct: 233 ALKRV 237
>At5g64420 putative protein
Length = 1306
Score = 33.9 bits (76), Expect = 0.24
Identities = 38/183 (20%), Positives = 72/183 (38%), Gaps = 29/183 (15%)
Query: 219 EKKEVVPLPVLRELQLLIEPKATVVNKSLRKAFVNLLTEFLFECSDMNSTPKSLLQILDV 278
+++E + VL + L + P ++ +S ++ F + C D+ T LL++L V
Sbjct: 840 DEEEPAVMDVLVDTLLSLLPHSSAPMRS------SIEQVFKYFCQDV--TNDGLLRMLRV 891
Query: 279 INKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSE 338
I K S H + ++++ + L++ DA E +
Sbjct: 892 IKKDLKPSRHQEDQDSDDLDDDEEDCLAIE---------------------DAEEENEEM 930
Query: 339 DSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEEI 398
ESD+ DDS+ + +V NSD + D ++ ++ R Y Q +E
Sbjct: 931 GETGESDEQTDDSEAVTGVVPMAVDREVPENSDDSDDDDGMDDDAMFRMDTYLAQIFKEK 990
Query: 399 TEQ 401
Q
Sbjct: 991 RNQ 993
>At5g10660 vacuolar calcium binding protein - like
Length = 407
Score = 33.9 bits (76), Expect = 0.24
Identities = 37/168 (22%), Positives = 66/168 (39%), Gaps = 17/168 (10%)
Query: 323 EFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLE 382
E D +D E + ED D + DE + + + S + D DK+ + I
Sbjct: 213 ETDVHISDHGEEPKEEDKDQFAQPDESGEEKETSPVAASTEEQKGELIDEDKSTEQIEEP 272
Query: 383 SKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHE---------FH 433
+ N NN +EE ++ S +N ++ A + D N+ + E
Sbjct: 273 KEPENIEENNSEEEEEVKKKSD--DEENSETVATTTDMNEAVNVEESKEEEKEEAEVKEE 330
Query: 434 ESYTEAAPRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDKTSMLAYN 481
E + AA + + + + +P + G KN+ + + K S AYN
Sbjct: 331 EGESSAAKEETTETMAQVEELPEE--GTKNEVVQGK----KESPTAYN 372
Score = 29.3 bits (64), Expect = 5.9
Identities = 37/204 (18%), Positives = 77/204 (37%), Gaps = 28/204 (13%)
Query: 340 SDNESDKDEDDSQCSENSLHCSVTP-SECLNSDSDKNCDTINLESKMRNTPYNNQCQEEI 398
+ + K E + S ++P S L S + + + +E+ + + + + +EE
Sbjct: 170 NSSSKSKKEGSGNVPKKSSGKEISPDSSPLASAHEDEEEIVKVETDVHISDHGEEPKEED 229
Query: 399 TEQFSTPMS---RKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRD-KSNFCEEKKPI 454
+QF+ P K A S +++ E I + + P + + N EE++ +
Sbjct: 230 KDQFAQPDESGEEKETSPVAASTEEQKGELIDEDKSTEQIEEPKEPENIEENNSEEEEEV 289
Query: 455 PTKYSGHKNQYLAAQDACDKTSMLAYNLIGRMLEEFAIAEDLNLDLSKRSYLNCDKQSED 514
K +N E A D+N ++ +K+ +
Sbjct: 290 KKKSDDEENS-----------------------ETVATTTDMNEAVNVEESKEEEKEEAE 326
Query: 515 IKETEEQSSSRKRKGGPPIVRVIE 538
+KE E +SS+ K + + +V E
Sbjct: 327 VKEEEGESSAAKEETTETMAQVEE 350
>At5g03060 putative protein
Length = 292
Score = 33.9 bits (76), Expect = 0.24
Identities = 34/181 (18%), Positives = 74/181 (40%), Gaps = 31/181 (17%)
Query: 198 YQFSPEVKSLQSDIMQAILSVEKKEVVPLPVLRELQLLIEPKATVVN------------- 244
+Q+ ++K L+ I+ + +EK+ ++E++ +I K ++N
Sbjct: 72 FQYQKQIKELEEKILSLLKDLEKERSEKEEYMKEMKGMISEKEAIINDLSVKNQELLIAK 131
Query: 245 -------KSLRKAFVNLLTEFLFECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHI 297
K + + L F D + S L+ L + + DV + E I
Sbjct: 132 EEEVEKLKKMENKYAELKERF-----DAAESQCSFLKSLFDAENLAGLGSSDVAFENEVI 186
Query: 298 EEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENS 357
E D +++ N++I ++ D+ + TD + D++N++ + + SENS
Sbjct: 187 VVE-DYNVTIK---NEVI--MVDDHNVNNTPTDTIVILDESDAENDNPHPRESNIISENS 240
Query: 358 L 358
+
Sbjct: 241 V 241
>At4g02720 unknown protein
Length = 422
Score = 33.9 bits (76), Expect = 0.24
Identities = 29/135 (21%), Positives = 53/135 (38%), Gaps = 7/135 (5%)
Query: 335 EQSEDSDNESDK-------DEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRN 387
+Q+EDSD +D+ + DD+ + S + SE S K + + R
Sbjct: 69 DQNEDSDENADEIQDKNGGERDDNSKGKERKGKSDSESESDGLRSRKRKSKSSRSKRRRK 128
Query: 388 TPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDKSNF 447
Y++ + E +E S R+ S+ K ++ H T+ + D+S+
Sbjct: 129 RSYDSDSESEGSESDSEEEDRRRRRKSSSKRKKSRSSRSFRKKRSHRRKTKYSDSDESSD 188
Query: 448 CEEKKPIPTKYSGHK 462
+ K I SG +
Sbjct: 189 EDSKAEISASSSGEE 203
>At3g19750 hypothetical protein
Length = 378
Score = 33.5 bits (75), Expect = 0.32
Identities = 23/100 (23%), Positives = 48/100 (48%), Gaps = 9/100 (9%)
Query: 317 DLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNC 376
D DY + E ++++S ESD +ED S SE + ++ SDS++ C
Sbjct: 259 DFADDYNEESESDGENAEAEADESTTESDAEEDSSAQSEED-----SEAKADGSDSEEAC 313
Query: 377 DTINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAV 416
++ E+ + P + ++ + E+ + + C+++AV
Sbjct: 314 LEVSEEAIKKIVPKLEEFEKLVAEE----LHGEACEAAAV 349
>At2g03470 putative MYB family transcription factor
Length = 450
Score = 33.5 bits (75), Expect = 0.32
Identities = 31/122 (25%), Positives = 50/122 (40%), Gaps = 15/122 (12%)
Query: 321 DYEFDQGFTDAYM--EEQSEDSDNESD-KDEDDSQCSENSLHCSVTP-------SECLNS 370
D E + G +Y EE+ E S N+ D ++E++ S N HC T E +N
Sbjct: 311 DEEDNNGNRSSYEDNEEEEETSSNDDDEEEEEEDDSSSNDAHCVDTDKASRDGFGEEVNV 370
Query: 371 DSDKNCDTINLESK----MRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENI 426
+ D +C + L+ N N +C + +S R D + D P NI
Sbjct: 371 EDD-SCMSFELQDSNLIFSHNPIKNRECHRSGEDSYSFDDQRFTSDCWNKNNDLLPTSNI 429
Query: 427 VK 428
++
Sbjct: 430 IE 431
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.131 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,532,881
Number of Sequences: 26719
Number of extensions: 562003
Number of successful extensions: 3134
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 79
Number of HSP's that attempted gapping in prelim test: 2901
Number of HSP's gapped (non-prelim): 178
length of query: 552
length of database: 11,318,596
effective HSP length: 104
effective length of query: 448
effective length of database: 8,539,820
effective search space: 3825839360
effective search space used: 3825839360
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC137825.14


