

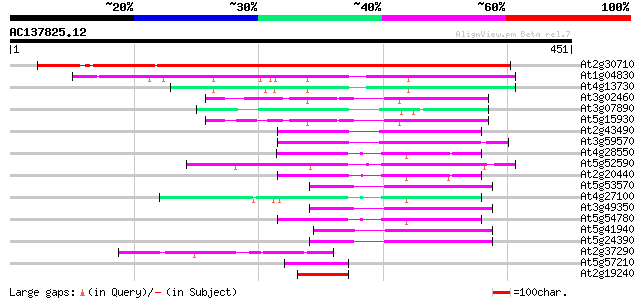
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137825.12 - phase: 0 /pseudo
(451 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g30710 unknown protein 568 e-162
At1g04830 unknown protein 110 2e-24
At4g13730 unknown protein 91 1e-18
At3g02460 putative plant adhesion molecule 80 2e-15
At3g07890 putative GTPase activator protein 80 3e-15
At5g15930 plant adhesion molecule 1 (PAM1) 77 1e-14
At2g43490 hypothetical protein 71 1e-12
At3g59570 putative protein 69 5e-12
At4g28550 putative protein 69 7e-12
At5g52590 putative protein 67 2e-11
At2g20440 unknown protein 61 1e-09
At5g53570 GTPase activator protein of Rab-like small GTPases-lik... 60 2e-09
At4g27100 unknown protein 56 4e-08
At3g49350 GTPase activating -like protein 53 3e-07
At5g54780 putative protein 52 5e-07
At5g41940 GTPase activator protein of Rab-like small GTPases-lik... 52 5e-07
At5g24390 GTPase activator-like protein of Rab-like small GTPases 51 1e-06
At2g37290 hypothetical protein 45 8e-05
At5g57210 microtubule-associated protein-like 44 2e-04
At2g19240 hypothetical protein 43 3e-04
>At2g30710 unknown protein
Length = 440
Score = 568 bits (1464), Expect = e-162
Identities = 277/380 (72%), Positives = 325/380 (84%), Gaps = 5/380 (1%)
Query: 23 DSRFSQTLRNVQGLLKGRSMPGKVLMSRRSEPSDNLNSKVSSTFYKRSFSHNDAGTSDQI 82
DSRF+QTL+NVQG LKGRS+PGKVL++RRS+P +S T Y+RS S NDAG ++
Sbjct: 13 DSRFNQTLKNVQGFLKGRSIPGKVLLTRRSDPPPY---PISPT-YQRSLSENDAGRNELF 68
Query: 83 SGAVEEEVQSTSKSVSAANVSKLKSSTSLGENLSEDIRKYTIGARATDSARVTKFNKVLS 142
VE E ++SK KL+S++S ++ E ++ IG R++DSARV KFNKVLS
Sbjct: 69 ESPVEVEDHNSSKKHDNTYAGKLRSNSSAERSVKE-VQNLKIGVRSSDSARVMKFNKVLS 127
Query: 143 GTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYD 202
T VIL+ LRELAW+GVP YMRP VWRLLLGY PPNSDR+E VL RKR EYL+ + Q+YD
Sbjct: 128 ETTVILEKLRELAWNGVPHYMRPDVWRLLLGYAPPNSDRREAVLRRKRLEYLESVGQFYD 187
Query: 203 IPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGI 262
+PDSERSDDE+NMLRQIAVDCPRTVPDV+FFQQ+QVQKSLERILY WAIRHPASGYVQGI
Sbjct: 188 LPDSERSDDEINMLRQIAVDCPRTVPDVSFFQQEQVQKSLERILYTWAIRHPASGYVQGI 247
Query: 263 NDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQP 322
NDLVTPFLV+F+SE+L+GG+D WSM DLS++K+S+VEADCYWCL+KLLDGMQDHYTFAQP
Sbjct: 248 NDLVTPFLVIFLSEYLDGGVDSWSMDDLSAEKVSDVEADCYWCLTKLLDGMQDHYTFAQP 307
Query: 323 GIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYL 382
GIQRLVFKLKELVRRIDEP+S+H+E+ GLEFLQFAFRW+NCLLIREIPF+LI RLWDTYL
Sbjct: 308 GIQRLVFKLKELVRRIDEPVSRHMEEHGLEFLQFAFRWYNCLLIREIPFNLINRLWDTYL 367
Query: 383 AEGDALPDFLVYIFASFLLT 402
AEGDALPDFLVYI+ASFLLT
Sbjct: 368 AEGDALPDFLVYIYASFLLT 387
>At1g04830 unknown protein
Length = 448
Score = 110 bits (275), Expect = 2e-24
Identities = 111/423 (26%), Positives = 178/423 (41%), Gaps = 81/423 (19%)
Query: 51 RSEPSDNLNSKVSSTFYKRSFSHNDAGTSDQISGAVEEEVQSTSKSVSAANVSKLKSSTS 110
R + + LNS + ST S S++D ++EE +S S + + S+TS
Sbjct: 3 RKKVPEWLNSTMWSTPPPPS-SYDDGLLRHSPVTKMKEEAESISVAPRLNSAPPPSSNTS 61
Query: 111 L---------GENLSEDIRKY--TIGARATDSARVTKFNKVLSGTVVILDNLRELAWSGV 159
+ G ++S +Y ++G A D +R + LS V+ + LR LA +
Sbjct: 62 VPSPSHRPRNGNSISGGSGEYGHSVGPSAEDFSRQAHVSAELSKKVINMKELRSLALQSL 121
Query: 160 PDY--MRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQY----------------- 200
PD +R VW+LLLGY PP L +KR +Y +
Sbjct: 122 PDSPGIRSTVWKLLLGYLPPERSLWSTELKQKRSQYKHYKDELLTSPSEITWKMVRSKGF 181
Query: 201 --YDIPDSER--------SDDE-----------------VNMLRQIAVDCPRTVPDVTFF 233
YD+ R +D++ + QI D RT PD+ FF
Sbjct: 182 DNYDLKSESRCMLARSRITDEDHPLSLGKASIWNTYFQDTETIEQIDRDVKRTHPDIPFF 241
Query: 234 QQQQV-----QKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMS 288
+ Q+S++ IL +A + YVQG+N+++ P VF +
Sbjct: 242 SGESSFARSNQESMKNILLVFAKLNQGIRYVQGMNEILAPIFYVF-------------RN 288
Query: 289 DLSSDKISNVEADCYWCLSKLLDGMQDHYTF----AQPGIQRLVFKLKELVRRIDEPISQ 344
D D S+ EAD ++C +LL G +D Y + GI+ + +L +LVR+ DE + +
Sbjct: 289 DPDEDSSSHAEADAFFCFVELLSGFRDFYCQQLDNSVVGIRSAITRLSQLVRKHDEELWR 348
Query: 345 HIEDQGLEFLQF-AFRWFNCLLIREIPFDLITRLWDTYLAEGDALPDFLVYIFASFLLTT 403
H+E QF AFRW LL +E F +WD L++ + + L+ I + L+
Sbjct: 349 HLEITTKVNPQFYAFRWITLLLTQEFSFFDSLHIWDALLSDPEGPLESLLGICCAMLVLV 408
Query: 404 NSR 406
R
Sbjct: 409 RRR 411
>At4g13730 unknown protein
Length = 449
Score = 90.9 bits (224), Expect = 1e-18
Identities = 83/333 (24%), Positives = 136/333 (39%), Gaps = 69/333 (20%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDY--MRPKVWRLLLGYEPPNSDRKEGVLG 187
D +R + LS V+ L LR++A G+PD +R VW+LLL Y P+ L
Sbjct: 96 DVSRKAQVVAELSKKVIDLKELRKIASQGLPDDAGIRSIVWKLLLDYLSPDRSLWSSELA 155
Query: 188 RKRGEYLDCISQYYDIP-------DSERSDD----------------------------- 211
+KR +Y + P D + D
Sbjct: 156 KKRSQYKQFKEELLMNPSEVTRKMDKSKGGDSNDPKIESPGALSRSEITHEDHPLSLGTT 215
Query: 212 --------EVNMLRQIAVDCPRTVPDVTFFQQQQV-----QKSLERILYAWAIRHPASGY 258
+ +L QI D RT PD+ FF Q +L+ IL +A +P Y
Sbjct: 216 SLWNNFFKDTEVLEQIERDVMRTHPDMHFFSGDSAVAKSNQDALKNILTIFAKLNPGIRY 275
Query: 259 VQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYT 318
VQG+N+++ P +F +D + E+D ++C +L+ G +D++
Sbjct: 276 VQGMNEILAPIFYIF-------------KNDPDKGNAAYAESDAFFCFVELMSGFRDNFC 322
Query: 319 F----AQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQF-AFRWFNCLLIREIPFDL 373
+ GI+ + +L L++ DE + +H+E QF AFRW LL +E F
Sbjct: 323 QQLDNSVVGIRYTITRLSLLLKHHDEELWRHLEVTTKINPQFYAFRWITLLLTQEFNFVE 382
Query: 374 ITRLWDTYLAEGDALPDFLVYIFASFLLTTNSR 406
+WDT L++ + + L+ I + L+ R
Sbjct: 383 SLHIWDTLLSDPEGPQETLLRICCAMLILVRRR 415
>At3g02460 putative plant adhesion molecule
Length = 353
Score = 80.1 bits (196), Expect = 2e-15
Identities = 62/235 (26%), Positives = 104/235 (43%), Gaps = 50/235 (21%)
Query: 158 GVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSERSDDEVNMLR 217
G+PD +R VW+L+ G R L Y + E S E++++R
Sbjct: 88 GIPDCLRGLVWQLISG---------------SRDLLLMNPGVYEQLVIYETSASELDIIR 132
Query: 218 QIAVDCPRTVPDVTFFQQQQV--QKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFIS 275
I+ RT P FFQ++ Q+SL +L A+++ GYVQG+ + L++++S
Sbjct: 133 DIS----RTFPSHVFFQKRHGPGQRSLYNVLKAYSVYDRDVGYVQGMG-FIAGLLLLYMS 187
Query: 276 EHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDG-----MQDHYTFAQPGIQRLVFK 330
E D +W L LL G M+ Y P +Q+ +F+
Sbjct: 188 EE-----------------------DAFWLLVALLKGAVHAPMEGLYHAGLPLVQQYLFQ 224
Query: 331 LKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEG 385
L+ LV+ + + +H + + +A +WF + PF L R+WD +L+EG
Sbjct: 225 LESLVKELIPKLGEHFTQEMINPSMYASQWFITVFSYSFPFPLALRIWDVFLSEG 279
>At3g07890 putative GTPase activator protein
Length = 400
Score = 79.7 bits (195), Expect = 3e-15
Identities = 64/239 (26%), Positives = 94/239 (38%), Gaps = 44/239 (18%)
Query: 151 LRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSERSD 210
L+ L G+P +RPKVW L G S E YY
Sbjct: 109 LKRLIRKGIPPVLRPKVWFSLSGAAKKKSTVPES---------------YYSDLTKAVEG 153
Query: 211 DEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFL 270
RQI D PRT P + + +L R+L ++ R GY QG+N + L
Sbjct: 154 MVTPATRQIDHDLPRTFPGHPWLDTPEGHAALRRVLVGYSFRDSDVGYCQGLNYVAALLL 213
Query: 271 VVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGM--QDHYTFAQPG--IQR 326
+V E D +W L+ LL+ + +D YT G +++
Sbjct: 214 LVM-----------------------KTEEDAFWMLAVLLENVLVRDCYTTNLSGCHVEQ 250
Query: 327 LVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEG 385
VFK +L+ + I+ H+ED G + A WF CL + +P + R+WD EG
Sbjct: 251 RVFK--DLLAQKCSRIATHLEDMGFDVSLVATEWFLCLFSKSLPSETTLRVWDVLFYEG 307
>At5g15930 plant adhesion molecule 1 (PAM1)
Length = 356
Score = 77.4 bits (189), Expect = 1e-14
Identities = 60/235 (25%), Positives = 104/235 (43%), Gaps = 50/235 (21%)
Query: 158 GVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSERSDDEVNMLR 217
G+PD +R VW+L+ G +L G Y+ + E S E++++R
Sbjct: 85 GIPDCLRGLVWQLISG--------SRDLLLMNPGVYVQLVIY-------ETSASELDIIR 129
Query: 218 QIAVDCPRTVPDVTFFQQQQV--QKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFIS 275
I+ RT P FFQ++ Q+SL +L A+++ GYVQG+ + L++++S
Sbjct: 130 DIS----RTFPSHVFFQKRHGPGQRSLYNVLKAYSVYDRDVGYVQGMG-FIAGLLLLYMS 184
Query: 276 EHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDG-----MQDHYTFAQPGIQRLVFK 330
E D +W L LL G ++ Y P +Q+ + +
Sbjct: 185 EE-----------------------DAFWLLVALLKGAVHSPIEGLYQAGLPLVQQYLLQ 221
Query: 331 LKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEG 385
+LVR + + +H + + +A +WF + +PF R+WD +LAEG
Sbjct: 222 FDQLVRELMPKLGEHFTQEMINPSMYASQWFITVFSYSLPFHSALRIWDVFLAEG 276
>At2g43490 hypothetical protein
Length = 756
Score = 70.9 bits (172), Expect = 1e-12
Identities = 47/165 (28%), Positives = 75/165 (44%), Gaps = 24/165 (14%)
Query: 216 LRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFIS 275
L +I VD RT + F++ + IL +A PA+GY QG++DLV+PF+V+F
Sbjct: 359 LHRIVVDVVRTDSHLEFYEDPGNLGRMSDILAVYAWVDPATGYCQGMSDLVSPFVVLF-- 416
Query: 276 EHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQP-GIQRLVFKLKEL 334
AD +WC L+ + ++ P G+ + L +
Sbjct: 417 ---------------------EDNADAFWCFEMLIRRTRANFQMEGPTGVMDQLQSLWHI 455
Query: 335 VRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWD 379
++ D+ I H+ G E L FAFR L RE+ F+ R+W+
Sbjct: 456 LQITDKDIFSHLSRIGAESLHFAFRMLLVLFRRELSFNEALRMWE 500
>At3g59570 putative protein
Length = 549
Score = 68.9 bits (167), Expect = 5e-12
Identities = 49/187 (26%), Positives = 83/187 (44%), Gaps = 27/187 (14%)
Query: 216 LRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFIS 275
L +I VD RT + F++ + IL +A PA+GY QG++DLV+PF+ +F
Sbjct: 389 LHRIVVDVVRTDSHLEFYEDPGNLGRMSDILAVYAWVDPATGYCQGMSDLVSPFVFLF-- 446
Query: 276 EHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQP-GIQRLVFKLKEL 334
AD +WC L+ + ++ P G+ + L +
Sbjct: 447 ---------------------EDNADAFWCFEMLIRRTRANFQMEGPTGVMDQLQSLWRI 485
Query: 335 VRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEGDALPDFLVY 394
++ D+ + H+ G E L FAFR L RE+ F+ R+W+ + F++
Sbjct: 486 LQLTDKEMFSHLSRIGAESLHFAFRMLLVLFRRELSFNKALRMWE---VGNSSAVCFVLP 542
Query: 395 IFASFLL 401
+ SF+L
Sbjct: 543 VSCSFIL 549
>At4g28550 putative protein
Length = 424
Score = 68.6 bits (166), Expect = 7e-12
Identities = 47/168 (27%), Positives = 82/168 (47%), Gaps = 27/168 (16%)
Query: 215 MLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFI 274
+L QI +D RT + F++ + Q L IL + +P GYVQG+ND+ +P +++
Sbjct: 166 VLSQIGLDVVRTDRYLCFYESESNQARLWDILSIYTWLNPDIGYVQGMNDICSPMIIL-- 223
Query: 275 SEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHY--TFAQPGIQRLVFKLK 332
++D EAD +WC + + +++++ T G+Q + L
Sbjct: 224 -------LED--------------EADAFWCFERAMRRLRENFRTTATSMGVQTQLGMLS 262
Query: 333 ELVRRIDEPISQHIED-QGLEFLQFAFRWFNCLLIREIPFDLITRLWD 379
++++ +D + QH+ED G E+L FA R L RE F LW+
Sbjct: 263 QVIKTVDPRLHQHLEDLDGGEYL-FAIRMLMVLFRREFSFLDALYLWE 309
>At5g52590 putative protein
Length = 338
Score = 67.4 bits (163), Expect = 2e-11
Identities = 66/275 (24%), Positives = 112/275 (40%), Gaps = 39/275 (14%)
Query: 143 GTVVILDNLRE-LAWSGVPDYMRPKVWRLLLGYEPPNSD--RKEGVLGRKRGEYLDCISQ 199
G VV LRE + + G+ +R +VW LLGY +S +E + KR EY Q
Sbjct: 23 GRVVESKALRERVFYGGIEHQLRREVWPFLLGYYAYDSTYAEREYLRSVKRMEYATLKQQ 82
Query: 200 YYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQK----SLERILYAWAIRHPA 255
+ I + R+ +D D F + S+ IL ++ +
Sbjct: 83 WQSISPEQAKRFTKYRERKGLIDKDVVRTDRAFEYYEGDDNLHVNSMRDILLTYSFYNFD 142
Query: 256 SGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQD 315
GY QG++D ++P L V M D E++ +WC L++ +
Sbjct: 143 LGYCQGMSDYLSPILFV--------------MED---------ESESFWCFVALMERLGP 179
Query: 316 HYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLIT 375
++ Q G+ +F L +LV +D P+ + ++ F FRW RE ++
Sbjct: 180 NFNRDQNGMHTQLFALSKLVELLDSPLHNYFKENDCLNYFFCFRWILIQFKREFEYEKTM 239
Query: 376 RLWDT----YLAEGDALPDFLVYIFASFLLTTNSR 406
+LW+ YL+E F +Y+ + L S+
Sbjct: 240 QLWEVMWTHYLSE-----HFHLYVCVAVLKRCRSK 269
>At2g20440 unknown protein
Length = 371
Score = 60.8 bits (146), Expect = 1e-09
Identities = 44/169 (26%), Positives = 78/169 (46%), Gaps = 29/169 (17%)
Query: 216 LRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFIS 275
L QI +D RT + F++ + Q L +L + + GYVQG+ND+ +P +++F
Sbjct: 51 LHQIGLDVARTDRYLCFYENDRNQSKLWDVLAIYTWLNLDIGYVQGMNDICSPMIILF-- 108
Query: 276 EHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHY--TFAQPGIQRLVFKLKE 333
DD E D +WC + + +++++ T G+Q + L +
Sbjct: 109 -------DD--------------EGDAFWCFERAMRRLRENFRATATSMGVQTQLGVLSQ 147
Query: 334 LVRRIDEPISQHIEDQGL---EFLQFAFRWFNCLLIREIPFDLITRLWD 379
+++ +D + QH+ + L E+L FA R L RE F LW+
Sbjct: 148 VIKTVDPRLHQHLGKKDLDGGEYL-FAIRMLMVLFRREFSFLDALYLWE 195
>At5g53570 GTPase activator protein of Rab-like small GTPases-like
protein
Length = 550
Score = 60.5 bits (145), Expect = 2e-09
Identities = 35/147 (23%), Positives = 66/147 (44%), Gaps = 23/147 (15%)
Query: 242 LERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEAD 301
L IL A+A+ P GY QG++DL++P L V +H +
Sbjct: 340 LVAILEAYAMYDPEIGYCQGMSDLLSPILAVISEDH-----------------------E 376
Query: 302 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWF 361
+WC + + ++ + GIQR + + ++++ D + +H+E+ E F +R
Sbjct: 377 AFWCFVGFMKKARHNFRLDEAGIQRQLSIVSKIIKNKDSQLYKHLENLQAEDCSFVYRMV 436
Query: 362 NCLLIREIPFDLITRLWDTYLAEGDAL 388
+ RE+ F+ LW+ A+ A+
Sbjct: 437 LVMFRRELSFEQTLCLWEVMWADQAAI 463
Score = 33.5 bits (75), Expect = 0.25
Identities = 53/206 (25%), Positives = 75/206 (35%), Gaps = 28/206 (13%)
Query: 49 SRRSEPSDNLNSKVSSTFYKRSFSHNDAGTSDQISGAVEEEVQST----SKSVSAANVSK 104
S S S +L S SS+ S S + S+ S + S+ + AN+S
Sbjct: 13 SGNSSSSSSLPSSSSSSLPSSSSSSPPSSNSNSYSNSNSSSSSSSWIHLRSVLFVANLSS 72
Query: 105 LKSSTSLGENL----SEDIRKYTIGARATDSARVTKFNKVLSGTVVILDNLRELAWSGVP 160
S TS S RK+ + S T K+ G V L +R GV
Sbjct: 73 PSSVTSSDRRRKSPWSRRKRKWALTPHQWRSL-FTPEGKLRDGGVGFLKKVRS---RGVD 128
Query: 161 DYMRPKVWRLLLGYEPPN--SDRKEGVLGRKRGEY----------LDC----ISQYYDIP 204
+R +VW LLG N S+ +E V +KR EY L C ++P
Sbjct: 129 PSIRAEVWLFLLGVYDLNSTSEEREAVKTQKRKEYEKLQRRCQMLLKCGNGSTDNLEELP 188
Query: 205 DSERSDDEVNMLRQIAVDCPRTVPDV 230
E + V + + P T DV
Sbjct: 189 SDEANSQCVRFVDDYKITGPMTSQDV 214
>At4g27100 unknown protein
Length = 436
Score = 56.2 bits (134), Expect = 4e-08
Identities = 68/314 (21%), Positives = 123/314 (38%), Gaps = 79/314 (25%)
Query: 121 KYTIGARATDSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLG-YEPPNS 179
++ I T S R + V G++ I LR + G+ +R +VW LLG Y+P ++
Sbjct: 26 RFKIKPGKTLSVRKWQAVFVQEGSLHIGKTLRRIRRGGIHPSIRGEVWEFLLGCYDPMST 85
Query: 180 -DRKEGVLGRKRGEYL----DCISQYYDIPDSERSD------------------DEVNM- 215
+ +E + R+R +Y +C Q + + S R E+N+
Sbjct: 86 FEEREQIRQRRRLQYASWKEEC-KQMFPVIGSGRFTTAPVITENGQPNYDPLVLQEINLG 144
Query: 216 ----------------------------LRQIAVDCPRTVPDVTFFQQQQVQKSLERILY 247
L QI +D RT + F+++++ L IL
Sbjct: 145 TNSNGSVFFKELTSRGPLDKKIIQWLLTLHQIGLDVNRTDRALVFYEKKENLSKLWDILS 204
Query: 248 AWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLS 307
+A GY QG++DL +P +++ ++D EAD +WC
Sbjct: 205 VYAWIDNDVGYCQGMSDLCSPMIIL---------LED--------------EADAFWCFE 241
Query: 308 KLLDGMQDHY--TFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLL 365
+L+ ++ ++ T G++ + L + + +D + QH++ G FA R
Sbjct: 242 RLMRRLRGNFRSTGRSVGVEAQLTHLSSITQVVDPKLHQHLDKLGGGDYLFAIRMLMVQF 301
Query: 366 IREIPFDLITRLWD 379
RE F LW+
Sbjct: 302 RREFSFCDSLYLWE 315
>At3g49350 GTPase activating -like protein
Length = 554
Score = 53.1 bits (126), Expect = 3e-07
Identities = 33/147 (22%), Positives = 64/147 (43%), Gaps = 23/147 (15%)
Query: 242 LERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEAD 301
L +L A+A+ P GY QG++DL++P L V +H +
Sbjct: 347 LVAVLEAYALYDPDIGYCQGMSDLLSPILSVIPDDH-----------------------E 383
Query: 302 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWF 361
+WC + + ++ + GI+R + + ++++ D + +H+E E F +R
Sbjct: 384 VFWCFVGFMKKARHNFRLDEVGIRRQLNIVSKIIKSKDSQLYRHLEKLQAEDCFFVYRMV 443
Query: 362 NCLLIREIPFDLITRLWDTYLAEGDAL 388
+ RE+ D LW+ A+ A+
Sbjct: 444 VVMFRRELTLDQTLCLWEVMWADQAAI 470
>At5g54780 putative protein
Length = 435
Score = 52.4 bits (124), Expect = 5e-07
Identities = 41/166 (24%), Positives = 72/166 (42%), Gaps = 25/166 (15%)
Query: 216 LRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFIS 275
L QI +D RT + F+++++ L IL +A GY QG++DL +P +++
Sbjct: 171 LHQIGLDVNRTDRTLVFYEKKENLSKLWDILALYAWIDNDVGYCQGMSDLCSPMIML--- 227
Query: 276 EHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHY--TFAQPGIQRLVFKLKE 333
++D EAD +WC +L+ ++ ++ T G++ + L
Sbjct: 228 ------LED--------------EADAFWCFERLMRRLRGNFRDTGRSVGVEAQLTHLAS 267
Query: 334 LVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWD 379
+ + ID + H+E G FA R RE F LW+
Sbjct: 268 ITQIIDPKLHHHLEKLGGGDYLFAIRMIMVQFRREFSFCDSLYLWE 313
>At5g41940 GTPase activator protein of Rab-like small GTPases-like
protein
Length = 506
Score = 52.4 bits (124), Expect = 5e-07
Identities = 33/144 (22%), Positives = 64/144 (43%), Gaps = 23/144 (15%)
Query: 245 ILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYW 304
IL A+A+ P GY QG++DL++P + V + L +W
Sbjct: 307 ILEAYAVYDPEIGYCQGMSDLLSPLIAVMEDDVL-----------------------AFW 343
Query: 305 CLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCL 364
C + + ++ + GI+R + + ++++ D + +H+E+ E F +R L
Sbjct: 344 CFVGFMSKARHNFRLDEVGIRRQLSMVSKIIKFKDIHLYRHLENLEAEDCFFVYRMVVVL 403
Query: 365 LIREIPFDLITRLWDTYLAEGDAL 388
RE+ F+ LW+ A+ A+
Sbjct: 404 FRRELTFEQTLCLWEVMWADQAAI 427
>At5g24390 GTPase activator-like protein of Rab-like small GTPases
Length = 528
Score = 51.2 bits (121), Expect = 1e-06
Identities = 31/147 (21%), Positives = 63/147 (42%), Gaps = 23/147 (15%)
Query: 242 LERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEAD 301
L +L A+A+ P GY QG++DL++P L V ++ +
Sbjct: 319 LVAVLEAYALHDPEIGYCQGMSDLLSPILSVIPDDY-----------------------E 355
Query: 302 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWF 361
+WC + + ++ + GI R + + ++++ D + +H+E E F +R
Sbjct: 356 AFWCFVGFMKKARQNFRVDEVGITRQLNIVSKIIKSKDSQLYKHLEKVKAEDCFFVYRMV 415
Query: 362 NCLLIREIPFDLITRLWDTYLAEGDAL 388
+ RE+ + LW+ A+ A+
Sbjct: 416 LVMFRRELTLEQTLHLWEVIWADQAAI 442
>At2g37290 hypothetical protein
Length = 544
Score = 45.1 bits (105), Expect = 8e-05
Identities = 46/179 (25%), Positives = 74/179 (40%), Gaps = 21/179 (11%)
Query: 88 EEVQSTSKSVSAANVSKLKSSTSLGENLSEDIRKYTIGARATDSARVTKFNKVLSGTVVI 147
+ ++ K++ + S +K S S E E+ R +T + K G+V
Sbjct: 114 KSTKNGQKNIVDDHASSIKESLSSIEESGENDRD---SETSTSRSHSIKEENEAQGSVSP 170
Query: 148 ------LDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYY 201
+ L L GVP +R +VW+ +G + +R Y D ++Q
Sbjct: 171 EPFFPWYEELEVLVRLGVPKDLRGEVWQAFVGVKARRVERY----------YQDLLAQIT 220
Query: 202 DIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQ 260
+ D SD + +QI D PRT P + + SL RIL A+A +P+ GY Q
Sbjct: 221 N-SDENSSDVQRKWKKQIEKDIPRTFPGHPALNENG-RDSLRRILLAYACHNPSVGYCQ 277
>At5g57210 microtubule-associated protein-like
Length = 737
Score = 43.5 bits (101), Expect = 2e-04
Identities = 21/52 (40%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Query: 222 DCPRTVPDV-TFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVV 272
D R P+ ++FQ Q L RIL W ++HP GY QG+++L+ P L V
Sbjct: 120 DLSRLYPEHGSYFQSSGCQGMLRRILLLWCLKHPEIGYRQGMHELLAPLLYV 171
Score = 35.4 bits (80), Expect = 0.065
Identities = 16/49 (32%), Positives = 24/49 (48%)
Query: 331 LKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWD 379
L L+ +D + H+ + G+E FA RW L RE P + +WD
Sbjct: 321 LYHLLSLVDASLHSHLVELGVEPQYFALRWLRVLFGREFPLSNLLIVWD 369
>At2g19240 hypothetical protein
Length = 840
Score = 43.1 bits (100), Expect = 3e-04
Identities = 18/41 (43%), Positives = 26/41 (62%)
Query: 232 FFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVV 272
+FQ + Q L RIL W ++HP GY QG+++L+ P L V
Sbjct: 121 YFQTPRYQGMLRRILLLWCLKHPEYGYRQGMHELLAPLLYV 161
Score = 30.0 bits (66), Expect = 2.7
Identities = 16/54 (29%), Positives = 24/54 (43%), Gaps = 1/54 (1%)
Query: 338 IDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWD-TYLAEGDALPD 390
+D + H+ + G+E F RW L RE + +WD LA+ A D
Sbjct: 309 VDSSLHSHLVELGVEPQYFGLRWLRVLFGREFLLQDLLLVWDEIILADNSARTD 362
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,119,907
Number of Sequences: 26719
Number of extensions: 427344
Number of successful extensions: 1447
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 1351
Number of HSP's gapped (non-prelim): 83
length of query: 451
length of database: 11,318,596
effective HSP length: 103
effective length of query: 348
effective length of database: 8,566,539
effective search space: 2981155572
effective search space used: 2981155572
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC137825.12


