

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137823.5 - phase: 0
(121 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
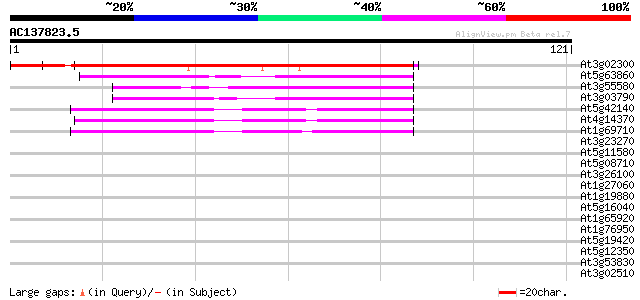
Score E
Sequences producing significant alignments: (bits) Value
At3g02300 unknown protein 141 6e-35
At5g63860 UVB-resistance protein UVR8 (gb|AAD43920.1) 45 7e-06
At3g55580 regulator of chromosome condensation-like protein 39 3e-04
At3g03790 unknown protein 39 5e-04
At5g42140 TMV resistance protein-like 38 7e-04
At4g14370 disease resistance N like protein 38 7e-04
At1g69710 putative regulator of chromosome condensation 38 7e-04
At3g23270 hypothetical protein 38 0.001
At5g11580 unknown protein 37 0.001
At5g08710 unknown protein 37 0.001
At3g26100 unknown protein (MPE11.29) 37 0.001
At1g27060 hypothetical protein 37 0.001
At1g19880 unknown protein 37 0.002
At5g16040 UVB-resistance protein-like 36 0.003
At1g65920 36 0.003
At1g76950 zinc finger protein (PRAF1) 36 0.003
At5g19420 putative protein 35 0.005
At5g12350 putative protein 35 0.005
At3g53830 putative protein 35 0.006
At3g02510 unknown protein 35 0.008
>At3g02300 unknown protein
Length = 471
Score = 141 bits (355), Expect = 6e-35
Identities = 65/87 (74%), Positives = 73/87 (83%), Gaps = 1/87 (1%)
Query: 1 MDIDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL 60
MDI I+G P + SIP KSAIYVWGYNQSGQTGR E++ LRIPKQLPP+LFGCPAG
Sbjct: 1 MDIGEIIGEVAP-SVSIPTKSAIYVWGYNQSGQTGRNEQEKLLRIPKQLPPELFGCPAGA 59
Query: 61 NTRWLDIACGREHTAAIASDGLLFTWG 87
N+RWLDI+CGREHTAA+ASDG LF WG
Sbjct: 60 NSRWLDISCGREHTAAVASDGSLFAWG 86
Score = 48.5 bits (114), Expect = 5e-07
Identities = 28/82 (34%), Positives = 43/82 (52%), Gaps = 3/82 (3%)
Query: 8 GTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQL-FGCPAGLNTRWLD 66
GT VA S + + WGYN+ GQ GR + L+ P+ + F A + +
Sbjct: 174 GTAHVVALS--EEGLLQAWGYNEQGQLGRGVTCEGLQAPRVINAYAKFLDEAPELVKIMQ 231
Query: 67 IACGREHTAAIASDGLLFTWGL 88
++CG HTAA++ G ++TWGL
Sbjct: 232 LSCGEYHTAALSDAGEVYTWGL 253
Score = 39.7 bits (91), Expect = 2e-04
Identities = 22/76 (28%), Positives = 37/76 (47%), Gaps = 11/76 (14%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRK--EKDDQLRIPKQLPPQLFGCPAGLN-TRWLDIACGR 71
A++ +Y WG GQ G + D+ IP+++ GL+ ++ACG
Sbjct: 240 AALSDAGEVYTWGLGSMGQLGHVSLQSGDKELIPRRV--------VGLDGVSMKEVACGG 291
Query: 72 EHTAAIASDGLLFTWG 87
HT A++ +G L+ WG
Sbjct: 292 VHTCALSLEGALYAWG 307
Score = 35.4 bits (80), Expect = 0.005
Identities = 19/67 (28%), Positives = 27/67 (39%), Gaps = 16/67 (23%)
Query: 21 SAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASD 80
S ++VWG NQ P+LF T ++CG H A++ +
Sbjct: 141 SRLWVWGQNQGSNL----------------PRLFSGAFPATTAIRQVSCGTAHVVALSEE 184
Query: 81 GLLFTWG 87
GLL WG
Sbjct: 185 GLLQAWG 191
Score = 28.5 bits (62), Expect = 0.56
Identities = 18/76 (23%), Positives = 30/76 (38%), Gaps = 14/76 (18%)
Query: 20 KSAIYVWGYNQSGQTGRKEK--------DDQLRIPKQLPPQLFGCPAGLNTRWLDIACGR 71
+ A+Y WG Q+GQ G + + + + +P + L +ACG
Sbjct: 300 EGALYAWGGGQAGQLGLGPQSGFFFSVSNGSEMLLRNVPVLVIPTDVRL------VACGH 353
Query: 72 EHTAAIASDGLLFTWG 87
HT +G + WG
Sbjct: 354 SHTLVYMREGRICGWG 369
>At5g63860 UVB-resistance protein UVR8 (gb|AAD43920.1)
Length = 440
Score = 44.7 bits (104), Expect = 7e-06
Identities = 24/72 (33%), Positives = 36/72 (49%), Gaps = 8/72 (11%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++ + + WG NQ+GQ G + +D L PQ G+ + +A G EHTA
Sbjct: 134 AVTMEGEVQSWGRNQNGQLGLGDTEDSL------VPQKIQAFEGIRIKM--VAAGAEHTA 185
Query: 76 AIASDGLLFTWG 87
A+ DG L+ WG
Sbjct: 186 AVTEDGDLYGWG 197
Score = 40.4 bits (93), Expect = 1e-04
Identities = 25/72 (34%), Positives = 38/72 (52%), Gaps = 8/72 (11%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
S+ A+Y +G+++ GQ G + +D L IP +L A N+ I+ G HT
Sbjct: 238 SVSYSGALYTYGWSKYGQLGHGDLEDHL-IPHKLE-------ALSNSFISQISGGWRHTM 289
Query: 76 AIASDGLLFTWG 87
A+ SDG L+ WG
Sbjct: 290 ALTSDGKLYGWG 301
Score = 35.4 bits (80), Expect = 0.005
Identities = 18/73 (24%), Positives = 38/73 (51%), Gaps = 8/73 (10%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
A++ +Y WG+ + G G ++ D+L +P+++ + + +ACG HT
Sbjct: 185 AAVTEDGDLYGWGWGRYGNLGLGDRTDRL-VPERVT-------STGGEKMSMVACGWRHT 236
Query: 75 AAIASDGLLFTWG 87
+++ G L+T+G
Sbjct: 237 ISVSYSGALYTYG 249
Score = 34.3 bits (77), Expect = 0.010
Identities = 18/65 (27%), Positives = 29/65 (43%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG+N+ GQ G DQ + P + + + ++CG HT A+
Sbjct: 297 LYGWGWNKFGQVGVGNNLDQCSPVQVRFPD--------DQKVVQVSCGWRHTLAVTERNN 348
Query: 83 LFTWG 87
+F WG
Sbjct: 349 VFAWG 353
Score = 32.0 bits (71), Expect = 0.050
Identities = 17/63 (26%), Positives = 30/63 (46%), Gaps = 9/63 (14%)
Query: 26 WGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL-LF 84
WG + GQ G + +D + P QL + + + CG +HT A + G+ ++
Sbjct: 39 WGRGEDGQLGHGDAED-----RPSPTQLSALDGH---QIVSVTCGADHTVAYSQSGMEVY 90
Query: 85 TWG 87
+WG
Sbjct: 91 SWG 93
Score = 30.0 bits (66), Expect = 0.19
Identities = 17/65 (26%), Positives = 26/65 (39%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG+ G+ G D P G+ + IACG H A+ +G
Sbjct: 89 VYSWGWGDFGRLGHGNSSDLFT------PLPIKALHGIRIK--QIACGDSHCLAVTMEGE 140
Query: 83 LFTWG 87
+ +WG
Sbjct: 141 VQSWG 145
>At3g55580 regulator of chromosome condensation-like protein
Length = 488
Score = 39.3 bits (90), Expect = 3e-04
Identities = 24/65 (36%), Positives = 31/65 (46%), Gaps = 6/65 (9%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y +G NQ GQ G DQ + P+L P N I+CG HTA I +G
Sbjct: 384 VYAFGGNQFGQLGTG--CDQA----ETLPKLLEAPNLENVNVKTISCGARHTAVITDEGR 437
Query: 83 LFTWG 87
+F WG
Sbjct: 438 VFCWG 442
Score = 34.3 bits (77), Expect = 0.010
Identities = 25/85 (29%), Positives = 33/85 (38%), Gaps = 19/85 (22%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLP---PQLFG--------------CPAGLNTRWL 65
++ WGY GQ G + + P +P P +G C L +
Sbjct: 255 VWGWGYGGEGQLGLGSRVRLVSSPHPIPCIEPSSYGKATSSGVNMSSVVQCGRVLGSYVK 314
Query: 66 DIACGREHTAAIASDGLLFT--WGL 88
IACG H+A I G L T WGL
Sbjct: 315 KIACGGRHSAVITDTGALLTFGWGL 339
Score = 30.0 bits (66), Expect = 0.19
Identities = 20/74 (27%), Positives = 33/74 (44%), Gaps = 8/74 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
+A I A+ +G+ GQ G+ DD+L P G+ ++A G H
Sbjct: 323 SAVITDTGALLTFGWGLYGQCGQGSTDDELS------PTCVSSLLGIRIE--EVAAGLWH 374
Query: 74 TAAIASDGLLFTWG 87
T +SDG ++ +G
Sbjct: 375 TTCASSDGDVYAFG 388
>At3g03790 unknown protein
Length = 1078
Score = 38.5 bits (88), Expect = 5e-04
Identities = 19/65 (29%), Positives = 32/65 (49%), Gaps = 9/65 (13%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG N+ GQ G D Q P+++ L + + ++ +HTA ++ G
Sbjct: 263 VYTWGSNREGQLGYTSVDTQAT-PRKV--------TSLKAKIVAVSAANKHTAVVSDCGE 313
Query: 83 LFTWG 87
+FTWG
Sbjct: 314 VFTWG 318
Score = 34.7 bits (78), Expect = 0.008
Identities = 20/83 (24%), Positives = 37/83 (44%), Gaps = 3/83 (3%)
Query: 6 ILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWL 65
++ K + +I +Y WG+ + G+ G E D + P+ +GL +R +
Sbjct: 187 LVSAAKFHSVAISTHGEVYTWGFGRGGRLGHPEFDIHSGQAAVITPRQV--ISGLGSRRV 244
Query: 66 D-IACGREHTAAIASDGLLFTWG 87
+A + HT G ++TWG
Sbjct: 245 KAVAAAKHHTVIATEGGDVYTWG 267
Score = 33.5 bits (75), Expect = 0.017
Identities = 18/65 (27%), Positives = 29/65 (43%), Gaps = 6/65 (9%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG N+ GQ G + +L L G + IA + HT + +DG
Sbjct: 314 VFTWGCNKEGQLGYGTSNSASNYSPRLVDYLKG------KVFTAIASSKYHTLVLRNDGE 367
Query: 83 LFTWG 87
++TWG
Sbjct: 368 VYTWG 372
Score = 25.8 bits (55), Expect = 3.6
Identities = 10/20 (50%), Positives = 12/20 (60%)
Query: 67 IACGREHTAAIASDGLLFTW 86
IA G H+ A+A DG F W
Sbjct: 408 IAAGMVHSLALAEDGAFFYW 427
>At5g42140 TMV resistance protein-like
Length = 1073
Score = 38.1 bits (87), Expect = 7e-04
Identities = 22/74 (29%), Positives = 33/74 (43%), Gaps = 8/74 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
AA + R+ ++ WG G+ G D PQL A + + +ACG H
Sbjct: 284 AALVSRQGEVFTWGEASGGRLGHGMGKDVTG------PQLIESLAATSIDF--VACGEFH 335
Query: 74 TAAIASDGLLFTWG 87
T A+ G ++TWG
Sbjct: 336 TCAVTMTGEIYTWG 349
Score = 32.0 bits (71), Expect = 0.050
Identities = 24/67 (35%), Positives = 31/67 (45%), Gaps = 9/67 (13%)
Query: 23 IYVWG--YNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASD 80
IY WG + +G G D IPK++ L G ++CG HTA I S
Sbjct: 345 IYTWGDGTHNAGLLGHGT-DVSHWIPKRISGPLEGLQIA------SVSCGPWHTALITST 397
Query: 81 GLLFTWG 87
G LFT+G
Sbjct: 398 GQLFTFG 404
Score = 30.4 bits (67), Expect = 0.15
Identities = 11/21 (52%), Positives = 15/21 (71%)
Query: 67 IACGREHTAAIASDGLLFTWG 87
IACG +H A ++ G +FTWG
Sbjct: 277 IACGVKHAALVSRQGEVFTWG 297
Score = 29.3 bits (64), Expect = 0.33
Identities = 11/22 (50%), Positives = 14/22 (63%)
Query: 66 DIACGREHTAAIASDGLLFTWG 87
+IACG H A + S +FTWG
Sbjct: 550 EIACGAYHVAVLTSRNEVFTWG 571
Score = 26.6 bits (57), Expect = 2.1
Identities = 15/36 (41%), Positives = 17/36 (46%), Gaps = 11/36 (30%)
Query: 63 RWLDIACGREHTAAIA-----------SDGLLFTWG 87
R + +ACG H AAI S G LFTWG
Sbjct: 432 RTIAVACGVWHAAAIVEVIVTHSSSSVSSGKLFTWG 467
>At4g14370 disease resistance N like protein
Length = 1996
Score = 38.1 bits (87), Expect = 7e-04
Identities = 23/73 (31%), Positives = 35/73 (47%), Gaps = 8/73 (10%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
A + R+ ++ W G+ G + D R P+L A N + +ACG HT
Sbjct: 1215 ALVTRQGEVFTWEEEAGGRLGHGIQVDVCR------PKLVEFLALTNIDF--VACGEYHT 1266
Query: 75 AAIASDGLLFTWG 87
A+++ G LFTWG
Sbjct: 1267 CAVSTSGDLFTWG 1279
Score = 30.0 bits (66), Expect = 0.19
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Query: 49 LPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLLFTWG 87
+P ++ G GL L +ACG H+A ++G LFT+G
Sbjct: 1298 IPKRVSGPVEGLQV--LSVACGTWHSALATANGKLFTFG 1334
Score = 30.0 bits (66), Expect = 0.19
Identities = 11/22 (50%), Positives = 14/22 (63%)
Query: 65 LDIACGREHTAAIASDGLLFTW 86
+DIACG H A + G +FTW
Sbjct: 1205 IDIACGVRHIALVTRQGEVFTW 1226
Score = 29.6 bits (65), Expect = 0.25
Identities = 19/65 (29%), Positives = 28/65 (42%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ G GQ G D +L P + +L G +I+CG H A + S
Sbjct: 1444 VFTMGGTSHGQLGSSNSDGKL--PCLVQDRLVGEFVE------EISCGDHHVAVLTSRSE 1495
Query: 83 LFTWG 87
+FTWG
Sbjct: 1496 VFTWG 1500
Score = 28.1 bits (61), Expect = 0.73
Identities = 19/66 (28%), Positives = 29/66 (43%), Gaps = 10/66 (15%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL-NTRWLDIACGREHTAAIASDG 81
++ WG + G K+ L LP C + L + + IACG T A+ + G
Sbjct: 1392 LFTWGDGDKNRLGHGNKETYL-----LPT----CVSSLIDYNFNQIACGHTFTVALTTSG 1442
Query: 82 LLFTWG 87
+FT G
Sbjct: 1443 HVFTMG 1448
Score = 27.3 bits (59), Expect = 1.2
Identities = 9/28 (32%), Positives = 17/28 (60%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQ 42
A + +S ++ WG +G+ G +KDD+
Sbjct: 1488 AVLTSRSEVFTWGKGSNGRLGHGDKDDR 1515
Score = 25.4 bits (54), Expect = 4.7
Identities = 17/40 (42%), Positives = 19/40 (47%), Gaps = 12/40 (30%)
Query: 58 AGLNTRWLDIACGREHTAAI----------ASDGLLFTWG 87
+GL T L +ACG HT AI S LFTWG
Sbjct: 1359 SGLKT--LKVACGVWHTVAIVEVMNQTGTSTSSRKLFTWG 1396
>At1g69710 putative regulator of chromosome condensation
Length = 1028
Score = 38.1 bits (87), Expect = 7e-04
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 8/74 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
A + ++ I+ WG + G+ G + D + P+ GL + +ACG H
Sbjct: 299 AVLVTKQGEIFSWGEGKGGKLGHGLETDAQK------PKFISSVRGLGFK--SLACGDFH 350
Query: 74 TAAIASDGLLFTWG 87
T AI G L++WG
Sbjct: 351 TCAITQSGDLYSWG 364
Score = 34.7 bits (78), Expect = 0.008
Identities = 18/39 (46%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query: 49 LPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLLFTWG 87
+P ++ G GL D+ACG HTA +AS G LFT+G
Sbjct: 383 IPKRVTGDLQGLYVS--DVACGPWHTAVVASSGQLFTFG 419
Score = 30.8 bits (68), Expect = 0.11
Identities = 19/69 (27%), Positives = 31/69 (44%), Gaps = 10/69 (14%)
Query: 20 KSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL-NTRWLDIACGREHTAAIA 78
+ ++ WG + GQ G + D +L +P+ C L N +ACG T +
Sbjct: 477 RGQVFTWGDGEKGQLGHGDNDTKL-LPE--------CVISLTNENICQVACGHSLTVSRT 527
Query: 79 SDGLLFTWG 87
S G ++T G
Sbjct: 528 SRGHVYTMG 536
Score = 28.9 bits (63), Expect = 0.43
Identities = 10/22 (45%), Positives = 14/22 (63%)
Query: 66 DIACGREHTAAIASDGLLFTWG 87
+IACG H A + S ++TWG
Sbjct: 567 EIACGSYHVAVLTSKSEIYTWG 588
Score = 28.5 bits (62), Expect = 0.56
Identities = 20/72 (27%), Positives = 34/72 (46%), Gaps = 15/72 (20%)
Query: 23 IYVWGYNQS-------GQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++VWG + S G + +D L +PK L + L+ + +IACG+ H
Sbjct: 249 VFVWGESISDGVLSGTGNSLNSTTEDAL-LPKALESTIV-----LDAQ--NIACGKCHAV 300
Query: 76 AIASDGLLFTWG 87
+ G +F+WG
Sbjct: 301 LVTKQGEIFSWG 312
Score = 24.6 bits (52), Expect = 8.0
Identities = 13/34 (38%), Positives = 17/34 (49%), Gaps = 13/34 (38%)
Query: 67 IACGREHTAAI-------------ASDGLLFTWG 87
+ACG HTAA+ +S G +FTWG
Sbjct: 451 VACGVWHTAAVVEVTNEASEAEVDSSRGQVFTWG 484
>At3g23270 hypothetical protein
Length = 1045
Score = 37.7 bits (86), Expect = 0.001
Identities = 25/87 (28%), Positives = 42/87 (47%), Gaps = 9/87 (10%)
Query: 1 MDIDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL 60
+D+ I+ + VA + R+ ++ WG G+ G + D R P+L A
Sbjct: 251 LDVHQIVCGVRHVAL-VTRQGEVFTWGEEVGGRLGHGIQVDISR------PKLVEFLALT 303
Query: 61 NTRWLDIACGREHTAAIASDGLLFTWG 87
N + +ACG HT +++ G LF+WG
Sbjct: 304 NIDF--VACGEYHTCVVSTSGDLFSWG 328
Score = 30.0 bits (66), Expect = 0.19
Identities = 11/22 (50%), Positives = 14/22 (63%)
Query: 66 DIACGREHTAAIASDGLLFTWG 87
+IACG H A + S +FTWG
Sbjct: 528 EIACGAHHVAVLTSRSEVFTWG 549
Score = 29.6 bits (65), Expect = 0.25
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Query: 49 LPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLLFTWG 87
+P ++ G GL L +ACG H+A ++G LFT+G
Sbjct: 347 IPKRVSGPLEGLQV--LSVACGTWHSALATANGKLFTFG 383
Score = 27.7 bits (60), Expect = 0.95
Identities = 19/66 (28%), Positives = 29/66 (43%), Gaps = 10/66 (15%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL-NTRWLDIACGREHTAAIASDG 81
++ WG + G K+ L LP C + L + + IACG T A+ + G
Sbjct: 441 LFTWGDGDKNRLGHGNKETYL-----LPT----CVSSLIDYNFHKIACGHTFTVALTTSG 491
Query: 82 LLFTWG 87
+FT G
Sbjct: 492 HVFTMG 497
>At5g11580 unknown protein
Length = 553
Score = 37.4 bits (85), Expect = 0.001
Identities = 20/66 (30%), Positives = 33/66 (49%), Gaps = 5/66 (7%)
Query: 23 IYVWGYNQSGQTGR-KEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDG 81
+Y WG G+ G KE D+ + + + P Q G R + +A G H+ A++ DG
Sbjct: 37 VYSWGRGMFGRLGTGKESDELVPVLVEFPNQA----EGDRIRIVGVAAGAYHSLAVSDDG 92
Query: 82 LLFTWG 87
++ WG
Sbjct: 93 SVWCWG 98
Score = 35.4 bits (80), Expect = 0.005
Identities = 18/70 (25%), Positives = 36/70 (50%), Gaps = 9/70 (12%)
Query: 21 SAIYVWGYNQSGQTG-RKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIAS 79
S + +G+ ++GQ G R K L P P+ + + + CG HT+ ++S
Sbjct: 289 SICWTFGFGENGQLGHRSNKSSSLPEPVSDLPE--------HAYLVSVDCGLFHTSVVSS 340
Query: 80 DGLLFTWGLD 89
+G +++WG++
Sbjct: 341 EGYVWSWGME 350
Score = 35.0 bits (79), Expect = 0.006
Identities = 21/57 (36%), Positives = 30/57 (51%), Gaps = 5/57 (8%)
Query: 21 SAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAI 77
S +Y WG N GQ G + + + R P+ + + F +GL DIACG HTA +
Sbjct: 227 SVLYSWGNNHHGQLGLGDGESRAR-PQTV--ETFNQKSGLTV--YDIACGAHHTALL 278
Score = 30.4 bits (67), Expect = 0.15
Identities = 10/28 (35%), Positives = 18/28 (63%)
Query: 60 LNTRWLDIACGREHTAAIASDGLLFTWG 87
L+ + + +A G HT A+ DG +++WG
Sbjct: 14 LSRKIISLAAGEAHTIALTGDGCVYSWG 41
>At5g08710 unknown protein
Length = 434
Score = 37.4 bits (85), Expect = 0.001
Identities = 23/75 (30%), Positives = 38/75 (50%), Gaps = 8/75 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQ-LRIPKQLPPQLFGCPAGLNTRWLDIACGRE 72
+A+I +Y+WG N SGQ G +K + +R+P ++ L G +A G E
Sbjct: 146 SAAITVDGELYMWGKNSSGQLGLGKKAARVVRVPTKV-EALHGITI------QSVALGSE 198
Query: 73 HTAAIASDGLLFTWG 87
H+ A+ G + +WG
Sbjct: 199 HSVAVTDGGEVLSWG 213
Score = 35.0 bits (79), Expect = 0.006
Identities = 22/72 (30%), Positives = 33/72 (45%), Gaps = 8/72 (11%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ G N GQ G + P ++ +GL+ L I+ G H+AAI DG
Sbjct: 103 VFATGLNDCGQLGVSDVKSHAMDPLEV--------SGLDKDILHISAGYYHSAAITVDGE 154
Query: 83 LFTWGLDFSYSL 94
L+ WG + S L
Sbjct: 155 LYMWGKNSSGQL 166
Score = 35.0 bits (79), Expect = 0.006
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 8/71 (11%)
Query: 17 IPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAA 76
+ R +Y WG N++G G P ++ G L + ++CG +HTAA
Sbjct: 314 VTRGGELYTWGSNENGCLGTDST-----YVSHSPVRVEG--PFLESTVSQVSCGWKHTAA 366
Query: 77 IASDGLLFTWG 87
I SD +FTWG
Sbjct: 367 I-SDNNVFTWG 376
Score = 30.0 bits (66), Expect = 0.19
Identities = 10/22 (45%), Positives = 14/22 (63%)
Query: 66 DIACGREHTAAIASDGLLFTWG 87
++ACG HT + G L+TWG
Sbjct: 303 EVACGGYHTCVVTRGGELYTWG 324
Score = 26.2 bits (56), Expect = 2.8
Identities = 17/51 (33%), Positives = 23/51 (44%), Gaps = 8/51 (15%)
Query: 27 GYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAI 77
G++ GQ G D R P + G N R + I+CG HTAA+
Sbjct: 388 GHSSGGQLGHGSDVDYAR------PAMVDL--GKNVRAVHISCGFNHTAAV 430
>At3g26100 unknown protein (MPE11.29)
Length = 532
Score = 37.4 bits (85), Expect = 0.001
Identities = 21/74 (28%), Positives = 32/74 (42%), Gaps = 8/74 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
+ ++ K +Y +GYN SGQ G +D+ RI Q R + A G
Sbjct: 155 SVAVTSKGEVYTFGYNNSGQLGHGHTEDEARIQPVRSLQ--------GVRIIQAAAGAAR 206
Query: 74 TAAIASDGLLFTWG 87
T I+ DG ++ G
Sbjct: 207 TMLISDDGKVYACG 220
Score = 28.5 bits (62), Expect = 0.56
Identities = 18/70 (25%), Positives = 30/70 (42%), Gaps = 6/70 (8%)
Query: 18 PRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAI 77
P ++Y G G+ G + D+ + P+ + LN + +A G H A +
Sbjct: 321 PNGMSVYSVGCGLGGKLGHGSRTDE-KYPRVIEQFQI-----LNLQPRVVAAGAWHAAVV 374
Query: 78 ASDGLLFTWG 87
DG + TWG
Sbjct: 375 GQDGRVCTWG 384
>At1g27060 hypothetical protein
Length = 376
Score = 37.4 bits (85), Expect = 0.001
Identities = 22/65 (33%), Positives = 30/65 (45%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG GQ G + D+L PQL + + L ACG H A+ S G
Sbjct: 17 VWSWGAGTDGQLGTTKLQDELL------PQLLSLTSLPSISML--ACGGAHVIALTSGGK 68
Query: 83 LFTWG 87
+FTWG
Sbjct: 69 VFTWG 73
Score = 29.6 bits (65), Expect = 0.25
Identities = 11/24 (45%), Positives = 17/24 (70%)
Query: 67 IACGREHTAAIASDGLLFTWGLDF 90
I+ +H+AAI++DG F+WG F
Sbjct: 201 ISANGDHSAAISADGQFFSWGRGF 224
Score = 29.3 bits (64), Expect = 0.33
Identities = 11/23 (47%), Positives = 16/23 (68%)
Query: 65 LDIACGREHTAAIASDGLLFTWG 87
+ IA G EH+AA+ +G + TWG
Sbjct: 298 MQIAAGAEHSAAVTENGEVKTWG 320
Score = 27.7 bits (60), Expect = 0.95
Identities = 16/65 (24%), Positives = 29/65 (44%), Gaps = 7/65 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPA-GLNTRWLDIACGRE 72
+A++ + WG+ + GQ G +D Q P+L + L T+ + + CG
Sbjct: 307 SAAVTENGEVKTWGWGEHGQLGLGNTND------QTSPELVSLGSIDLRTKEIKVYCGSG 360
Query: 73 HTAAI 77
T A+
Sbjct: 361 FTYAV 365
>At1g19880 unknown protein
Length = 538
Score = 36.6 bits (83), Expect = 0.002
Identities = 20/73 (27%), Positives = 33/73 (44%), Gaps = 6/73 (8%)
Query: 21 SAIYVWGYNQSGQTGR------KEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
++I G Q GQ G KD +R+ + P+ + + +ACG HT
Sbjct: 188 ASILTAGLPQYGQLGHGTDNEFNMKDSSVRLAYEAQPRPKAIASLAGETIVKVACGTNHT 247
Query: 75 AAIASDGLLFTWG 87
A+ +G ++TWG
Sbjct: 248 VAVDKNGYVYTWG 260
Score = 30.4 bits (67), Expect = 0.15
Identities = 20/65 (30%), Positives = 28/65 (42%), Gaps = 10/65 (15%)
Query: 24 YVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDG-- 81
Y WG N+ GQ G + I + P + G + + A GR HT ++ DG
Sbjct: 80 YTWGRNEKGQLGHGDM-----IQRDRPTVVSGLS---KHKIVKAAAGRNHTVVVSDDGQS 131
Query: 82 LLFTW 86
L F W
Sbjct: 132 LGFGW 136
Score = 27.7 bits (60), Expect = 0.95
Identities = 10/29 (34%), Positives = 16/29 (54%)
Query: 59 GLNTRWLDIACGREHTAAIASDGLLFTWG 87
G+N R++ C H A+ +G +TWG
Sbjct: 55 GVNIRFVATGCASFHCVALDVEGRCYTWG 83
>At5g16040 UVB-resistance protein-like
Length = 396
Score = 36.2 bits (82), Expect = 0.003
Identities = 18/74 (24%), Positives = 37/74 (49%), Gaps = 6/74 (8%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
+ ++ + +Y WG + G+ G + D ++ PQ AG + + ++CG H
Sbjct: 260 STALTNEGEVYGWGRGEHGRLGFGDNDKS----SKMLPQKVNLLAGEDI--IQVSCGGTH 313
Query: 74 TAAIASDGLLFTWG 87
+ A+ DG +F++G
Sbjct: 314 SVALTRDGRIFSFG 327
Score = 33.9 bits (76), Expect = 0.013
Identities = 18/72 (25%), Positives = 34/72 (47%), Gaps = 8/72 (11%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++ ++ WG N+ GQ G + P+ P + G + +DIA G H+
Sbjct: 210 ALKEDGTLWAWGNNEYGQLGTGDTQ-----PRSYPIPVQGLD---DLTLVDIAAGGWHST 261
Query: 76 AIASDGLLFTWG 87
A+ ++G ++ WG
Sbjct: 262 ALTNEGEVYGWG 273
Score = 31.2 bits (69), Expect = 0.086
Identities = 20/69 (28%), Positives = 30/69 (42%), Gaps = 12/69 (17%)
Query: 21 SAIYVWGYNQSGQTG--RKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIA 78
+++ WG + GQ G E+ + + L P N R + G ++ AI
Sbjct: 5 TSVIAWGSGEDGQLGLGTDEEKELASVVDALEP--------FNVR--SVVGGSRNSLAIC 54
Query: 79 SDGLLFTWG 87
DG LFTWG
Sbjct: 55 DDGKLFTWG 63
Score = 28.1 bits (61), Expect = 0.73
Identities = 15/65 (23%), Positives = 25/65 (38%), Gaps = 7/65 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG+NQ G G + P + + N + + A G H A+ G
Sbjct: 59 LFTWGWNQRGTLGHPPETKTESTPSLV-------KSLANVKIVQAAIGGWHCLAVDDQGR 111
Query: 83 LFTWG 87
+ WG
Sbjct: 112 AYAWG 116
Score = 26.2 bits (56), Expect = 2.8
Identities = 20/64 (31%), Positives = 28/64 (43%), Gaps = 12/64 (18%)
Query: 24 YVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLL 83
YVW + Q G + Q+ +P ++ GL L IA G H A+ DG L
Sbjct: 170 YVWTWGQPWPPGDIK---QISVPVRVQ--------GLENVRL-IAVGAFHNLALKEDGTL 217
Query: 84 FTWG 87
+ WG
Sbjct: 218 WAWG 221
Score = 25.4 bits (54), Expect = 4.7
Identities = 25/88 (28%), Positives = 35/88 (39%), Gaps = 20/88 (22%)
Query: 3 IDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEK---DDQLRIPKQLPPQLFGCPAG 59
I G T VA + R I+ +G G+ G K L +P +PP P G
Sbjct: 305 IQVSCGGTHSVA--LTRDGRIFSFGRGDHGRLGYGRKVTTGQPLELPIHIPP-----PEG 357
Query: 60 L--------NTRWL--DIACGREHTAAI 77
+ +W+ +ACG HT AI
Sbjct: 358 RFNHTDEEDDGKWIAKHVACGGRHTLAI 385
>At1g65920
Length = 1006
Score = 36.2 bits (82), Expect = 0.003
Identities = 21/76 (27%), Positives = 34/76 (44%), Gaps = 8/76 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
AA + R+ ++ WG SG+ G K D + PK++ + +AC
Sbjct: 304 AALVTRQGEVFCWGNGNSGKLGLKVNID-IDHPKRVESLE-------DVAVRSVACSDHQ 355
Query: 74 TAAIASDGLLFTWGLD 89
T A+ G L+ WG+D
Sbjct: 356 TCAVTESGELYLWGID 371
Score = 33.5 bits (75), Expect = 0.017
Identities = 20/72 (27%), Positives = 34/72 (46%), Gaps = 9/72 (12%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++ +Y+WG + G E+ + +++ L G + L +ACG HTA
Sbjct: 358 AVTESGELYLWGIDG----GTIEQSGSQFLTRKISDVLGG-----SLTVLSVACGAWHTA 408
Query: 76 AIASDGLLFTWG 87
+ S G LFT+G
Sbjct: 409 IVTSSGQLFTYG 420
Score = 30.4 bits (67), Expect = 0.15
Identities = 27/83 (32%), Positives = 35/83 (41%), Gaps = 16/83 (19%)
Query: 8 GTTKPVAASIPRKSAIYVWGYNQSGQTG-RKEKDDQLRIPKQLPPQLFGCPAGLNTRWL- 65
G T VA SI +Y G + GQ G + KD + + G TR
Sbjct: 519 GWTLTVALSI--SGTVYTMGSSIHGQLGCPRAKDKSVNVV-----------LGNLTRQFV 565
Query: 66 -DIACGREHTAAIASDGLLFTWG 87
DIA G H A + S G ++TWG
Sbjct: 566 KDIASGSHHVAVLTSFGNVYTWG 588
Score = 28.1 bits (61), Expect = 0.73
Identities = 22/78 (28%), Positives = 33/78 (42%), Gaps = 9/78 (11%)
Query: 10 TKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIAC 69
T+PV S K I +WG G K ++D L P+L + + I+
Sbjct: 249 TQPVTRSNVLKD-IMIWGAITGLIDGSKNQNDALS------PKLLESATMFDVQ--SISL 299
Query: 70 GREHTAAIASDGLLFTWG 87
G +H A + G +F WG
Sbjct: 300 GAKHAALVTRQGEVFCWG 317
Score = 25.8 bits (55), Expect = 3.6
Identities = 14/35 (40%), Positives = 17/35 (48%), Gaps = 12/35 (34%)
Query: 65 LDIACGREHTAAIA------------SDGLLFTWG 87
+ ++CG HTAAI S G LFTWG
Sbjct: 450 ISVSCGPWHTAAIVETANDRKFYNAKSCGKLFTWG 484
Score = 24.6 bits (52), Expect = 8.0
Identities = 19/56 (33%), Positives = 26/56 (45%), Gaps = 10/56 (17%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLD-IACGREHTAAI 77
+Y WG +GQ G + D+ P + P L R ++ IACG TAAI
Sbjct: 584 VYTWGKGMNGQLGLGDVRDR-NSPVLVEP--------LGDRLVESIACGLNLTAAI 630
>At1g76950 zinc finger protein (PRAF1)
Length = 1103
Score = 35.8 bits (81), Expect = 0.003
Identities = 23/75 (30%), Positives = 34/75 (44%), Gaps = 9/75 (12%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLD-IACGRE 72
AA + R+ I+ WG G+ G D P+L ++ +D +ACG
Sbjct: 293 AAFVTRQGEIFTWGEESGGRLGHGIGKDVFH------PRLVESLTATSS--VDFVACGEF 344
Query: 73 HTAAIASDGLLFTWG 87
HT A+ G L+TWG
Sbjct: 345 HTCAVTLAGELYTWG 359
Score = 31.6 bits (70), Expect = 0.066
Identities = 18/65 (27%), Positives = 30/65 (45%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG + G +KD +L+ P PA ++ + IACG T + + G
Sbjct: 474 LFTWGDGDKNRLGHGDKDPRLK------PTCV--PALIDYNFHKIACGHSLTVGLTTSGQ 525
Query: 83 LFTWG 87
+FT G
Sbjct: 526 VFTMG 530
Score = 30.8 bits (68), Expect = 0.11
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Query: 49 LPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLLFTWG 87
+P ++ G GL+ ++CG HTA I S G LFT+G
Sbjct: 378 IPKRIAGSLEGLHVA--SVSCGPWHTALITSYGRLFTFG 414
Score = 29.6 bits (65), Expect = 0.25
Identities = 11/21 (52%), Positives = 13/21 (61%)
Query: 67 IACGREHTAAIASDGLLFTWG 87
IACG H A + G +FTWG
Sbjct: 286 IACGVRHAAFVTRQGEIFTWG 306
Score = 28.9 bits (63), Expect = 0.43
Identities = 18/64 (28%), Positives = 33/64 (51%), Gaps = 10/64 (15%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLD-IACGREH 73
A++ ++ +Y WG +G+ G + +D+ ++P + L R + IACG +
Sbjct: 570 AALTSRNEVYTWGKGANGRLGHGDLEDR-KVPTIVE--------ALKDRHVKYIACGSNY 620
Query: 74 TAAI 77
TAAI
Sbjct: 621 TAAI 624
Score = 28.5 bits (62), Expect = 0.56
Identities = 10/22 (45%), Positives = 15/22 (67%)
Query: 66 DIACGREHTAAIASDGLLFTWG 87
+I+CG H AA+ S ++TWG
Sbjct: 561 EISCGAYHVAALTSRNEVYTWG 582
Score = 27.7 bits (60), Expect = 0.95
Identities = 23/85 (27%), Positives = 38/85 (44%), Gaps = 20/85 (23%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
A I ++ +G G G +K+ ++ P+++ +GL T + ++CG HT
Sbjct: 402 ALITSYGRLFTFGDGTFGVLGHGDKET-VQYPREVESL-----SGLRT--IAVSCGVWHT 453
Query: 75 AAIA------------SDGLLFTWG 87
AA+ S G LFTWG
Sbjct: 454 AAVVEIIVTQSNSSSVSSGKLFTWG 478
>At5g19420 putative protein
Length = 1121
Score = 35.4 bits (80), Expect = 0.005
Identities = 21/75 (28%), Positives = 35/75 (46%), Gaps = 10/75 (13%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLD-IACGRE 72
A + ++ + WG G+ G D ++ PK + LNT ++ +ACG
Sbjct: 340 AVLVTKQGESFSWGEESEGRLGHGV-DSNVQHPKLID--------ALNTTNIELVACGEY 390
Query: 73 HTAAIASDGLLFTWG 87
H+ A+ G L+TWG
Sbjct: 391 HSCAVTLSGDLYTWG 405
Score = 32.7 bits (73), Expect = 0.029
Identities = 21/65 (32%), Positives = 29/65 (44%), Gaps = 7/65 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG G G + +PK++ + G IACG HTA + S G
Sbjct: 401 LYTWGKGDFGILGHGNEVSHW-VPKRVNFLMEGIHVS------SIACGPYHTAVVTSAGQ 453
Query: 83 LFTWG 87
LFT+G
Sbjct: 454 LFTFG 458
Score = 28.1 bits (61), Expect = 0.73
Identities = 18/65 (27%), Positives = 29/65 (43%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y G GQ G D ++P ++ +L + +IACG H A + S
Sbjct: 570 VYTMGSPVYGQLGNPHADG--KVPTRVDGKLH------KSFVEEIACGAYHVAVLTSRTE 621
Query: 83 LFTWG 87
++TWG
Sbjct: 622 VYTWG 626
Score = 27.3 bits (59), Expect = 1.2
Identities = 9/22 (40%), Positives = 13/22 (58%)
Query: 66 DIACGREHTAAIASDGLLFTWG 87
+IACG +H + G F+WG
Sbjct: 332 NIACGGQHAVLVTKQGESFSWG 353
Score = 26.9 bits (58), Expect = 1.6
Identities = 18/63 (28%), Positives = 28/63 (43%), Gaps = 8/63 (12%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
A + ++ +Y WG +G+ G + DD R L L + + IACG T
Sbjct: 614 AVLTSRTEVYTWGKGSNGRLGHGDADD--RNSPTLVESL------KDKQVKSIACGSNFT 665
Query: 75 AAI 77
AA+
Sbjct: 666 AAV 668
Score = 25.8 bits (55), Expect = 3.6
Identities = 15/37 (40%), Positives = 17/37 (45%), Gaps = 12/37 (32%)
Query: 63 RWLDIACGREHTAAIA------------SDGLLFTWG 87
R + ACG HTAA+ S G LFTWG
Sbjct: 486 RTVRAACGVWHTAAVVEVMVGSSSSSNCSSGKLFTWG 522
>At5g12350 putative protein
Length = 1062
Score = 35.4 bits (80), Expect = 0.005
Identities = 21/75 (28%), Positives = 35/75 (46%), Gaps = 10/75 (13%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLD-IACGRE 72
A + ++ + WG G+ G D ++ PK + LNT ++ +ACG
Sbjct: 307 AVLVTKQGESFSWGEESEGRLGHGV-DSNIQQPKLID--------ALNTTNIELVACGEF 357
Query: 73 HTAAIASDGLLFTWG 87
H+ A+ G L+TWG
Sbjct: 358 HSCAVTLSGDLYTWG 372
Score = 33.9 bits (76), Expect = 0.013
Identities = 22/65 (33%), Positives = 29/65 (43%), Gaps = 7/65 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG G G + +PK++ L G IACG HTA + S G
Sbjct: 368 LYTWGKGDFGVLGHGNEVSHW-VPKRVNFLLEGIHVS------SIACGPYHTAVVTSAGQ 420
Query: 83 LFTWG 87
LFT+G
Sbjct: 421 LFTFG 425
Score = 27.3 bits (59), Expect = 1.2
Identities = 9/22 (40%), Positives = 13/22 (58%)
Query: 66 DIACGREHTAAIASDGLLFTWG 87
+IACG +H + G F+WG
Sbjct: 299 NIACGGQHAVLVTKQGESFSWG 320
Score = 27.3 bits (59), Expect = 1.2
Identities = 10/22 (45%), Positives = 14/22 (63%)
Query: 66 DIACGREHTAAIASDGLLFTWG 87
+IACG H A + S ++TWG
Sbjct: 572 EIACGAYHVAVLTSRTEVYTWG 593
Score = 26.6 bits (57), Expect = 2.1
Identities = 18/63 (28%), Positives = 28/63 (43%), Gaps = 8/63 (12%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
A + ++ +Y WG +G+ G + DD R L L + + IACG T
Sbjct: 581 AVLTSRTEVYTWGKGSNGRLGHGDVDD--RNSPTLVESL------KDKQVKSIACGTNFT 632
Query: 75 AAI 77
AA+
Sbjct: 633 AAV 635
Score = 25.8 bits (55), Expect = 3.6
Identities = 15/37 (40%), Positives = 17/37 (45%), Gaps = 12/37 (32%)
Query: 63 RWLDIACGREHTAAIA------------SDGLLFTWG 87
R + ACG HTAA+ S G LFTWG
Sbjct: 453 RTVRAACGVWHTAAVVEVMVGSSSSSNCSSGKLFTWG 489
>At3g53830 putative protein
Length = 487
Score = 35.0 bits (79), Expect = 0.006
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y +G NQ GQ G ++ +P+ L Q G + + ++CG H+A +A DG
Sbjct: 383 VYAFGGNQFGQLGTGTDHAEI-LPRLLDGQNL---EGKHAK--AVSCGARHSAVLAEDGQ 436
Query: 83 LFTWG 87
L WG
Sbjct: 437 LLCWG 441
Score = 30.8 bits (68), Expect = 0.11
Identities = 20/74 (27%), Positives = 32/74 (43%), Gaps = 8/74 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
+A+I + +G+ GQ G +DQLR + R +A G H
Sbjct: 322 SAAITDAGGLITFGWGLYGQCGHGNTNDQLRP--------MAVSEVKSVRMESVAAGLWH 373
Query: 74 TAAIASDGLLFTWG 87
T I+SDG ++ +G
Sbjct: 374 TICISSDGKVYAFG 387
Score = 26.9 bits (58), Expect = 1.6
Identities = 24/90 (26%), Positives = 33/90 (36%), Gaps = 25/90 (27%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWL----------------- 65
I+ WGY GQ G + + P L P L +G ++
Sbjct: 250 IWGWGYGGEGQLGLGSRIKMVSSP-HLIPCLESIGSGKERSFILHQGGTTTTSAQASREP 308
Query: 66 -----DIACGREHTAAIASDG--LLFTWGL 88
I+CG H+AAI G + F WGL
Sbjct: 309 GQYIKAISCGGRHSAAITDAGGLITFGWGL 338
Score = 24.6 bits (52), Expect = 8.0
Identities = 20/82 (24%), Positives = 29/82 (34%), Gaps = 23/82 (28%)
Query: 14 AASIPRKSAIYVWG--------YNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWL 65
A +I K + WG Y SG+ G E + +P + P +
Sbjct: 64 AMAISEKGKLITWGSTDDEGQSYVASGKHG--ETPEPFPLPTEAPV-------------V 108
Query: 66 DIACGREHTAAIASDGLLFTWG 87
+ G H A + G FTWG
Sbjct: 109 QASSGWAHCAVVTETGEAFTWG 130
>At3g02510 unknown protein
Length = 393
Score = 34.7 bits (78), Expect = 0.008
Identities = 18/74 (24%), Positives = 36/74 (48%), Gaps = 6/74 (8%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
+ ++ K +Y WG + G+ G + D ++ PQ A + + ++CG H
Sbjct: 260 STALTDKGEVYGWGRGEHGRLGLGDNDKS----SKMVPQKVNLLAEEDI--IQVSCGGTH 313
Query: 74 TAAIASDGLLFTWG 87
+ A+ DG +F++G
Sbjct: 314 SVALTRDGRIFSFG 327
Score = 32.3 bits (72), Expect = 0.038
Identities = 17/65 (26%), Positives = 27/65 (41%), Gaps = 7/65 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG+NQ G G + + IP ++ A N + A G H A+ G
Sbjct: 59 MFTWGWNQRGTLGHQPETKTENIPSRV-------KALANVKITQAAIGGWHCLAVDDQGR 111
Query: 83 LFTWG 87
+ WG
Sbjct: 112 AYAWG 116
Score = 30.0 bits (66), Expect = 0.19
Identities = 18/69 (26%), Positives = 29/69 (41%), Gaps = 12/69 (17%)
Query: 21 SAIYVWGYNQSGQTG--RKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIA 78
++I WG + GQ G E+ + + + L P + G ++ AI
Sbjct: 5 TSIIAWGSGEDGQLGIGTNEEKEWACVVEALEPYSVR----------SVVSGSRNSLAIC 54
Query: 79 SDGLLFTWG 87
DG +FTWG
Sbjct: 55 DDGTMFTWG 63
Score = 29.3 bits (64), Expect = 0.33
Identities = 18/65 (27%), Positives = 28/65 (42%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+ WG N+ GQ G + P P + G + +DIA G H+ A+ G
Sbjct: 217 LLAWGNNEYGQLGTGDTQ-----PTSHPVPVQGLD---DLTLVDIAAGGWHSTALTDKGE 268
Query: 83 LFTWG 87
++ WG
Sbjct: 269 VYGWG 273
Score = 25.4 bits (54), Expect = 4.7
Identities = 28/110 (25%), Positives = 36/110 (32%), Gaps = 47/110 (42%)
Query: 24 YVWGYNQSGQTGRKEKDDQLR--------IPKQLPPQL---------------------- 53
Y WG N+ GQ G + D++ IPK+ P+L
Sbjct: 113 YAWGGNEYGQCGEEPLKDEMGRPVRRDIVIPKRCAPKLTVRQVAAGGTHSVVLTREGHVW 172
Query: 54 -FGCP---------------AGLNTRWLDIACGREHTAAIASDGLLFTWG 87
+G P GL L IA G H A+ DG L WG
Sbjct: 173 TWGQPWPPGDIKQISVPVRVQGLENVRL-IAVGAFHNLALEEDGRLLAWG 221
Score = 24.6 bits (52), Expect = 8.0
Identities = 26/88 (29%), Positives = 34/88 (38%), Gaps = 20/88 (22%)
Query: 3 IDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEK---DDQLRIPKQLPPQLFGCPAG 59
I G T VA + R I+ +G G+ G K L +P ++PP P G
Sbjct: 305 IQVSCGGTHSVA--LTRDGRIFSFGRGDHGRLGYGRKVTTGQPLELPIKIPP-----PEG 357
Query: 60 L--------NTRWL--DIACGREHTAAI 77
+W IACG HT AI
Sbjct: 358 SFNHTDEEEEGKWSAKSIACGGRHTLAI 385
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.143 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,808,097
Number of Sequences: 26719
Number of extensions: 109640
Number of successful extensions: 383
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 222
Number of HSP's gapped (non-prelim): 142
length of query: 121
length of database: 11,318,596
effective HSP length: 97
effective length of query: 24
effective length of database: 8,726,853
effective search space: 209444472
effective search space used: 209444472
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC137823.5


