

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137822.17 - phase: 0
(418 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
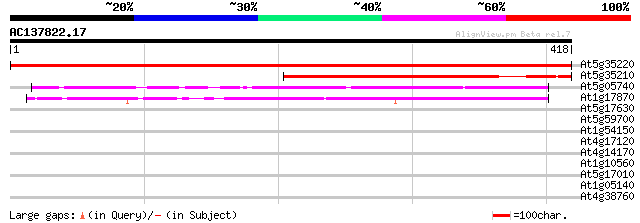
Score E
Sequences producing significant alignments: (bits) Value
At5g35220 putative protein 705 0.0
At5g35210 putative protein 302 3e-82
At5g05740 putative protein 181 7e-46
At1g17870 hypothetical protein 144 1e-34
At5g17630 glucose 6 phosphate/phosphate translocator-like protein 32 0.85
At5g59700 receptor-like protein kinase precursor - like 31 1.1
At1g54150 unknown protein 31 1.5
At4g17120 hypothetical protein 30 1.9
At4g14170 hypothetical protein 30 2.5
At1g10560 unknown protein 29 4.2
At5g17010 sugar transporter - like protein 28 7.2
At1g05140 Unknown protein (YUP8H12.25) 28 7.2
At4g38760 putative protein 28 9.4
>At5g35220 putative protein
Length = 548
Score = 705 bits (1820), Expect = 0.0
Identities = 342/418 (81%), Positives = 380/418 (90%)
Query: 1 MELLGPEKVDPADVKKIKDKLFGYSTFWVTKEEPFGELGEGILFIGNLRGKREDIFSILQ 60
MELLGPEKVDPADVK IKDKLFGYSTFWVTKEEPFG+LGEGILF+GNLRGK+ED+F+ LQ
Sbjct: 131 MELLGPEKVDPADVKLIKDKLFGYSTFWVTKEEPFGDLGEGILFLGNLRGKKEDVFAKLQ 190
Query: 61 NRLVEATGDKYNLFMVEEPDSDSPDPRGGPRVSFGLLRKEVSEPEETTLWQYVVASLLFL 120
+LVE DKYNLFM+EEP+S+ PDPRGG RVSFGLLRKEVSEP TTLWQYV+A +LFL
Sbjct: 191 RKLVEVASDKYNLFMIEEPNSEGPDPRGGARVSFGLLRKEVSEPGPTTLWQYVIALILFL 250
Query: 121 LTIGTSVEVGIASQINRLPPELVKFLTDPNYTEAPDMEMLYPFVESALPLAYGVLGVLLF 180
LTIG+SVE+GIASQINRLPPE+VK+ TDPN E PDME+LYPFV++ALPLAYGVLG+LLF
Sbjct: 251 LTIGSSVELGIASQINRLPPEVVKYFTDPNAVEPPDMELLYPFVDAALPLAYGVLGILLF 310
Query: 181 HEVGHFLAAFPKQVKLSIPFFIPHITLGSFGAITQFKSILPDRSTQVDISLAGPFAGAVL 240
HE+GHFLAA PK+VKLSIP+FIP+ITLGSFGAITQFKSILPDRST+VDISLAGPFAGA L
Sbjct: 311 HELGHFLAAVPKKVKLSIPYFIPNITLGSFGAITQFKSILPDRSTKVDISLAGPFAGAAL 370
Query: 241 SFSMFAVGLLLSSNPDVAGDLVQVPSMLFQGSLLLGLISRATLGYAAVHAATVPIHPLVI 300
S SMFAVGL LS+ PD A DLVQVPSMLFQGSLLLGLISRATLGYAA+HAATV IHPLVI
Sbjct: 371 SVSMFAVGLFLSTEPDAANDLVQVPSMLFQGSLLLGLISRATLGYAALHAATVSIHPLVI 430
Query: 301 AGWCGLTIQAFNMLPVGCLDGGRSVQGAFGKGATMVFGLTTYTLLGLGVLGGPLSLAWGF 360
AGWCGLT AFNMLPVGCLDGGR+VQGAFGK A + FGL+TY +LGL VLGGPL+L WG
Sbjct: 431 AGWCGLTTTAFNMLPVGCLDGGRAVQGAFGKNALVTFGLSTYVMLGLRVLGGPLALPWGL 490
Query: 361 FVIFSQRSPEKPCLNDVTEVGTWRQTFVGVAIFLAVLTLLPVWDELAEELGIGLVTTF 418
+V+ QR+PEKPCLNDVTEVGTWR+ VG+A+ L VLTLLPVWDELAEE+GIGLVTTF
Sbjct: 491 YVLICQRTPEKPCLNDVTEVGTWRKALVGIALILVVLTLLPVWDELAEEVGIGLVTTF 548
>At5g35210 putative protein
Length = 1606
Score = 302 bits (773), Expect = 3e-82
Identities = 158/214 (73%), Positives = 172/214 (79%), Gaps = 20/214 (9%)
Query: 205 ITLGSFGAITQFKSILPDRSTQVDISLAGPFAGAVLSFSMFAVGLLLSSNPDVAGDLVQV 264
ITLGSFGAITQFKSILPDRST+VDISLAGPFAGA LS SMFAVGL LS+ PD A DLVQV
Sbjct: 1413 ITLGSFGAITQFKSILPDRSTKVDISLAGPFAGAALSVSMFAVGLFLSTEPDAANDLVQV 1472
Query: 265 PSMLFQGSLLLGLISRATLGYAAVHAATVPIHPLVIAGWCGLTIQAFNMLPVGCLDGGRS 324
PSMLFQGSLLLGLISRATLGYAA+HAATV IHPLVIAGWCGLT AFNMLPVGCLDGGR+
Sbjct: 1473 PSMLFQGSLLLGLISRATLGYAALHAATVSIHPLVIAGWCGLTTTAFNMLPVGCLDGGRA 1532
Query: 325 VQGAFGKGATMVFGLTTYTLLGLGVLGGPLSLAWGFFVIFSQRSPEKPCLNDVTEVGTWR 384
VQGAFGK A + FGL+TY +LGL VLGGPL+L WG +V+
Sbjct: 1533 VQGAFGKNALVTFGLSTYVMLGLRVLGGPLALPWGLYVLIC------------------- 1573
Query: 385 QTFVGVAIFLAVLTLLPVWDELAEELGIGLVTTF 418
+ VG+A+ L VLTLLPVWDELA E+GIGLVTTF
Sbjct: 1574 RALVGIALILVVLTLLPVWDELA-EVGIGLVTTF 1606
>At5g05740 putative protein
Length = 556
Score = 181 bits (459), Expect = 7e-46
Identities = 127/386 (32%), Positives = 193/386 (49%), Gaps = 32/386 (8%)
Query: 17 IKDKLFGYSTFWVTKEEPFGELGEGILFIGNLRGKREDIFSILQNRLVEATGDKYNLFMV 76
++ ++FG+ TF+VT +EP+ G+LF GNLRGK + ++ R+ GD+Y LF++
Sbjct: 187 LRGQVFGFDTFFVTSQEPYEG---GVLFKGNLRGKPATSYEKIKTRMENNFGDQYKLFLL 243
Query: 77 EEPDSDSPDPRGGPRVSFGLLRKEVSEPEETTLWQYVVASLLFLLTIGTSVEVGIASQIN 136
P+ D P PR S EPE T + ++ A L+ + T +
Sbjct: 244 TNPEDDKPVAVVVPRRSL--------EPETTAVPEWFAAGSFGLVALFTL----FLRNVP 291
Query: 137 RLPPELVKFLTDPNYTEAPDMEMLYPFVESALPLAYGVLGVLLFHEVGHFLAAFPKQVKL 196
L +L+ + +E+L + AL A VLGV HE+GH L A +KL
Sbjct: 292 ALQSDLLSAFDN--------LELLKDGLPGALVTAL-VLGV---HELGHILVANSLGIKL 339
Query: 197 SIPFFIPHITLGSFGAITQFKSILPDRSTQVDISLAGPFAGAVLSFSMFAVGLLLSSNPD 256
+PFF+P +GSFGAIT+ K+I+ R + ++ AGP AG L +F +GL + P
Sbjct: 340 GVPFFVPSWQIGSFGAITRIKNIVAKREDLLKVAAAGPLAGFSLGLILFLIGLFV---PP 396
Query: 257 VAGDLVQVPSMLFQGSLLLGLISRATLGYAAVHAATVPIHPLVIAGWCGLTIQAFNMLPV 316
G V V + +F S L G I++ LG A ++ ++PLVI W GL I N +P
Sbjct: 397 SDGIGVVVDASVFHESFLAGGIAKLLLGDALKEGTSISLNPLVIWAWAGLLINGINSIPA 456
Query: 317 GCLDGGRSVQGAFG-KGATMVFGLTTYTLLGLGVLGGPLSLAWGFFVIFSQRSPEKPCLN 375
G LDGG+ +G K AT + G + LLGL L ++ W + F QR P P
Sbjct: 457 GELDGGKIAFSIWGRKTATRLTG-ASIALLGLSALFSDVAFYWVVLIFFLQRGPIAPLAE 515
Query: 376 DVTEVGTWRQTFVGVAIFLAVLTLLP 401
++T + + +FL++L LP
Sbjct: 516 EITVPDDKYVSLGILVLFLSLLVCLP 541
>At1g17870 hypothetical protein
Length = 573
Score = 144 bits (362), Expect = 1e-34
Identities = 118/402 (29%), Positives = 178/402 (43%), Gaps = 41/402 (10%)
Query: 13 DVKKIKDKLFGYSTFWVTKEEPFGELGEGILFIGNLRGKREDIFSILQNRLVEATGDKYN 72
D+ K+K FG+ TF+ T FG+ G +FIGNLR +++ L+ +L EA G
Sbjct: 179 DLNKLKS-CFGFDTFFATDVRRFGDGG---IFIGNLRKPIDEVTPKLEAKLSEAAGRDVV 234
Query: 73 LFMVEEPDSDSPDP--RGGPRVSFGLLRKEVSEPEETTLWQYVVASLLFLLTIGTSVEVG 130
++ +EE ++ P+ L + +T W YV A L + T GT +
Sbjct: 235 VWFMEERSNEITKQVCMVQPKAEIDL---QFESTRLSTPWGYVSAIALCVTTFGT---IA 288
Query: 131 IASQINRLPPELVKFLTDPNYTEAPDMEMLYPFVESALPLAYGVLGVLLFHEVGHFLAAF 190
+ S F P+ T ++ + +PL G L +L E+ + A
Sbjct: 289 LMSG----------FFLKPDAT-------FDDYIANVVPLFGGFLSILGVSEIATRVTAA 331
Query: 191 PKQVKLSIPFFIPHITLGSFGAITQFKSILPDRSTQVDISLAGPFAGAVLSFSMFAVGLL 250
VKLS F +P G G + ++S+LP++ DI +A A A L+ + A
Sbjct: 332 RHGVKLSPSFLVPSNWTGCLGVMNNYESLLPNKKALFDIPVART-ASAYLTSLLLAAAAF 390
Query: 251 LSSNPDVAGD-LVQVPSMLFQGSLLLGLISRATLGYA---------AVHAATVPIHPLVI 300
+S GD + + F + LL + YA AV VP+ PL
Sbjct: 391 ISDGSFNGGDNALYIRPQFFDNNPLLSFVQFVVGPYADDLGNVLPNAVEGVGVPVDPLAF 450
Query: 301 AGWCGLTIQAFNMLPVGCLDGGRSVQGAFGKGATMVFGLTTYTLLGLGVLGGP-LSLAWG 359
AG G+ + + N+LP G L+GGR Q FG+ + TT LLG+G L G L LAWG
Sbjct: 451 AGLLGMVVTSLNLLPCGRLEGGRIAQAMFGRSTAAILSFTTSLLLGIGGLSGSVLCLAWG 510
Query: 360 FFVIFSQRSPEKPCLNDVTEVGTWRQTFVGVAIFLAVLTLLP 401
F F + E P +++T VG R + V + LTL P
Sbjct: 511 LFATFFRGGEETPAKDEITPVGDDRFAWGIVLGLICFLTLFP 552
>At5g17630 glucose 6 phosphate/phosphate translocator-like protein
Length = 417
Score = 31.6 bits (70), Expect = 0.85
Identities = 30/100 (30%), Positives = 44/100 (44%), Gaps = 19/100 (19%)
Query: 160 LYPFVESALPLAYGVLGVLLFHEVGHFLA---------AFPKQVKLSIPFF--IPHITLG 208
LYP + + P +LG LFH +GH A +F +K + P F I LG
Sbjct: 163 LYPCPKISKPFIIALLGPALFHTIGHISACVSFSKVAVSFTHVIKSAEPVFSVIFSSLLG 222
Query: 209 SFGAITQFKSILP-------DRSTQVDISLAGPFAGAVLS 241
+ + SILP T+V +L G +GA++S
Sbjct: 223 DSYPLAVWLSILPIVMGCSLAAVTEVSFNLGG-LSGAMIS 261
>At5g59700 receptor-like protein kinase precursor - like
Length = 829
Score = 31.2 bits (69), Expect = 1.1
Identities = 40/142 (28%), Positives = 61/142 (42%), Gaps = 25/142 (17%)
Query: 240 LSFSMFAVGLLLSSNPDVAGDLVQVPSMLFQ-----GSLLLGLISRATLGYAAVHAATVP 294
L F+++ + + N D++ L S + GS L R ++G ++VH
Sbjct: 309 LYFNLYVDSMDVVENLDLSSYLSNTLSGAYAMDFVTGSAKLTKRIRVSIGRSSVHTD--- 365
Query: 295 IHPLVIAGWCGLTIQAFN----MLPVGC-LDGGRSVQGAFGKGATMVFGLTTYTLLGLGV 349
+P I GL I N L +G L G S K M+ GLT +LL L V
Sbjct: 366 -YPTAILN--GLEIMKMNNSKSQLSIGTFLPSGSS--STTKKNVGMIIGLTIGSLLALVV 420
Query: 350 LGGPLSLAWGFFVIFSQRSPEK 371
LG GFFV++ +R ++
Sbjct: 421 LG-------GFFVLYKKRGRDQ 435
>At1g54150 unknown protein
Length = 383
Score = 30.8 bits (68), Expect = 1.5
Identities = 13/29 (44%), Positives = 21/29 (71%)
Query: 260 DLVQVPSMLFQGSLLLGLISRATLGYAAV 288
DL++ + +F GS++LG++S L YAAV
Sbjct: 263 DLMEQTNFIFLGSVILGIVSVGILSYAAV 291
>At4g17120 hypothetical protein
Length = 1661
Score = 30.4 bits (67), Expect = 1.9
Identities = 18/50 (36%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Query: 181 HEVGHFLAAFPKQVKL-SIPFFIPHITLGSFGAITQFKSILPDRSTQVDI 229
+ VG FL A P + L P+ + HI GS + + S + DR+T DI
Sbjct: 44 NSVGSFLPADPSTLNLLGRPYELRHILFGSTAVLPKESSYVDDRTTPDDI 93
>At4g14170 hypothetical protein
Length = 428
Score = 30.0 bits (66), Expect = 2.5
Identities = 20/67 (29%), Positives = 31/67 (45%), Gaps = 4/67 (5%)
Query: 241 SFSMFAVGLLLSSNPDVAGDLVQVPSM---LFQGSLLLGLISRATLGYAAVHAATVPIHP 297
S S+F L+ D+ G L+ P + QG +LGL +GY+ +V +H
Sbjct: 147 SSSLFVSSALVIMYVDM-GKLLHAPMFGGYVQQGEAMLGLAMFREMGYSGFALDSVVMHG 205
Query: 298 LVIAGWC 304
+ GWC
Sbjct: 206 KSVHGWC 212
>At1g10560 unknown protein
Length = 697
Score = 29.3 bits (64), Expect = 4.2
Identities = 24/79 (30%), Positives = 36/79 (45%), Gaps = 10/79 (12%)
Query: 275 LGLISRATLGYAAVHAATVPIHPLVIAGWC-GLTIQAFNMLPVGCLDGGRSVQGAFGKGA 333
+G+I R L A ++ P+ + C GL + CL+GGR V G K +
Sbjct: 593 IGVIRRGGLKLAVKILSSSEDSPVAVKQHCVGLILNL-------CLNGGRDVVGVLVKNS 645
Query: 334 TMVFGLTTYTLLGLGVLGG 352
++ L YT+L G GG
Sbjct: 646 LVMGSL--YTVLSNGEYGG 662
>At5g17010 sugar transporter - like protein
Length = 503
Score = 28.5 bits (62), Expect = 7.2
Identities = 19/67 (28%), Positives = 31/67 (45%), Gaps = 7/67 (10%)
Query: 245 FAVGLLLSSNPDVAGDLVQVPSMLFQGSLLLGLISRATLGYAAVHAATVPIHPLVIAGWC 304
+A +L ++ AGD +V S+LLGL+ G A V + PL++ G
Sbjct: 327 YAPSILQTAGFSAAGDATRV-------SILLGLLKLIMTGVAVVVIDRLGRRPLLLGGVG 379
Query: 305 GLTIQAF 311
G+ + F
Sbjct: 380 GMVVSLF 386
>At1g05140 Unknown protein (YUP8H12.25)
Length = 441
Score = 28.5 bits (62), Expect = 7.2
Identities = 12/34 (35%), Positives = 22/34 (64%)
Query: 164 VESALPLAYGVLGVLLFHEVGHFLAAFPKQVKLS 197
+ES L + + +++ HE GHFLAA + +++S
Sbjct: 79 LESVLEASAVLTAIIVVHETGHFLAASLQGIRVS 112
>At4g38760 putative protein
Length = 561
Score = 28.1 bits (61), Expect = 9.4
Identities = 18/51 (35%), Positives = 23/51 (44%), Gaps = 5/51 (9%)
Query: 352 GPLSLAWGFFVIFSQRSP---EKPCLNDVTEVGTWRQTF--VGVAIFLAVL 397
GPL LAW F+ P E P L D+ V Q F ++ FL +L
Sbjct: 232 GPLVLAWAVFLCLISSLPGKEESPFLMDIDHVSYVHQAFEAASLSYFLEIL 282
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.143 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,273,557
Number of Sequences: 26719
Number of extensions: 394514
Number of successful extensions: 855
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 836
Number of HSP's gapped (non-prelim): 14
length of query: 418
length of database: 11,318,596
effective HSP length: 102
effective length of query: 316
effective length of database: 8,593,258
effective search space: 2715469528
effective search space used: 2715469528
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137822.17


