

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137822.11 - phase: 0
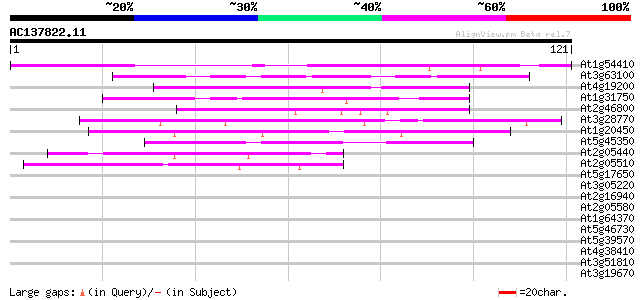
(121 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g54410 unknown protein 93 2e-20
At3g63100 putative protein 49 3e-07
At4g19200 unknown protein 45 6e-06
At1g31750 unknown protein 42 6e-05
At2g46800 putative zinc transporter 41 8e-05
At3g28770 hypothetical protein 40 2e-04
At1g20450 putative cold-acclimation protein 39 3e-04
At5g45350 unknown protein 39 5e-04
At2g05440 putative glycine-rich protein 39 5e-04
At2g05510 putative glycine-rich protein 38 7e-04
At5g17650 glycine/proline-rich protein 38 0.001
At3g05220 unknown protein 38 0.001
At2g16940 splicing factor like protein 38 0.001
At2g05580 unknown protein 38 0.001
At1g64370 unknown protein 37 0.001
At5g46730 unknown protein 37 0.002
At5g39570 unknown protein 37 0.002
At4g38410 cold-regulated protein - like 36 0.003
At3g51810 embryonic abundant protein AtEm1 36 0.003
At3g19670 unknown protein 36 0.003
>At1g54410 unknown protein
Length = 98
Score = 92.8 bits (229), Expect = 2e-20
Identities = 62/134 (46%), Positives = 68/134 (50%), Gaps = 51/134 (38%)
Query: 1 MAGIMNKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHG 60
MAG++NKIGDALHIGG KEGEHK E+ EHK
Sbjct: 1 MAGLINKIGDALHIGGGNKEGEHKKEE-------------------------EHK----- 30
Query: 61 FGHGDHKEGHHGEEHKEGFVDKIKDKIHG----------EGADGEKKKKK---EKKKHGE 107
H + H EHKEG VDKIKDKIHG + DGEKKKKK EKK H +
Sbjct: 31 ----KHVDEHKSGEHKEGIVDKIKDKIHGGEGKSHDGEGKSHDGEKKKKKDKKEKKHHDD 86
Query: 108 GHEHGHDSSSSDSD 121
GH SSSSDSD
Sbjct: 87 GHH----SSSSDSD 96
>At3g63100 putative protein
Length = 199
Score = 49.3 bits (116), Expect = 3e-07
Identities = 32/90 (35%), Positives = 39/90 (42%), Gaps = 15/90 (16%)
Query: 23 HKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEGFVDK 82
H HGH G H E HG GHG G HG GHG H HG +H+ G
Sbjct: 70 HCDHCHGHGYGHGHRE-----HGHDRGHG---HGRGHGHGHG-HGHRRHGRDHRHG---- 116
Query: 83 IKDKIHGEGADGEKKKKKEKKKHGEGHEHG 112
+D+ H G G + ++ G GH HG
Sbjct: 117 -RDRGHHRG-HGHHHHRGHRRGRGRGHGHG 144
Score = 36.2 bits (82), Expect = 0.003
Identities = 20/61 (32%), Positives = 24/61 (38%), Gaps = 24/61 (39%)
Query: 59 HGFGHGDHKEGH-----HGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGH 113
HG+GHG + GH HG H G HG G ++HG H HG
Sbjct: 77 HGYGHGHREHGHDRGHGHGRGHGHG---------HGHG----------HRRHGRDHRHGR 117
Query: 114 D 114
D
Sbjct: 118 D 118
Score = 36.2 bits (82), Expect = 0.003
Identities = 28/99 (28%), Positives = 41/99 (41%), Gaps = 15/99 (15%)
Query: 16 GDKKEGEHKGEQHG--HVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGE 73
G ++ G +G HG H G HG +HG HG + +G G GH HH
Sbjct: 82 GHREHGHDRGHGHGRGHGHGHGHGH---RRHGRDHRHGRD-RGHHRGHGH------HHHR 131
Query: 74 EHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHG 112
H+ G + + HG G G ++ E+ + G G
Sbjct: 132 GHRRG---RGRGHGHGRGRGGHVQEAGERWEQEVGGRGG 167
Score = 33.9 bits (76), Expect = 0.013
Identities = 18/56 (32%), Positives = 22/56 (39%), Gaps = 15/56 (26%)
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGH 70
G D + G +G GH G HH H G +G G GHG + GH
Sbjct: 110 GRDHRHGRDRGHHRGH--GHHH-------------HRGHRRGRGRGHGHGRGRGGH 150
Score = 29.3 bits (64), Expect = 0.33
Identities = 19/65 (29%), Positives = 23/65 (35%)
Query: 49 GHGGEHKGEQHGFGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEG 108
G GG G G G + G H G D HG G ++ + HG G
Sbjct: 36 GGGGGGGGVDGGVDRGCCRHCCGGRRHDYGRDCCHCDHCHGHGYGHGHREHGHDRGHGHG 95
Query: 109 HEHGH 113
HGH
Sbjct: 96 RGHGH 100
>At4g19200 unknown protein
Length = 179
Score = 45.1 bits (105), Expect = 6e-06
Identities = 28/71 (39%), Positives = 30/71 (41%), Gaps = 5/71 (7%)
Query: 32 GGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDH---KEGHHGEEHKEGFVDKIKDKIH 88
G H G +G GHGG HGFGHG H K G HG + K G K H
Sbjct: 109 GAHHVGHASHNPYGHAVGHGGYGHAPAHGFGHGGHGKFKHGKHGGKFKHG--KHGKHGKH 166
Query: 89 GEGADGEKKKK 99
G G K KK
Sbjct: 167 GMFGGGGKFKK 177
Score = 28.5 bits (62), Expect = 0.56
Identities = 17/44 (38%), Positives = 21/44 (47%), Gaps = 4/44 (9%)
Query: 23 HKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDH 66
H G H G HG + +H G HGG+ K +HG HG H
Sbjct: 127 HGGYGHAPAHGFGHGGHGKFKH---GKHGGKFKHGKHG-KHGKH 166
>At1g31750 unknown protein
Length = 176
Score = 41.6 bits (96), Expect = 6e-05
Identities = 30/83 (36%), Positives = 37/83 (44%), Gaps = 12/83 (14%)
Query: 21 GEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHH----GEEHK 76
G H HG G HG+YK GF GG G +G+ FG G +K G H G+ K
Sbjct: 102 GGHHAGHHGGYGHHGHGKYK---RGFFGG-GKYKRGKHSMFGGGKYKRGKHGMFGGKRSK 157
Query: 77 EGFVDKIKDKIHGEGADGEKKKK 99
G + G+G G KK K
Sbjct: 158 HGMFGGKR----GKGMFGRKKWK 176
Score = 32.7 bits (73), Expect = 0.029
Identities = 26/84 (30%), Positives = 31/84 (35%), Gaps = 8/84 (9%)
Query: 29 GHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEGFVDKIKDKIH 88
G VGG G G H G H G H GHG +K G G K K H
Sbjct: 84 GGVGGLIAGAATAAAAAMGGHHAGHHGGYGH-HGHGKYKRGFFGG-------GKYKRGKH 135
Query: 89 GEGADGEKKKKKEKKKHGEGHEHG 112
G+ K+ K G+ +HG
Sbjct: 136 SMFGGGKYKRGKHGMFGGKRSKHG 159
>At2g46800 putative zinc transporter
Length = 398
Score = 41.2 bits (95), Expect = 8e-05
Identities = 27/75 (36%), Positives = 36/75 (48%), Gaps = 12/75 (16%)
Query: 37 GEYKGEQHGFVGGHGGEHKGEQHG------FGHGDHKEGH---HGEE--HKEGFV-DKIK 84
G G HG GHG +H HG H DH+ GH HGE+ H G V +++
Sbjct: 181 GHDHGHSHGHGHGHGHDHHNHSHGVTVTTHHHHHDHEHGHSHGHGEDKHHAHGDVTEQLL 240
Query: 85 DKIHGEGADGEKKKK 99
DK + A EK+K+
Sbjct: 241 DKSKTQVAAKEKRKR 255
Score = 33.5 bits (75), Expect = 0.017
Identities = 19/70 (27%), Positives = 22/70 (31%), Gaps = 16/70 (22%)
Query: 52 GEHKGEQHGFGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEH 111
G G HG GHG HG +H H G +HG H H
Sbjct: 181 GHDHGHSHGHGHG------HGHDHHN----------HSHGVTVTTHHHHHDHEHGHSHGH 224
Query: 112 GHDSSSSDSD 121
G D + D
Sbjct: 225 GEDKHHAHGD 234
Score = 31.6 bits (70), Expect = 0.066
Identities = 23/76 (30%), Positives = 25/76 (32%), Gaps = 26/76 (34%)
Query: 41 GEQHGFVGGHGGEHKGEQHGFGHGDHKEGH--------------HGEEHKEGFVDKIKDK 86
G HG GHG HG GH H H HG H G +DK
Sbjct: 181 GHDHGHSHGHG-------HGHGHDHHNHSHGVTVTTHHHHHDHEHGHSHGHG-----EDK 228
Query: 87 IHGEGADGEKKKKKEK 102
H G E+ K K
Sbjct: 229 HHAHGDVTEQLLDKSK 244
Score = 24.6 bits (52), Expect = 8.0
Identities = 12/25 (48%), Positives = 13/25 (52%), Gaps = 2/25 (8%)
Query: 23 HKGEQHGHVGGEHHGEYKGEQHGFV 47
H +HGH G HGE K HG V
Sbjct: 213 HHDHEHGHSHG--HGEDKHHAHGDV 235
>At3g28770 hypothetical protein
Length = 2081
Score = 40.0 bits (92), Expect = 2e-04
Identities = 34/112 (30%), Positives = 55/112 (48%), Gaps = 12/112 (10%)
Query: 16 GDKKEGEHKGEQHGHV----GGEHHGEYKGEQHG-FVGGHGGEHKGEQHGFGHGDHKEGH 70
G K+ +G + ++ GG+ +G + G V +GGE + G G +EG
Sbjct: 1624 GGKEVSTEEGSKDSNIVERNGGKEDSIKEGSEDGKTVEINGGEELSTEEGSKDGKIEEGK 1683
Query: 71 HGEEH--KEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHE-HGHDSSSSD 119
G+E+ KEG D DKI EG +G++ KE K G+ +E HG ++ +
Sbjct: 1684 EGKENSTKEGSKD---DKIE-EGMEGKENSTKESSKDGKINEIHGDKEATME 1731
Score = 29.6 bits (65), Expect = 0.25
Identities = 26/91 (28%), Positives = 40/91 (43%), Gaps = 4/91 (4%)
Query: 17 DKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHK 76
DKK+ E +Q+ + E E K QH + + K ++ + KE + K
Sbjct: 1116 DKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQK 1175
Query: 77 EGFVDKIKDKIHGEGADGEKKKKKEKKKHGE 107
VDK + K D +KKK+KE K+ E
Sbjct: 1176 NE-VDKKEKK---SSKDQQKKKEKEMKESEE 1202
Score = 28.5 bits (62), Expect = 0.56
Identities = 20/57 (35%), Positives = 24/57 (42%), Gaps = 5/57 (8%)
Query: 67 KEGHHGEEHKEGFVDKIKDKIHGEGADGEKKK--KKEKKKHGEGHEHGHDSSSSDSD 121
KE E HK K +DK E KK+ KKEKKKH E + D +
Sbjct: 1068 KEKKESENHKS---KKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDME 1121
Score = 26.2 bits (56), Expect = 2.8
Identities = 13/32 (40%), Positives = 19/32 (58%)
Query: 73 EEHKEGFVDKIKDKIHGEGADGEKKKKKEKKK 104
+E KEG ++ KD I+ K KKK+KK+
Sbjct: 919 DEKKEGNKEENKDTINTSSKQKGKDKKKKKKE 950
>At1g20450 putative cold-acclimation protein
Length = 260
Score = 39.3 bits (90), Expect = 3e-04
Identities = 35/100 (35%), Positives = 45/100 (45%), Gaps = 12/100 (12%)
Query: 18 KKEGEHKGEQHGHVGGE-HHGEYKGEQHGFVGGHGGE-HKGEQHGFGHGDHKEGHHGEEH 75
KK+ E K ++ + E H E FV H E HK H H+E EE+
Sbjct: 37 KKKEEVKPQETTTLASEFEHKTQISEPESFVAKHEEEEHKPTLLEQLHQKHEEE---EEN 93
Query: 76 KEGFVDKI-------KDKIHGEGADGEKKKKKEKKKHGEG 108
K +DK+ EG DGEKKKK++KKK EG
Sbjct: 94 KPSLLDKLHRSNSSSSSSSDEEGEDGEKKKKEKKKKIVEG 133
Score = 30.8 bits (68), Expect = 0.11
Identities = 15/30 (50%), Positives = 23/30 (76%), Gaps = 2/30 (6%)
Query: 74 EHKEGFVDKIKDKIHGEGA--DGEKKKKKE 101
E K+GF+DKIK+K+ G A GE++KK++
Sbjct: 228 EEKKGFMDKIKEKLPGYHAKTTGEEEKKEK 257
Score = 29.6 bits (65), Expect = 0.25
Identities = 15/31 (48%), Positives = 19/31 (60%), Gaps = 3/31 (9%)
Query: 65 DHKEGHHGEEHKEGFVDKIKDKIHGEGADGE 95
DHK EE K+GF+DKIK+K+ G E
Sbjct: 179 DHKPE---EEEKKGFMDKIKEKLPGHSKKPE 206
>At5g45350 unknown protein
Length = 177
Score = 38.5 bits (88), Expect = 5e-04
Identities = 26/71 (36%), Positives = 29/71 (40%), Gaps = 12/71 (16%)
Query: 30 HVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEGFVDKIKDKIHG 89
HV HG Y +G GHG G +G GHG K G HG K K HG
Sbjct: 119 HVAHSSHGPYGHAAYGHGFGHG---HGYGYGHGHGKFKHGKHG---------KFKHGKHG 166
Query: 90 EGADGEKKKKK 100
G+ KK K
Sbjct: 167 MFGGGKFKKWK 177
Score = 33.1 bits (74), Expect = 0.023
Identities = 19/45 (42%), Positives = 22/45 (48%), Gaps = 3/45 (6%)
Query: 28 HGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHG 72
HG G +G G HG+ GHG H +HG HG K G HG
Sbjct: 125 HGPYGHAAYGHGFGHGHGYGYGHG--HGKFKHG-KHGKFKHGKHG 166
Score = 27.7 bits (60), Expect = 0.95
Identities = 18/54 (33%), Positives = 25/54 (45%), Gaps = 8/54 (14%)
Query: 16 GDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHG-FGHGDHKE 68
G G G HG+ G HG++K +H G+ K +HG FG G K+
Sbjct: 129 GHAAYGHGFGHGHGYGYGHGHGKFKHGKH-------GKFKHGKHGMFGGGKFKK 175
>At2g05440 putative glycine-rich protein
Length = 154
Score = 38.5 bits (88), Expect = 5e-04
Identities = 28/68 (41%), Positives = 31/68 (45%), Gaps = 10/68 (14%)
Query: 9 GDALHIGGDKKEGEHKGEQHGHVGGE--HHGEYKGEQHGFVGGH--GGEHKGEQHGFGHG 64
G H GG G H G GH GG H+G G G GGH GG + G G+G G
Sbjct: 77 GGGGHYGGG---GGHYGGGGGHYGGGGGHYGGGGGHYGGGGGGHGGGGHYGGGGGGYGGG 133
Query: 65 DHKEGHHG 72
GHHG
Sbjct: 134 G---GHHG 138
Score = 36.2 bits (82), Expect = 0.003
Identities = 26/75 (34%), Positives = 35/75 (46%), Gaps = 6/75 (8%)
Query: 4 IMNKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGH 63
+++++ A G K E E + G+ GG HG + G HG GGHG H G G GH
Sbjct: 17 VVSEVSAARQSGMVKPESEETVQPEGYGGG--HGGHGG--HGGGGGHG--HGGHNGGGGH 70
Query: 64 GDHKEGHHGEEHKEG 78
G G G + G
Sbjct: 71 GLDGYGGGGGHYGGG 85
Score = 33.5 bits (75), Expect = 0.017
Identities = 22/56 (39%), Positives = 23/56 (40%), Gaps = 3/56 (5%)
Query: 9 GDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHG 64
G H GG G H G GH GG G G +G GG G G G GHG
Sbjct: 91 GGGGHYGGG---GGHYGGGGGHYGGGGGGHGGGGHYGGGGGGYGGGGGHHGGGGHG 143
Score = 32.3 bits (72), Expect = 0.038
Identities = 24/60 (40%), Positives = 27/60 (45%), Gaps = 5/60 (8%)
Query: 15 GGDKKEGEHKGEQHG-HVGGEHHG-EYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHG 72
GG G G HG H GG HG + G G GG GG + G G GH GH+G
Sbjct: 48 GGHGGHGGGGGHGHGGHNGGGGHGLDGYGGGGGHYGGGGGHYGG---GGGHYGGGGGHYG 104
Score = 31.6 bits (70), Expect = 0.066
Identities = 21/57 (36%), Positives = 25/57 (43%), Gaps = 2/57 (3%)
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHH 71
GG +G G H GG H+G G G G +GG G +G G G H G H
Sbjct: 68 GGHGLDGYGGGGGHYGGGGGHYGGGGGHYGGGGGHYGGG--GGHYGGGGGGHGGGGH 122
Score = 31.2 bits (69), Expect = 0.086
Identities = 21/58 (36%), Positives = 25/58 (42%), Gaps = 3/58 (5%)
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHG 72
GG G + G HG G G + G G GG GG + G G GH GH+G
Sbjct: 57 GGHGHGGHNGGGGHGLDGYGGGGGHYGGGGGHYGGGGGHYGG---GGGHYGGGGGHYG 111
Score = 29.6 bits (65), Expect = 0.25
Identities = 23/56 (41%), Positives = 24/56 (42%), Gaps = 6/56 (10%)
Query: 21 GEHKGEQHGHVGG---EHHGEYKGEQHGFVG-GHGGEHKGEQHGFGHGDHKEGHHG 72
G G GH GG H G G HG G G GG H G G GH GH+G
Sbjct: 44 GGGHGGHGGHGGGGGHGHGGHNGGGGHGLDGYGGGGGHYG--GGGGHYGGGGGHYG 97
>At2g05510 putative glycine-rich protein
Length = 127
Score = 38.1 bits (87), Expect = 7e-04
Identities = 25/71 (35%), Positives = 36/71 (50%), Gaps = 3/71 (4%)
Query: 4 IMNKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVG-GHGGEHKGEQHGF- 61
+++++ A G K E E + G+ GG H G G +G G GHGG + G HG
Sbjct: 17 VVSEVSAARQSGMVKPESEATVQPEGYHGG-HGGHGGGGHYGGGGHGHGGHNGGGGHGLD 75
Query: 62 GHGDHKEGHHG 72
G+G GH+G
Sbjct: 76 GYGGGHGGHYG 86
Score = 37.7 bits (86), Expect = 0.001
Identities = 26/64 (40%), Positives = 28/64 (43%), Gaps = 6/64 (9%)
Query: 15 GGDKKEGEHKGEQHGHVG----GEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDH--KE 68
GG G + G HGH G G H + G HG G GG H G G G G H
Sbjct: 48 GGHGGGGHYGGGGHGHGGHNGGGGHGLDGYGGGHGGHYGGGGGHYGGGGGHGGGGHYGGG 107
Query: 69 GHHG 72
GHHG
Sbjct: 108 GHHG 111
Score = 34.3 bits (77), Expect = 0.010
Identities = 22/59 (37%), Positives = 25/59 (42%), Gaps = 1/59 (1%)
Query: 21 GEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQ-HGFGHGDHKEGHHGEEHKEG 78
G H G H GG HG + G + G+GG H G G GH GH G H G
Sbjct: 48 GGHGGGGHYGGGGHGHGGHNGGGGHGLDGYGGGHGGHYGGGGGHYGGGGGHGGGGHYGG 106
Score = 33.9 bits (76), Expect = 0.013
Identities = 26/63 (41%), Positives = 28/63 (44%), Gaps = 10/63 (15%)
Query: 15 GGDKKEGEHKGEQHG--HVGGEHHGEYKGE--QHGFVGGHGGE-HKGEQHGFGHGDHKEG 69
GG G + G HG GG H G Y G +G GGHGG H G G G H G
Sbjct: 59 GGHGHGGHNGGGGHGLDGYGGGHGGHYGGGGGHYGGGGGHGGGGHYG-----GGGHHGGG 113
Query: 70 HHG 72
HG
Sbjct: 114 GHG 116
>At5g17650 glycine/proline-rich protein
Length = 173
Score = 37.7 bits (86), Expect = 0.001
Identities = 26/90 (28%), Positives = 33/90 (35%), Gaps = 13/90 (14%)
Query: 23 HKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEGFVDK 82
H HG+ H G + G + G G H H H HHG + G+
Sbjct: 83 HGYPSHGYPRPSHSGHHHGGIGAIIAGGVAAAAGAHHMSHHHGHYGHHHGHGYGYGY--- 139
Query: 83 IKDKIHGEGADGEKKKKKEKKKHGEGHEHG 112
HG G K K K KHG+ +HG
Sbjct: 140 -----HGHG-----KFKHGKFKHGKFGKHG 159
Score = 35.8 bits (81), Expect = 0.003
Identities = 25/80 (31%), Positives = 29/80 (36%), Gaps = 23/80 (28%)
Query: 23 HKGEQHGHVGG----------------EHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDH 66
H G HG +G HHG Y G HG G+G HG G H
Sbjct: 95 HSGHHHGGIGAIIAGGVAAAAGAHHMSHHHGHY-GHHHGHGYGYG------YHGHGKFKH 147
Query: 67 KEGHHGEEHKEGFVDKIKDK 86
+ HG+ K G K K K
Sbjct: 148 GKFKHGKFGKHGMFGKHKGK 167
Score = 35.0 bits (79), Expect = 0.006
Identities = 25/64 (39%), Positives = 29/64 (45%), Gaps = 11/64 (17%)
Query: 23 HKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHG--EEHKEGFV 80
H HGH G HHG HG+ G+ G K + F HG K G HG +HK F
Sbjct: 119 HMSHHHGHYG-HHHG------HGYGYGYHGHGKFKHGKFKHG--KFGKHGMFGKHKGKFF 169
Query: 81 DKIK 84
K K
Sbjct: 170 KKWK 173
Score = 30.0 bits (66), Expect = 0.19
Identities = 12/36 (33%), Positives = 18/36 (49%)
Query: 34 EHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEG 69
+H+ +G H G GG++ HG+GH H G
Sbjct: 5 QHNHSDRGFFHNLAGFAGGQYPPHGHGYGHHGHGYG 40
>At3g05220 unknown protein
Length = 541
Score = 37.7 bits (86), Expect = 0.001
Identities = 37/138 (26%), Positives = 48/138 (33%), Gaps = 53/138 (38%)
Query: 17 DKKEGEHKGEQHGHVGGE---------------------HHGEYKGEQ---------HGF 46
D+ + + G HGH GG HHG G + +GF
Sbjct: 171 DEDDDDEMGHGHGHGGGGGNHMPPNKMMMPNKMMPQMGGHHGNGGGPKGPNEIMMMMNGF 230
Query: 47 VGGHGGEHKGEQHGF-------GHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKKK 99
GG GG KG GF G G+ K G G++ G GEK KK
Sbjct: 231 KGGGGGGKKGGGGGFEIPVQMKGMGEGKNGKDGKK----------------GKGGEKGKK 274
Query: 100 KEKKKHGEGHEHGHDSSS 117
+ K+ G G D+ S
Sbjct: 275 EGKENKGGGKTGKTDAKS 292
Score = 30.4 bits (67), Expect = 0.15
Identities = 35/121 (28%), Positives = 51/121 (41%), Gaps = 20/121 (16%)
Query: 16 GDKKEGEHKGEQHG--HVGGEHHG--EYKGEQHGFVG-------GHGGEHKGEQHGFGHG 64
G K +G KG++ G + GG G + K G +G G+G E K GHG
Sbjct: 263 GKKGKGGEKGKKEGKENKGGGKTGKTDAKSGGGGLLGFFKKGKSGNGDEKKSAGKKDGHG 322
Query: 65 DHKEGHH----GEEHKEGFVDK----IKDKIHGEGADGEKKKKKEKKKHGEGHEHGHDSS 116
+K H G +H + K K HG G D ++ K K G G++ H+ S
Sbjct: 323 GNKVKSHGGGGGVQHYDSGPKKGGGGTKGGGHG-GLDIDELMKHSKGGGGGGNKGNHNHS 381
Query: 117 S 117
+
Sbjct: 382 A 382
Score = 24.6 bits (52), Expect = 8.0
Identities = 16/56 (28%), Positives = 21/56 (36%)
Query: 9 GDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHG 64
G H K+G + GH G + K + G GG+ G H G G G
Sbjct: 333 GGVQHYDSGPKKGGGGTKGGGHGGLDIDELMKHSKGGGGGGNKGNHNHSAKGIGGG 388
>At2g16940 splicing factor like protein
Length = 561
Score = 37.7 bits (86), Expect = 0.001
Identities = 27/107 (25%), Positives = 46/107 (42%), Gaps = 7/107 (6%)
Query: 6 NKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGD 65
N++G+ GGD+K + +G++ H G+ K E+ G + H D
Sbjct: 20 NEVGNG---GGDEKV-KSEGKERSR-SSRHRGDKKKERDEDEDGRRSKRSRSHHRSRSRD 74
Query: 66 HKEGHH--GEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHE 110
+ H EH++ +K DK G D E+ + K++ G HE
Sbjct: 75 RERDRHRSSREHRDRDREKDVDKEERNGKDRERDRDKDRDSKGRDHE 121
>At2g05580 unknown protein
Length = 302
Score = 37.7 bits (86), Expect = 0.001
Identities = 30/76 (39%), Positives = 34/76 (44%), Gaps = 14/76 (18%)
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGH------GGE--HKGEQHGFGHGDH 66
GG K G G++ G GG G KG G GGH GG+ HKG G G G H
Sbjct: 178 GGQKGGGGQGGQKGG--GGRGQGGMKGGGGGGQGGHKGGGGGGGQGGHKGGGGGGGQGGH 235
Query: 67 KEGHHGEE----HKEG 78
K G G + HK G
Sbjct: 236 KGGGGGGQGGGGHKGG 251
Score = 37.4 bits (85), Expect = 0.001
Identities = 29/70 (41%), Positives = 33/70 (46%), Gaps = 6/70 (8%)
Query: 15 GGDKKEGEHK----GEQHGHVGGEHHGEYKGEQHGFVGGHGGEHK-GEQHGFGHGDHKEG 69
GG + +G K G Q GH GG G G + G GG G HK G G G G HK G
Sbjct: 192 GGGRGQGGMKGGGGGGQGGHKGGGGGGGQGGHKGGGGGGGQGGHKGGGGGGQGGGGHKGG 251
Query: 70 HHGE-EHKEG 78
G+ HK G
Sbjct: 252 GGGQGGHKGG 261
Score = 35.8 bits (81), Expect = 0.003
Identities = 24/58 (41%), Positives = 25/58 (42%), Gaps = 3/58 (5%)
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHG 72
GG +G HKG G GG H G Q G GG GG H G G G G G G
Sbjct: 227 GGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGHKGGGGGGHVG---GGGRGGGGGGRGG 281
Score = 33.9 bits (76), Expect = 0.013
Identities = 28/66 (42%), Positives = 30/66 (45%), Gaps = 12/66 (18%)
Query: 15 GGDKKEGEHKGE-----QHGHVGGEH---HGEYKGEQHGFVGGHGGEHKGEQHGFGHGDH 66
GG +G HKG Q GH GG G +KG G G GG HKG G G G H
Sbjct: 203 GGGGGQGGHKGGGGGGGQGGHKGGGGGGGQGGHKGGGGG--GQGGGGHKG--GGGGQGGH 258
Query: 67 KEGHHG 72
K G G
Sbjct: 259 KGGGGG 264
Score = 33.9 bits (76), Expect = 0.013
Identities = 24/57 (42%), Positives = 26/57 (45%), Gaps = 4/57 (7%)
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGH--GGEHKGEQHGFGHGDHKEG 69
GG +G HKG G G H G G Q G GGH GG +G G G G H G
Sbjct: 215 GGGGGQGGHKGGGGGGGQGGHKGGGGGGQGG--GGHKGGGGGQGGHKGGGGGGHVGG 269
Score = 32.0 bits (71), Expect = 0.050
Identities = 24/61 (39%), Positives = 25/61 (40%), Gaps = 5/61 (8%)
Query: 15 GGDKKEGEHK---GEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHH 71
GG + G HK G Q GH GG G G G GG GG G G G G G
Sbjct: 240 GGGQGGGGHKGGGGGQGGHKGGGGGGHVGGGGRG--GGGGGRGGGGSGGGGGGGSGRGGG 297
Query: 72 G 72
G
Sbjct: 298 G 298
Score = 31.6 bits (70), Expect = 0.066
Identities = 32/101 (31%), Positives = 42/101 (40%), Gaps = 5/101 (4%)
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEY-KGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGE 73
GG +G KG G GG+ G +G Q G GG GG+ G G G G K G G
Sbjct: 90 GGGGGQGGQKGGGGGGQGGQKGGGGGQGGQKGGGGGQGGQKGG--GGGGQGGQKGGGGGG 147
Query: 74 E--HKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHG 112
+ K G + G G G+K + + G G + G
Sbjct: 148 QGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQGG 188
Score = 31.6 bits (70), Expect = 0.066
Identities = 24/58 (41%), Positives = 26/58 (44%), Gaps = 2/58 (3%)
Query: 9 GDALHIGGDKKEGEHKGEQHG-HVGGEHHGEYKGEQ-HGFVGGHGGEHKGEQHGFGHG 64
G H GG +G HKG G HVGG G G + G GG GG G G G G
Sbjct: 244 GGGGHKGGGGGQGGHKGGGGGGHVGGGGRGGGGGGRGGGGSGGGGGGGSGRGGGGGGG 301
Score = 29.3 bits (64), Expect = 0.33
Identities = 22/66 (33%), Positives = 29/66 (43%), Gaps = 4/66 (6%)
Query: 17 DKKEGEHKGEQHGHVG---GEHHGEYKGEQHGFVGGHGGEHKGEQHGFG-HGDHKEGHHG 72
D +E E G +G G G G Q G GG GG G++ G G G K G G
Sbjct: 66 DGEEEEESKNNQGGIGRGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGQGGQKGGGGG 125
Query: 73 EEHKEG 78
+ ++G
Sbjct: 126 QGGQKG 131
>At1g64370 unknown protein
Length = 178
Score = 37.4 bits (85), Expect = 0.001
Identities = 35/110 (31%), Positives = 49/110 (43%), Gaps = 22/110 (20%)
Query: 28 HGHVGGEHHGEYKGEQHGFVGGHG-----GEHKGEQ------HG--FGHGDHKEGHHGEE 74
H GG G +KG Q+G G G G+HK ++ HG G D K+ H
Sbjct: 75 HRSHGGFLDGLFKG-QNGQKGQSGLGTFLGQHKSQEAKKSQGHGKLLGQHDQKKTHETNS 133
Query: 75 HKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHD---SSSSDSD 121
G I + G ++K + KKK+ +GH G++ SS SDSD
Sbjct: 134 GLNGLGMFINN-----GEKKHRRKSEHKKKNKDGHGSGNESGSSSGSDSD 178
>At5g46730 unknown protein
Length = 268
Score = 37.0 bits (84), Expect = 0.002
Identities = 25/67 (37%), Positives = 29/67 (42%), Gaps = 5/67 (7%)
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHG---FGHGDHKEGHH 71
GG G H G +G GGE G G HG GG+GG G G +G G G +
Sbjct: 176 GGGSAGGAHGGSGYG--GGEGGGAGGGGSHGGAGGYGGGGGGGSGGGGAYGGGGAHGGGY 233
Query: 72 GEEHKEG 78
G EG
Sbjct: 234 GSGGGEG 240
Score = 34.3 bits (77), Expect = 0.010
Identities = 24/66 (36%), Positives = 27/66 (40%), Gaps = 4/66 (6%)
Query: 9 GDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKE 68
G A GG+ G G G GG Y G +G GGHGG G G HG
Sbjct: 132 GSAYGAGGEHASGYGNGAGEG--GGAGASGYGGGAYGGGGGHGGGGGGGSAGGAHGG--S 187
Query: 69 GHHGEE 74
G+ G E
Sbjct: 188 GYGGGE 193
Score = 31.6 bits (70), Expect = 0.066
Identities = 30/106 (28%), Positives = 35/106 (32%), Gaps = 17/106 (16%)
Query: 12 LHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHH 71
L+ G G H G G GG G Y GE G GG GE G +G G G
Sbjct: 28 LNYGQASGGGGHGGG--GGSGGVSSGGYGGESGGGYGGGSGEGAGGGYGGAEGYASGGGS 85
Query: 72 GEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHDSSS 117
G HG G G GEG G+ ++
Sbjct: 86 G---------------HGGGGGGAASSGGYASGAGEGGGGGYGGAA 116
Score = 30.8 bits (68), Expect = 0.11
Identities = 20/48 (41%), Positives = 22/48 (45%), Gaps = 3/48 (6%)
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFG 62
GG G G +G GGE G G G+ GGHGG G HG G
Sbjct: 220 GGAYGGGGAHGGGYGSGGGEGGGYGGGAAGGYGGGHGG---GGGHGGG 264
Score = 30.4 bits (67), Expect = 0.15
Identities = 28/86 (32%), Positives = 30/86 (34%), Gaps = 6/86 (6%)
Query: 9 GDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKE 68
G A GG G G G GGE G Y G GG G +G G G G H
Sbjct: 31 GQASGGGGHGGGGGSGGVSSGGYGGESGGGYGGGSGEGAGGGYGGAEGYASGGGSG-HGG 89
Query: 69 GHHGEEHKEGFVDKIKDKIHGEGADG 94
G G G+ GEG G
Sbjct: 90 GGGGAASSGGYASGA-----GEGGGG 110
Score = 30.4 bits (67), Expect = 0.15
Identities = 25/64 (39%), Positives = 28/64 (43%), Gaps = 5/64 (7%)
Query: 9 GDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKE 68
G A GG G G +G GG H G Y G G GG+GG G G+G G
Sbjct: 204 GGAGGYGGGGGGGSGGGGAYGG-GGAHGGGY-GSGGGEGGGYGGGAAG---GYGGGHGGG 258
Query: 69 GHHG 72
G HG
Sbjct: 259 GGHG 262
Score = 28.9 bits (63), Expect = 0.43
Identities = 22/66 (33%), Positives = 26/66 (39%), Gaps = 10/66 (15%)
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHK--------GEQHGFGHGDH 66
GG G +G G+ G E + G HG GG GG GE G G+G
Sbjct: 58 GGGYGGGSGEGAGGGYGGAEGYASGGGSGHG--GGGGGAASSGGYASGAGEGGGGGYGGA 115
Query: 67 KEGHHG 72
GH G
Sbjct: 116 AGGHAG 121
Score = 28.1 bits (61), Expect = 0.73
Identities = 20/57 (35%), Positives = 25/57 (43%), Gaps = 5/57 (8%)
Query: 16 GDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHG 72
G+ G + G GH GG G G G G GGEH G+G+G + G G
Sbjct: 105 GEGGGGGYGGAAGGHAGGGGGGS--GGGGGSAYGAGGEHAS---GYGNGAGEGGGAG 156
Score = 27.7 bits (60), Expect = 0.95
Identities = 23/63 (36%), Positives = 24/63 (37%), Gaps = 5/63 (7%)
Query: 2 AGIMNKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGF 61
AG G A GG G G G GG H G G G G GG H G G+
Sbjct: 155 AGASGYGGGAYGGGG----GHGGGGGGGSAGGAHGGSGYGGGEGGGAGGGGSH-GGAGGY 209
Query: 62 GHG 64
G G
Sbjct: 210 GGG 212
Score = 26.9 bits (58), Expect = 1.6
Identities = 19/44 (43%), Positives = 20/44 (45%), Gaps = 5/44 (11%)
Query: 9 GDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGG 52
G H GG G GE G+ GG G G HG GGHGG
Sbjct: 225 GGGAHGGG---YGSGGGEGGGYGGGAAGGY--GGGHGGGGGHGG 263
Score = 26.2 bits (56), Expect = 2.8
Identities = 16/48 (33%), Positives = 19/48 (39%)
Query: 32 GGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEGF 79
GG G +G G+ G GG G G G G G EH G+
Sbjct: 98 GGYASGAGEGGGGGYGGAAGGHAGGGGGGSGGGGGSAYGAGGEHASGY 145
>At5g39570 unknown protein
Length = 381
Score = 37.0 bits (84), Expect = 0.002
Identities = 20/89 (22%), Positives = 41/89 (45%), Gaps = 11/89 (12%)
Query: 17 DKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHK 76
+++EG ++ +G + G Y+ + G + + ++ G GD +EG +G
Sbjct: 293 EQEEGSYRKPSYGRSEEQEEGSYRKQPSYGRGNDDDDDEQRRNRSGSGDDEEGSYG---- 348
Query: 77 EGFVDKIKDKIHGEGADGEKKKKKEKKKH 105
+ K G +D +++KKK + KH
Sbjct: 349 -------RKKYGGNDSDEDEEKKKHRHKH 370
>At4g38410 cold-regulated protein - like
Length = 163
Score = 36.2 bits (82), Expect = 0.003
Identities = 20/70 (28%), Positives = 35/70 (49%), Gaps = 11/70 (15%)
Query: 29 GHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEGFVDKIKDKIH 88
GH G+ G + + GGH G+ + E H + + ++GF++KIK+K+
Sbjct: 104 GHEDGKEKGFMEKIKDKLPGGHNGKPEAEPH-----------NDKAKEKGFMEKIKEKLP 152
Query: 89 GEGADGEKKK 98
G D +KK+
Sbjct: 153 GHTNDEKKKE 162
Score = 26.9 bits (58), Expect = 1.6
Identities = 11/17 (64%), Positives = 14/17 (81%)
Query: 91 GADGEKKKKKEKKKHGE 107
G +GEKK+KK+KKK E
Sbjct: 71 GENGEKKEKKKKKKKNE 87
Score = 25.4 bits (54), Expect = 4.7
Identities = 14/49 (28%), Positives = 23/49 (46%), Gaps = 2/49 (4%)
Query: 73 EEHKEGFVDK--IKDKIHGEGADGEKKKKKEKKKHGEGHEHGHDSSSSD 119
EE K ++ + D + GE +KKEKKK + +E D ++
Sbjct: 48 EEEKPSLAERFHLSDSSSSDEEAGENGEKKEKKKKKKKNEVAEDQCETE 96
Score = 25.0 bits (53), Expect = 6.1
Identities = 16/44 (36%), Positives = 23/44 (51%), Gaps = 1/44 (2%)
Query: 79 FVDKIKDKIHG-EGADGEKKKKKEKKKHGEGHEHGHDSSSSDSD 121
F+ K + +H E A K+ K+E+K H DSSSSD +
Sbjct: 26 FLKKKPEDVHSSENARVTKEPKEEEKPSLAERFHLSDSSSSDEE 69
>At3g51810 embryonic abundant protein AtEm1
Length = 152
Score = 35.8 bits (81), Expect = 0.003
Identities = 35/106 (33%), Positives = 48/106 (45%), Gaps = 13/106 (12%)
Query: 16 GDKKEGEHKGEQHGHVG----GEHHGEYKGEQHGFVG----GH-GGEHKGEQHGFGHGDH 66
G K G+ + EQ GH G G GE + EQ G G GH GGE + EQ GH +
Sbjct: 40 GRSKGGQTRKEQLGHEGYQEIGHKGGEARKEQLGHEGYQEMGHKGGEARKEQ--LGHEGY 97
Query: 67 KEGHH--GEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHE 110
+E H GE KE + ++ +G +K ++ EG E
Sbjct: 98 QEMGHKGGEARKEQLGHEGYKEMGRKGGLSTMEKSGGERAEEEGIE 143
>At3g19670 unknown protein
Length = 960
Score = 35.8 bits (81), Expect = 0.003
Identities = 30/120 (25%), Positives = 43/120 (35%), Gaps = 11/120 (9%)
Query: 13 HIGGDKKEGEHKGEQHGHVGGEHH-----GEYK---GEQHGFVGGHGGEHKGEQHGFGHG 64
H G K G K +HH G+Y E HG G +H H
Sbjct: 837 HDKGRDKYGREKDRVRERDSDDHHKKGAAGKYNHDMNEPHGKERRRSGRDSHNRHRERHT 896
Query: 65 DHKEG---HHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHDSSSSDSD 121
KE H E HK G K G ++ E + K+++++ E EH + D +
Sbjct: 897 SVKENDTDHFKESHKAGGGHKKSRHQRGWVSEAEVEGKEKRRRKEEAREHTKEEELEDGE 956
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.307 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,854,255
Number of Sequences: 26719
Number of extensions: 207293
Number of successful extensions: 1888
Number of sequences better than 10.0: 333
Number of HSP's better than 10.0 without gapping: 211
Number of HSP's successfully gapped in prelim test: 128
Number of HSP's that attempted gapping in prelim test: 774
Number of HSP's gapped (non-prelim): 852
length of query: 121
length of database: 11,318,596
effective HSP length: 97
effective length of query: 24
effective length of database: 8,726,853
effective search space: 209444472
effective search space used: 209444472
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC137822.11


