

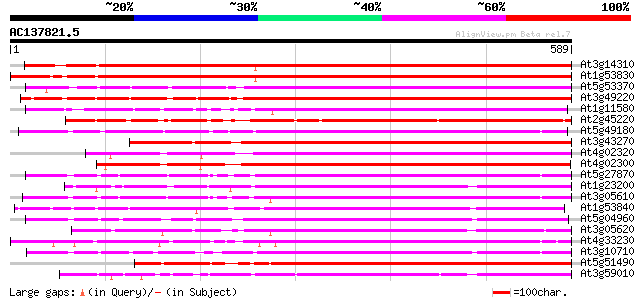
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.5 - phase: 0
(589 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g14310 pectin methylesterase like protein 818 0.0
At1g53830 unknown protein 805 0.0
At5g53370 pectinesterase 458 e-129
At3g49220 pectinesterase - like protein 449 e-126
At1g11580 unknown protein 440 e-124
At2g45220 pectinesterase like protein 426 e-119
At5g49180 pectin methylesterase 419 e-117
At3g43270 pectinesterase like protein 419 e-117
At4g02320 pectinesterase - like protein 414 e-116
At4g02300 putative pectinesterase 407 e-114
At5g27870 pectin methyl-esterase - like protein 404 e-113
At1g23200 putative pectinesterase 404 e-112
At3g05610 putative pectinesterase 399 e-111
At1g53840 unknown protein 393 e-109
At5g04960 pectinesterase 383 e-106
At3g05620 putative pectinesterase 382 e-106
At4g33230 pectinesterase - like protein 380 e-105
At3g10710 putative pectinesterase 379 e-105
At5g51490 pectinesterase 375 e-104
At3g59010 pectinesterase precursor-like protein 374 e-104
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 818 bits (2114), Expect = 0.0
Identities = 416/588 (70%), Positives = 467/588 (78%), Gaps = 26/588 (4%)
Query: 16 NKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTT 75
NKK +L+S LL VA++ I A +K + K +LS SHA+L+S+C++T
Sbjct: 17 NKKLVLLSAAVALLFVAAVAGISAGASKANEKR----------TLSPSSHAVLRSSCSST 66
Query: 76 LYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLL-RKSLTKREK 134
YPELC SA+ + + ++T+ KDVI S+N+T AVEHNYFTV+KL+ RK LT REK
Sbjct: 67 RYPELCISAVVTAGGV--ELTSQKDVIEASVNLTITAVEHNYFTVKKLIKKRKGLTPREK 124
Query: 135 IALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSH 194
ALHDCLETIDETLDEL E DL LYP+KKTL +HA DLKTLISSAITNQ TCLDGFSH
Sbjct: 125 TALHDCLETIDETLDELHETVEDLHLYPTKKTLREHAGDLKTLISSAITNQETCLDGFSH 184
Query: 195 DDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEEN 254
DDADK+VRK L +GQIHVEHMCSNALAM KNMTD DIA FEQ + +N+ K +E
Sbjct: 185 DDADKQVRKALLKGQIHVEHMCSNALAMIKNMTDTDIANFEQKAKITSNNRKLKEENQET 244
Query: 255 GV-------------GWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPL 301
V GWP W+SAGDRRLLQGS VKAD VAADGSG FKTV+ AVAAAP
Sbjct: 245 TVAVDIAGAGELDSEGWPTWLSAGDRRLLQGSGVKADATVAADGSGTFKTVAAAVAAAPE 304
Query: 302 KSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAI 361
S+KRYVI IKAGVY+ENVEV KKK NIMF+GDGRT TIITGSRNVVDGSTTFHSATVA
Sbjct: 305 NSNKRYVIHIKAGVYRENVEVAKKKKNIMFMGDGRTRTIITGSRNVVDGSTTFHSATVAA 364
Query: 362 VGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCF 421
VG FLARDITFQNTAGP+KHQAVALRVG+D SAFYNCD++AYQDTLYVH+NRQFFV C
Sbjct: 365 VGERFLARDITFQNTAGPSKHQAVALRVGSDFSAFYNCDMLAYQDTLYVHSNRQFFVKCL 424
Query: 422 ISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKD 481
I+GTVDFIFGN+AVV Q+CDIHARRPNSGQKNMVTAQGR DPNQNTGIVIQKCRIGAT D
Sbjct: 425 IAGTVDFIFGNAAVVLQDCDIHARRPNSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSD 484
Query: 482 LEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGP 541
L+ VKG+FPTYLGRPWKEYS+TV MQS+ISDVI P GW EW G FALNTL YREY NTG
Sbjct: 485 LQSVKGSFPTYLGRPWKEYSQTVIMQSAISDVIRPEGWSEWTGTFALNTLTYREYSNTGA 544
Query: 542 GAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
GAGT+ RV W+GFKVIT+AAEAQ +T G FIGG WL STGFPFSLGL
Sbjct: 545 GAGTANRVKWRGFKVITAAAEAQKYTAGQFIGGGGWLSSTGFPFSLGL 592
>At1g53830 unknown protein
Length = 587
Score = 805 bits (2078), Expect = 0.0
Identities = 416/596 (69%), Positives = 474/596 (78%), Gaps = 16/596 (2%)
Query: 1 MNKIKQSLAGISNSGNKKKLLISLFT-TLLIVASIVAIVATTTKNSNKSKNNSIASSSLS 59
M IK+ ++ S+ N KKL++S LL++ASIV I ATTT N++KN I +
Sbjct: 1 MAPIKEFISKFSDFKNNKKLILSSAAIALLLLASIVGIAATTT---NQNKNQKITT---- 53
Query: 60 LSHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFT 119
LS SHAILKS C++TLYPELCFSA+++ ++T+ K+VI SLN+TT+AV+HNYF
Sbjct: 54 LSSTSHAILKSVCSSTLYPELCFSAVAATGG--KELTSQKEVIEASLNLTTKAVKHNYFA 111
Query: 120 VEKLLL-RKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLI 178
V+KL+ RK LT RE ALHDCLETIDETLDEL A DL YP +K+L +HADDLKTLI
Sbjct: 112 VKKLIAKRKGLTPREVTALHDCLETIDETLDELHVAVEDLHQYPKQKSLRKHADDLKTLI 171
Query: 179 SSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFE--Q 236
SSAITNQ TCLDGFS+DDAD++VRK L +GQ+HVEHMCSNALAM KNMT+ DIA FE
Sbjct: 172 SSAITNQGTCLDGFSYDDADRKVRKALLKGQVHVEHMCSNALAMIKNMTETDIANFELRD 231
Query: 237 TNMVLGSNKNRKLLEEENGV---GWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVS 293
+ +N NRKL E + GWP+W+S GDRRLLQGST+KAD VA DGSG+F TV+
Sbjct: 232 KSSTFTNNNNRKLKEVTGDLDSDGWPKWLSVGDRRLLQGSTIKADATVADDGSGDFTTVA 291
Query: 294 EAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTT 353
AVAAAP KS+KR+VI IKAGVY+ENVEV KKKTNIMFLGDGR TIITGSRNVVDGSTT
Sbjct: 292 AAVAAAPEKSNKRFVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTT 351
Query: 354 FHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNN 413
FHSATVA VG FLARDITFQNTAGP+KHQAVALRVG+D SAFY CD+ AYQDTLYVH+N
Sbjct: 352 FHSATVAAVGERFLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSN 411
Query: 414 RQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQK 473
RQFFV C I+GTVDFIFGN+A V Q+CDI+ARRPNSGQKNMVTAQGR DPNQNTGIVIQ
Sbjct: 412 RQFFVKCHITGTVDFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQN 471
Query: 474 CRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVY 533
CRIG T DL VKG FPTYLGRPWKEYSRTV MQS ISDVI P GWHEW+G+FAL+TL Y
Sbjct: 472 CRIGGTSDLLAVKGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFALDTLTY 531
Query: 534 REYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
REY N G GAGT+ RV WKG+KVITS EAQ FT G FIGG WL STGFPFSL L
Sbjct: 532 REYLNRGGGAGTANRVKWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFSLSL 587
>At5g53370 pectinesterase
Length = 587
Score = 458 bits (1179), Expect = e-129
Identities = 255/580 (43%), Positives = 345/580 (58%), Gaps = 30/580 (5%)
Query: 17 KKKLLISLFTTLLIVASIVAI---VATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACT 73
K K + LFT ++V +V +S K++ + ++S C+
Sbjct: 29 KTKTKLILFTLAVLVVGVVCFGIFAGIRAVDSGKTEPKLTRKPTQAISR--------TCS 80
Query: 74 TTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTKRE 133
+LYP LC + P + ++I +S N T + +T + + + R
Sbjct: 81 KSLYPNLCIDTLLDFPGSL--TADENELIHISFNATLQKFSKALYTSSTITYTQ-MPPRV 137
Query: 134 KIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFS 193
+ A CLE +D+++D L A + +V+ ++ D+ T +SSA+TN TC DGF
Sbjct: 138 RSAYDSCLELLDDSVDALTRALSSVVVVSGDES----HSDVMTWLSSAMTNHDTCTDGFD 193
Query: 194 HDDADK-EVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEE 252
+ EV+ + + M SN LA+ KD++ N NRKLL
Sbjct: 194 EIEGQGGEVKDQVIGAVKDLSEMVSNCLAIFAGKV-KDLSGVPVVN-------NRKLLGT 245
Query: 253 ENGVGWPEWISAGDRRLL--QGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIK 310
E P W+ DR LL S ++AD+ V+ DGSG FKT++EA+ AP SS+R+VI
Sbjct: 246 EETEELPNWLKREDRELLGTPTSAIQADITVSKDGSGTFKTIAEAIKKAPEHSSRRFVIY 305
Query: 311 IKAGVYKE-NVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLAR 369
+KAG Y+E N++V +KKTN+MF+GDG+ T+ITG +++ D TTFH+AT A G F+ R
Sbjct: 306 VKAGRYEEENLKVGRKKTNLMFIGDGKGKTVITGGKSIADDLTTFHTATFAATGAGFIVR 365
Query: 370 DITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFI 429
D+TF+N AGPAKHQAVALRVG D + Y C+II YQD LYVH+NRQFF C I GTVDFI
Sbjct: 366 DMTFENYAGPAKHQAVALRVGGDHAVVYRCNIIGYQDALYVHSNRQFFRECEIYGTVDFI 425
Query: 430 FGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNF 489
FGN+AV+ Q+C+I+AR+P + QK +TAQ R DPNQNTGI I C++ AT DLE KG++
Sbjct: 426 FGNAAVILQSCNIYARKPMAQQKITITAQNRKDPNQNTGISIHACKLLATPDLEASKGSY 485
Query: 490 PTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRV 549
PTYLGRPWK YSR V+M S + D IDP GW EWNG FAL++L Y EY N G G+G +RV
Sbjct: 486 PTYLGRPWKLYSRVVYMMSDMGDHIDPRGWLEWNGPFALDSLYYGEYMNKGLGSGIGQRV 545
Query: 550 TWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
W G+ VITS EA FT FI GSSWL STG F GL
Sbjct: 546 KWPGYHVITSTVEASKFTVAQFISGSSWLPSTGVSFFSGL 585
>At3g49220 pectinesterase - like protein
Length = 598
Score = 449 bits (1154), Expect = e-126
Identities = 251/582 (43%), Positives = 355/582 (60%), Gaps = 26/582 (4%)
Query: 12 SNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSA 71
+N +KKKL++S + +L ++ I+A S N S+ L+ + A
Sbjct: 37 NNRRSKKKLVVS--SIVLAISLILAAAIFAGVRSRLKLNQSVPG----LARKPSQAISKA 90
Query: 72 CTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTK 131
C T +PELC ++ P + ++ KD+I +++N+T H ++ L +
Sbjct: 91 CELTRFPELCVDSLMDFPG-SLAASSSKDLIHVTVNMTLHHFSHALYSSASLSF-VDMPP 148
Query: 132 REKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDG 191
R + A C+E +D+++D L A + +V +K D+ T +S+A+TN TC +G
Sbjct: 149 RARSAYDSCVELLDDSVDALSRALSSVVSSSAKP------QDVTTWLSAALTNHDTCTEG 202
Query: 192 FSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLE 251
F D D V+ + ++ + SN LA+ D D +F + +NR+LL
Sbjct: 203 FDGVD-DGGVKDHMTAALQNLSELVSNCLAIFSASHDGD--DFAGVPI-----QNRRLLG 254
Query: 252 -EENGVGWPEWISAGDRRLLQG--STVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYV 308
EE +P W+ +R +L+ S ++AD++V+ DG+G KT+SEA+ AP S++R +
Sbjct: 255 VEEREEKFPRWMRPKEREILEMPVSQIQADIIVSKDGNGTCKTISEAIKKAPQNSTRRII 314
Query: 309 IKIKAGVYKEN-VEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFL 367
I +KAG Y+EN ++V +KK N+MF+GDG+ T+I+G +++ D TTFH+A+ A G F+
Sbjct: 315 IYVKAGRYEENNLKVGRKKINLMFVGDGKGKTVISGGKSIFDNITTFHTASFAATGAGFI 374
Query: 368 ARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVD 427
ARDITF+N AGPAKHQAVALR+GAD + Y C+II YQDTLYVH+NRQFF C I GTVD
Sbjct: 375 ARDITFENWAGPAKHQAVALRIGADHAVIYRCNIIGYQDTLYVHSNRQFFRECDIYGTVD 434
Query: 428 FIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKG 487
FIFGN+AVV QNC I+AR+P QKN +TAQ R DPNQNTGI I R+ A DL+ G
Sbjct: 435 FIFGNAAVVLQNCSIYARKPMDFQKNTITAQNRKDPNQNTGISIHASRVLAASDLQATNG 494
Query: 488 NFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSK 547
+ TYLGRPWK +SRTV+M S I + GW EWN FAL+TL Y EY N+GPG+G +
Sbjct: 495 STQTYLGRPWKLFSRTVYMMSYIGGHVHTRGWLEWNTTFALDTLYYGEYLNSGPGSGLGQ 554
Query: 548 RVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
RV+W G++VI S AEA FT FI GSSWL STG F GL
Sbjct: 555 RVSWPGYRVINSTAEANRFTVAEFIYGSSWLPSTGVSFLAGL 596
>At1g11580 unknown protein
Length = 557
Score = 440 bits (1132), Expect = e-124
Identities = 257/573 (44%), Positives = 344/573 (59%), Gaps = 39/573 (6%)
Query: 17 KKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTL 76
K K L + + + I+ S+ A + + ++S+ ++S + H +H
Sbjct: 16 KHKNLCLVLSFVAILGSVAFFTAQLISVNTNNNDDSLLTTS-QICHGAHD---------- 64
Query: 77 YPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTKREKIA 136
+ C + +S ++ N D++ + L + +E V + +R + R+K
Sbjct: 65 -QDSCQALLSEFTTLSLSKLNRLDLLHVFLKNSVWRLESTMTMVSEARIRSNGV-RDKAG 122
Query: 137 LHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHDD 196
DC E +D + D + + +L + ++ T +SS +TN +TCL+ S D
Sbjct: 123 FADCEEMMDVSKDRMMSSMEEL---RGGNYNLESYSNVHTWLSSVLTNYMTCLESIS--D 177
Query: 197 ADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEENGV 256
+++++ +E + S A +A F VL + + K++
Sbjct: 178 VSVNSKQIVKP---QLEDLVSRARVA--------LAIFVS---VLPARDDLKMIISNR-- 221
Query: 257 GWPEWISAGDRRLLQGST----VKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIK 312
+P W++A DR+LL+ S V A+VVVA DG+G FKTV+EAVAAAP S+ RYVI +K
Sbjct: 222 -FPSWLTALDRKLLESSPKTLKVTANVVVAKDGTGKFKTVNEAVAAAPENSNTRYVIYVK 280
Query: 313 AGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDIT 372
GVYKE +++ KKK N+M +GDG+ TIITGS NV+DGSTTF SATVA G F+A+DI
Sbjct: 281 KGVYKETIDIGKKKKNLMLVGDGKDATIITGSLNVIDGSTTFRSATVAANGDGFMAQDIW 340
Query: 373 FQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGN 432
FQNTAGPAKHQAVALRV AD + C I AYQDTLY H RQF+ + +I+GTVDFIFGN
Sbjct: 341 FQNTAGPAKHQAVALRVSADQTVINRCRIDAYQDTLYTHTLRQFYRDSYITGTVDFIFGN 400
Query: 433 SAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTY 492
SAVVFQNCDI AR P +GQKNM+TAQGR D NQNT I IQKC+I A+ DL VKG+ T+
Sbjct: 401 SAVVFQNCDIVARNPGAGQKNMLTAQGREDQNQNTAISIQKCKITASSDLAPVKGSVKTF 460
Query: 493 LGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWK 552
LGRPWK YSRTV MQS I + IDP GW W+G FAL+TL Y EY NTGPGA TSKRV WK
Sbjct: 461 LGRPWKLYSRTVIMQSFIDNHIDPAGWFPWDGEFALSTLYYGEYANTGPGADTSKRVNWK 520
Query: 553 GFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPF 585
GFKVI + EA+ FT I G WL TG F
Sbjct: 521 GFKVIKDSKEAEQFTVAKLIQGGLWLKPTGVTF 553
>At2g45220 pectinesterase like protein
Length = 511
Score = 426 bits (1096), Expect = e-119
Identities = 247/534 (46%), Positives = 329/534 (61%), Gaps = 46/534 (8%)
Query: 59 SLSHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITT-RAV--EH 115
++S ++ +K+ C+ T P+ C ++ N I + + + +S+ + RA+ +
Sbjct: 21 TVSGYNQKDVKAWCSQTPNPKPCEYFLTHNSN-NEPIKSESEFLKISMKLVLDRAILAKT 79
Query: 116 NYFTVEKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLK 175
+ FT+ K REK A DC++ D T+ ++ E + P+ K D +
Sbjct: 80 HAFTLGP----KCRDTREKAAWEDCIKLYDLTVSKINETMD-----PNVKC---SKLDAQ 127
Query: 176 TLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFE 235
T +S+A+TN TC GF V ++ +V ++ N LA+ K F
Sbjct: 128 TWLSTALTNLDTCRAGFLELGVTDIVLPLMSN---NVSNLLCNTLAINK-------VPFN 177
Query: 236 QTNMVLGSNKNRKLLEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEA 295
T G+P W+ GDR+LLQ ST K + VVA DGSGNFKT+ EA
Sbjct: 178 YT--------------PPEKDGFPSWVKPGDRKLLQSSTPKDNAVVAKDGSGNFKTIKEA 223
Query: 296 VAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFH 355
+ AA S R+VI +K GVY EN+E+ KK N+M GDG TIITGS++V G+TTF+
Sbjct: 224 IDAA--SGSGRFVIYVKQGVYSENLEIRKK--NVMLRGDGIGKTIITGSKSVGGGTTTFN 279
Query: 356 SATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQ 415
SATVA VG F+AR ITF+NTAG + QAVALR G+DLS FY C AYQDTLYVH+NRQ
Sbjct: 280 SATVAAVGDGFIARGITFRNTAGASNEQAVALRSGSDLSVFYQCSFEAYQDTLYVHSNRQ 339
Query: 416 FFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCR 475
F+ +C + GTVDFIFGN+A V QNC+I ARRP S + N +TAQGR DPNQNTGI+I R
Sbjct: 340 FYRDCDVYGTVDFIFGNAAAVLQNCNIFARRPRS-KTNTITAQGRSDPNQNTGIIIHNSR 398
Query: 476 IGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYRE 535
+ A DL V G+ TYLGRPW++YSRTVFM++S+ +IDP GW EW+GNFAL TL Y E
Sbjct: 399 VTAASDLRPVLGSTKTYLGRPWRQYSRTVFMKTSLDSLIDPRGWLEWDGNFALKTLFYAE 458
Query: 536 YQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
+QNTGPGA TS RVTW GF+V+ SA+EA FT G F+ G SW+ S+ PF+ GL
Sbjct: 459 FQNTGPGASTSGRVTWPGFRVLGSASEASKFTVGTFLAGGSWIPSS-VPFTSGL 511
>At5g49180 pectin methylesterase
Length = 571
Score = 419 bits (1076), Expect = e-117
Identities = 241/583 (41%), Positives = 349/583 (59%), Gaps = 25/583 (4%)
Query: 10 GISNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILK 69
G+ KKK +I+ T L+V +VA+ TT++N++ S+ + + ++
Sbjct: 2 GVDGELKKKKCIIAGVITALLVLMVVAVGITTSRNTSHSEKIVPVQIKTATT-----AVE 56
Query: 70 SACTTTLYPELCFSAI-SSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKS 128
+ C T Y E C +++ + P+ T + D+I L N+T R++E + L K+
Sbjct: 57 AVCAPTDYKETCVNSLMKASPDSTQPL----DLIKLGFNVTIRSIEDSIKKASVELTAKA 112
Query: 129 LTKRE-KIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVT 187
++ K AL C + +++ D+LK+ ++ + S + +DL+ +S +I Q T
Sbjct: 113 ANDKDTKGALELCEKLMNDATDDLKKCLDNFDGF-SIPQIEDFVEDLRVWLSGSIAYQQT 171
Query: 188 CLDGFSHDDA--DKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNK 245
C+D F ++ ++++K+ + + + SN LAM N+++ + EF T + K
Sbjct: 172 CMDTFEETNSKLSQDMQKIFKTSR----ELTSNGLAMITNISNL-LGEFNVTGVTGDLGK 226
Query: 246 N-RKLLEEENGVGWPEWISAGDRRLLQGST-VKADVVVAADGSGNFKTVSEAVAAAPLKS 303
RKLL E+G+ P W+ RRL+ VKA+VVVA DGSG +KT++EA+ A P +
Sbjct: 227 YARKLLSAEDGI--PSWVGPNTRRLMATKGGVKANVVVAHDGSGQYKTINEALNAVPKAN 284
Query: 304 SKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGST-TFHSATVAIV 362
K +VI IK GVY E V+V KK T++ F+GDG T T ITGS N G T+ +ATVAI
Sbjct: 285 QKPFVIYIKQGVYNEKVDVTKKMTHVTFIGDGPTKTKITGSLNYYIGKVKTYLTATVAIN 344
Query: 363 GGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFI 422
G NF A++I F+NTAGP HQAVALRV ADL+ FYNC I YQDTLYVH++RQFF +C +
Sbjct: 345 GDNFTAKNIGFENTAGPEGHQAVALRVSADLAVFYNCQIDGYQDTLYVHSHRQFFRDCTV 404
Query: 423 SGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDL 482
SGTVDFIFG+ VV QNC+I R+P Q M+TAQGR D ++TG+V+Q C I
Sbjct: 405 SGTVDFIFGDGIVVLQNCNIVVRKPMKSQSCMITAQGRSDKRESTGLVLQNCHITGEPAY 464
Query: 483 EGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPG 542
VK YLGRPWKE+SRT+ M ++I DVIDP GW WNG+FALNTL Y EY+N GPG
Sbjct: 465 IPVKSINKAYLGRPWKEFSRTIIMGTTIDDVIDPAGWLPWNGDFALNTLYYAEYENNGPG 524
Query: 543 AGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPF 585
+ ++RV W G K + S +A FTP F+ G+ W+ P+
Sbjct: 525 SNQAQRVKWPGIKKL-SPKQALRFTPARFLRGNLWIPPNRVPY 566
>At3g43270 pectinesterase like protein
Length = 527
Score = 419 bits (1076), Expect = e-117
Identities = 228/467 (48%), Positives = 293/467 (61%), Gaps = 14/467 (2%)
Query: 126 RKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSK-KTLYQHADDLKTLISSAITN 184
+K+ R A+ DC++ +D +EL + K + DL+T IS+A++N
Sbjct: 70 KKAGKSRVSNAIVDCVDLLDSAAEELSWIISASQSPNGKDNSTGDVGSDLRTWISAALSN 129
Query: 185 QVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSN 244
Q TCLDGF + + ++K++ G V N L M + K +
Sbjct: 130 QDTCLDGF--EGTNGIIKKIVAGGLSKVGTTVRNLLTMVHSPPSKPKPK---------PI 178
Query: 245 KNRKLLEEENGVG-WPEWISAGDRRLLQGSTVK-ADVVVAADGSGNFKTVSEAVAAAPLK 302
K + + + +G +P W+ GDR+LLQ + AD VVAADG+GNF T+S+AV AAP
Sbjct: 179 KAQTMTKAHSGFSKFPSWVKPGDRKLLQTDNITVADAVVAADGTGNFTTISDAVLAAPDY 238
Query: 303 SSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIV 362
S+KRYVI +K GVY ENVE+ KKK NIM +GDG T+ITG+R+ +DG TTF SAT A+
Sbjct: 239 STKRYVIHVKRGVYVENVEIKKKKWNIMMVGDGIDATVITGNRSFIDGWTTFRSATFAVS 298
Query: 363 GGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFI 422
G F+ARDITFQNTAGP KHQAVA+R DL FY C + YQDTLY H+ RQFF C I
Sbjct: 299 GRGFIARDITFQNTAGPEKHQAVAIRSDTDLGVFYRCAMRGYQDTLYAHSMRQFFRECII 358
Query: 423 SGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDL 482
+GTVDFIFG++ VFQ+C I A++ QKN +TAQGR DPN+ TG IQ I A DL
Sbjct: 359 TGTVDFIFGDATAVFQSCQIKAKQGLPNQKNSITAQGRKDPNEPTGFTIQFSNIAADTDL 418
Query: 483 EGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPG 542
TYLGRPWK YSRTVFMQ+ +SD I+PVGW EWNGNFAL+TL Y EY N+GPG
Sbjct: 419 LLNLNTTATYLGRPWKLYSRTVFMQNYMSDAINPVGWLEWNGNFALDTLYYGEYMNSGPG 478
Query: 543 AGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
A +RV W G+ V+ ++AEA +FT I G+ WL STG F GL
Sbjct: 479 ASLDRRVKWPGYHVLNTSAEANNFTVSQLIQGNLWLPSTGITFIAGL 525
>At4g02320 pectinesterase - like protein
Length = 518
Score = 414 bits (1065), Expect = e-116
Identities = 228/527 (43%), Positives = 317/527 (59%), Gaps = 41/527 (7%)
Query: 80 LCFSAISSEPNITHKITNHKDVISL---SLNITTRAVEHNYFTVEKLLLR--KSLTKREK 134
L F + ++T + T+ D+ L +LN T V + LL R +L+ R+
Sbjct: 16 LLFLSTVFSSHVTQRFTSSDDITELVVTALNQTISNVNLSSSNFSDLLQRLGSNLSHRDL 75
Query: 135 IALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSH 194
A DCLE +D+T+ +L A + L ++ ++K L+S+A+TN TCLDGF+
Sbjct: 76 CAFDDCLELLDDTVFDLTTAISKL------RSHSPELHNVKMLLSAAMTNTRTCLDGFAS 129
Query: 195 DDADKE----------VRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSN 244
D D+ V + L+E ++ S++LAM +N+ + ++
Sbjct: 130 SDNDENLNNNDNKTYGVAESLKESLFNISSHVSDSLAMLENIPGHIPGKVKED------- 182
Query: 245 KNRKLLEEENGVGWPEWISAGDRRLLQGST--VKADVVVAADGSGNFKTVSEAVAAAPLK 302
VG+P W+S DR LLQ K ++VVA +G+GN+ T+ EA++AAP
Sbjct: 183 -----------VGFPMWVSGSDRNLLQDPVDETKVNLVVAQNGTGNYTTIGEAISAAPNS 231
Query: 303 SSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIV 362
S R+VI IK G Y EN+E+P++KT IMF+GDG T+I +R+ DG T FHSATV +
Sbjct: 232 SETRFVIYIKCGEYFENIEIPREKTMIMFIGDGIGRTVIKANRSYADGWTAFHSATVGVR 291
Query: 363 GGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFI 422
G F+A+D++F N AGP KHQAVALR +DLSA+Y C +YQDT+YVH+++QF+ C I
Sbjct: 292 GSGFIAKDLSFVNYAGPEKHQAVALRSSSDLSAYYRCSFESYQDTIYVHSHKQFYRECDI 351
Query: 423 SGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDL 482
GTVDFIFG+++VVFQNC ++ARRPN QK + TAQGR + + TGI I RI A DL
Sbjct: 352 YGTVDFIFGDASVVFQNCSLYARRPNPNQKIIYTAQGRENSREPTGISIISSRILAAPDL 411
Query: 483 EGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPG 542
V+ NF YLGRPW+ YSRTV M+S I D++DP GW +W +FAL TL Y EY N GPG
Sbjct: 412 IPVQANFKAYLGRPWQLYSRTVIMKSFIDDLVDPAGWLKWKDDFALETLYYGEYMNEGPG 471
Query: 543 AGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
+ + RV W GFK I + EA F+ G FI G+ WL ST PF+L L
Sbjct: 472 SNMTNRVQWPGFKRIETVEEASQFSVGPFIDGNKWLNSTRIPFTLDL 518
>At4g02300 putative pectinesterase
Length = 532
Score = 407 bits (1047), Expect = e-114
Identities = 230/510 (45%), Positives = 312/510 (61%), Gaps = 35/510 (6%)
Query: 92 THKITNHK---DVISLSLNITTRAVE--HNYFTVEKLLLRKSLTKREKIALHDCLETIDE 146
T ++T K ++I LN+T V + F+ + L +LT E+ A DCL +D+
Sbjct: 44 TQRLTETKTIPELIIADLNLTILKVNLASSNFSDLQTRLFPNLTHYERCAFEDCLGLLDD 103
Query: 147 TLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHDDADK------E 200
T+ +L+ A +DL ++ +D+ L+++ +T Q TCLDGFS D + E
Sbjct: 104 TISDLETAVSDL------RSSSLEFNDISMLLTNVMTYQDTCLDGFSTSDNENNNDMTYE 157
Query: 201 VRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEENGVGWPE 260
+ + L+E + + + SN+L M + + S K E V +P
Sbjct: 158 LPENLKEIILDISNNLSNSLHMLQVI----------------SRKKPSPKSSEVDVEYPS 201
Query: 261 WISAGDRRLLQGSTVKAD--VVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYKE 318
W+S D+RLL+ + + + VA DG+GNF T+++AV AAP S R++I IK G Y E
Sbjct: 202 WLSENDQRLLEAPVQETNYNLSVAIDGTGNFTTINDAVFAAPNMSETRFIIYIKGGEYFE 261
Query: 319 NVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAG 378
NVE+PKKKT IMF+GDG T+I +R+ +DG +TF + TV + G ++A+DI+F N+AG
Sbjct: 262 NVELPKKKTMIMFIGDGIGKTVIKANRSRIDGWSTFQTPTVGVKGKGYIAKDISFVNSAG 321
Query: 379 PAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQ 438
PAK QAVA R G+D SAFY C+ YQDTLYVH+ +QF+ C I GT+DFIFGN+AVVFQ
Sbjct: 322 PAKAQAVAFRSGSDHSAFYRCEFDGYQDTLYVHSAKQFYRECDIYGTIDFIFGNAAVVFQ 381
Query: 439 NCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWK 498
N ++AR+PN G K TAQ R +Q TGI I CRI A DL VK NF YLGRPW+
Sbjct: 382 NSSLYARKPNPGHKIAFTAQSRNQSDQPTGISILNCRILAAPDLIPVKENFKAYLGRPWR 441
Query: 499 EYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVIT 558
+YSRTV ++S I D+I P GW E +FAL TL Y EY N GPGA +KRVTW GF+ I
Sbjct: 442 KYSRTVIIKSFIDDLIHPAGWLEGKKDFALETLYYGEYMNEGPGANMAKRVTWPGFRRIE 501
Query: 559 SAAEAQSFTPGNFIGGSSWLGSTGFPFSLG 588
+ EA FT G FI GS+WL STG PFSLG
Sbjct: 502 NQTEATQFTVGPFIDGSTWLNSTGIPFSLG 531
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 404 bits (1039), Expect = e-113
Identities = 225/577 (38%), Positives = 338/577 (57%), Gaps = 31/577 (5%)
Query: 17 KKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTL 76
KK+ +I +++L+++ +VA+ + N + + + ++S+ +K C T
Sbjct: 13 KKRYVIISISSVLLISMVVAVTIGVSVNKSDNAGDEEITTSVKA-------IKDVCAPTD 65
Query: 77 YPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTKREKIA 136
Y E C + + T ++ +++ + N T + + + + ++ R K+A
Sbjct: 66 YKETCEDTLRKDAKDT---SDPLELVKTAFNATMKQIS-DVAKKSQTMIELQKDPRAKMA 121
Query: 137 LHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGF--SH 194
L C E +D + EL ++ +L + K + + L+ +S+ I+++ TCLDGF +
Sbjct: 122 LDQCKELMDYAIGELSKSFEELGKFEFHK-VDEALVKLRIWLSATISHEQTCLDGFQGTQ 180
Query: 195 DDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEEN 254
+A + ++K L+ + + H N LAM M++ M + +R+LL +E
Sbjct: 181 GNAGETIKKALKTA-VQLTH---NGLAMVTEMSNY------LGQMQIPEMNSRRLLSQE- 229
Query: 255 GVGWPEWISAGDRRLLQG--STVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIK 312
+P W+ A RRLL S VK D+VVA DGSG +KT++EA+ P K + +V+ IK
Sbjct: 230 ---FPSWMDARARRLLNAPMSEVKPDIVVAQDGSGQYKTINEALNFVPKKKNTTFVVHIK 286
Query: 313 AGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDIT 372
G+YKE V+V + T+++F+GDG T+I+GS++ DG TT+ +ATVAIVG +F+A++I
Sbjct: 287 EGIYKEYVQVNRSMTHLVFIGDGPDKTVISGSKSYKDGITTYKTATVAIVGDHFIAKNIA 346
Query: 373 FQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGN 432
F+NTAG KHQAVA+RV AD S FYNC YQDTLY H++RQF+ +C ISGT+DF+FG+
Sbjct: 347 FENTAGAIKHQAVAIRVLADESIFYNCKFDGYQDTLYAHSHRQFYRDCTISGTIDFLFGD 406
Query: 433 SAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTY 492
+A VFQNC + R+P Q +TA GR DP ++TG V+Q C I D VK TY
Sbjct: 407 AAAVFQNCTLLVRKPLLNQACPITAHGRKDPRESTGFVLQGCTIVGEPDYLAVKEQSKTY 466
Query: 493 LGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWK 552
LGRPWKEYSRT+ M + I D + P GW W G F LNTL Y E QNTGPGA +KRVTW
Sbjct: 467 LGRPWKEYSRTIIMNTFIPDFVPPEGWQPWLGEFGLNTLFYSEVQNTGPGAAITKRVTWP 526
Query: 553 GFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
G K + S E FTP +I G +W+ G P+ LGL
Sbjct: 527 GIKKL-SDEEILKFTPAQYIQGDAWIPGKGVPYILGL 562
>At1g23200 putative pectinesterase
Length = 554
Score = 404 bits (1037), Expect = e-112
Identities = 236/556 (42%), Positives = 332/556 (59%), Gaps = 50/556 (8%)
Query: 58 LSLSH-HSHAILKSACTTTLYPELCFSAISSEP-----NITHKITNHKDVISLSLNITTR 111
+S+SH ++H I ++C T YP +C +S+ P + T T H V+S +++ +
Sbjct: 25 VSISHLNAHFI--TSCKQTPYPSVCDHHMSNSPLKTLDDQTDGFTFHDLVVSSTMD---Q 79
Query: 112 AVEHNYFTVEKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHA 171
AV+ + V L SL K AL DCLE ++T+D+L ++ Y S
Sbjct: 80 AVQLHRL-VSSLKQHHSLHKHATSALFDCLELYEDTIDQLNHSRRSYGQYSSPH------ 132
Query: 172 DDLKTLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIH--VEHMCSNALAMTKNMTDK 229
D +T +S+AI NQ TC +GF K Q H + SN+LA+TK +
Sbjct: 133 -DRQTSLSAAIANQDTCRNGFRDFKLTSSYSKYFPV-QFHRNLTKSISNSLAVTKAAAEA 190
Query: 230 D----------IAEFEQTNMVLGSNKNRKLL--EEENGVGWPEWISAGDRRLLQGS--TV 275
+ +F + G +R+LL +E +P W DR+LL+ S T
Sbjct: 191 EAVAEKYPSTGFTKFSKQRSSAGGGSHRRLLLFSDEK---FPSWFPLSDRKLLEDSKTTA 247
Query: 276 KADVVVAADGSGNFKTVSEAV-AAAPL-KSSKRYVIKIKAGVYKENVEVPKKKTNIMFLG 333
KAD+VVA DGSG++ ++ +AV AAA L + ++R VI +KAGVY+ENV + K N+M +G
Sbjct: 248 KADLVVAKDGSGHYTSIQQAVNAAAKLPRRNQRLVIYVKAGVYRENVVIKKSIKNVMVIG 307
Query: 334 DGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADL 393
DG +TI+TG+RNV DG+TTF SAT A+ G F+A+ ITF+NTAGP KHQAVALR +D
Sbjct: 308 DGIDSTIVTGNRNVQDGTTTFRSATFAVSGNGFIAQGITFENTAGPEKHQAVALRSSSDF 367
Query: 394 SAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKN 453
S FY C YQDTLY+H++RQF NC I GTVDFIFG++ + QNC+I+AR+P SGQKN
Sbjct: 368 SVFYACSFKGYQDTLYLHSSRQFLRNCNIYGTVDFIFGDATAILQNCNIYARKPMSGQKN 427
Query: 454 MVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDV 513
+TAQ R +P++ TG VIQ + + TYLGRPW+ +SRTVFM+ ++ +
Sbjct: 428 TITAQSRKEPDETTGFVIQSSTVATASE---------TYLGRPWRSHSRTVFMKCNLGAL 478
Query: 514 IDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIG 573
+ P GW W+G+FAL+TL Y EY NTG GA S RV W G+ VI + EA+ FT NF+
Sbjct: 479 VSPAGWLPWSGSFALSTLYYGEYGNTGAGASVSGRVKWPGYHVIKTVTEAEKFTVENFLD 538
Query: 574 GSSWLGSTGFPFSLGL 589
G+ W+ +TG P + GL
Sbjct: 539 GNYWITATGVPVNDGL 554
>At3g05610 putative pectinesterase
Length = 669
Score = 399 bits (1024), Expect = e-111
Identities = 222/582 (38%), Positives = 334/582 (57%), Gaps = 31/582 (5%)
Query: 14 SGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACT 73
S K++ ++ +++L+++ +VA+ + N + + A + S+ +K C
Sbjct: 9 SKRKRRYIVITISSVLLISMVVAVTVGVSLNKHDGDSKGKAEVNASVK-----AVKDVCA 63
Query: 74 TTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTKRE 133
T Y + C + T T+ +++ + N+T + + + + ++ R
Sbjct: 64 PTDYRKTCEDTLIKNGKNT---TDPMELVKTAFNVTMKQIT-DAAKKSQTIMELQKDSRT 119
Query: 134 KIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGF- 192
++AL C E +D LDEL + +L + L + +L+ +S+AI+++ TCL+GF
Sbjct: 120 RMALDQCKELMDYALDELSNSFEELGKFEFH-LLDEALINLRIWLSAAISHEETCLEGFQ 178
Query: 193 -SHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLE 251
+ +A + ++K L+ I + H N LA+ M++ F + G N R L E
Sbjct: 179 GTQGNAGETMKKALKTA-IELTH---NGLAIISEMSN-----FVGQMQIPGLNSRRLLAE 229
Query: 252 EENGVGWPEWISAGDRRLLQG----STVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRY 307
G+P W+ R+LLQ S VK D+VVA DGSG +KT++EA+ P K + +
Sbjct: 230 -----GFPSWVDQRGRKLLQAAAAYSDVKPDIVVAQDGSGQYKTINEALQFVPKKRNTTF 284
Query: 308 VIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFL 367
V+ IKAG+YKE V+V K ++++F+GDG TII+G++N DG TT+ +ATVAIVG F+
Sbjct: 285 VVHIKAGLYKEYVQVNKTMSHLVFIGDGPDKTIISGNKNYKDGITTYRTATVAIVGNYFI 344
Query: 368 ARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVD 427
A++I F+NTAG KHQAVA+RV +D S F+NC YQDTLY H++RQFF +C ISGT+D
Sbjct: 345 AKNIGFENTAGAIKHQAVAVRVQSDESIFFNCRFDGYQDTLYTHSHRQFFRDCTISGTID 404
Query: 428 FIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKG 487
F+FG++A VFQNC + R+P Q +TA GR DP ++TG V Q C I D VK
Sbjct: 405 FLFGDAAAVFQNCTLLVRKPLPNQACPITAHGRKDPRESTGFVFQGCTIAGEPDYLAVKE 464
Query: 488 NFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSK 547
YLGRPWKEYSRT+ M + I D + P GW W G+F L TL Y E QNTGPG+ +
Sbjct: 465 TSKAYLGRPWKEYSRTIIMNTFIPDFVQPQGWQPWLGDFGLKTLFYSEVQNTGPGSALAN 524
Query: 548 RVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
RVTW G K + S + FTP +I G W+ G P++ GL
Sbjct: 525 RVTWAGIKTL-SEEDILKFTPAQYIQGDDWIPGKGVPYTTGL 565
>At1g53840 unknown protein
Length = 586
Score = 393 bits (1009), Expect = e-109
Identities = 232/587 (39%), Positives = 342/587 (57%), Gaps = 33/587 (5%)
Query: 6 QSLAGISNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSH 65
Q LA + K+ LL+S+ +LI I A+VAT +K+KN S S L+ +
Sbjct: 17 QDLA-LKKKTRKRLLLLSISVVVLIAVIIAAVVATVV---HKNKNESTPSPPPELTPSTS 72
Query: 66 AILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLL 125
LK+ C+ T +PE C S+IS P+ T+ + + LSL + ++ EKL
Sbjct: 73 --LKAICSVTRFPESCISSISKLPS--SNTTDPETLFKLSLKVIIDELDSISDLPEKLS- 127
Query: 126 RKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHA-DDLKTLISSAITN 184
+++ +R K AL C + I++ LD L + + + KKTL +DLKT +S+ +T+
Sbjct: 128 KETEDERIKSALRVCGDLIEDALDRLNDTVSAIDDEEKKKTLSSSKIEDLKTWLSATVTD 187
Query: 185 QVTCLDGFSH------DDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTN 238
TC D + A+ + + L+ SN+LA+ + ++
Sbjct: 188 HETCFDSLDELKQNKTEYANSTITQNLKSAMSRSTEFTSNSLAIVSKILSA------LSD 241
Query: 239 MVLGSNKNRKLLEE--ENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAV 296
+ + ++ R+L+ + V + +W RRLLQ + +K DV VA DG+G+ TV+EAV
Sbjct: 242 LGIPIHRRRRLMSHHHQQSVDFEKWAR---RRLLQTAGLKPDVTVAGDGTGDVLTVNEAV 298
Query: 297 AAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHS 356
A P KS K +VI +K+G Y ENV + K K N+M GDG+ TII+GS+N VDG+ T+ +
Sbjct: 299 AKVPKKSLKMFVIYVKSGTYVENVVMDKSKWNVMIYGDGKGKTIISGSKNFVDGTPTYET 358
Query: 357 ATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQF 416
AT AI G F+ +DI NTAG AKHQAVA R G+D S +Y C +QDTLY H+NRQF
Sbjct: 359 ATFAIQGKGFIMKDIGIINTAGAAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQF 418
Query: 417 FVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRI 476
+ +C ++GT+DFIFG++AVVFQ C I R+P S Q N +TAQG+ DPNQ++G+ IQ+C I
Sbjct: 419 YRDCDVTGTIDFIFGSAAVVFQGCKIMPRQPLSNQFNTITAQGKKDPNQSSGMSIQRCTI 478
Query: 477 GATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEW-NGNFALNTLVYRE 535
A ++ PTYLGRPWKE+S TV M++ I V+ P GW W +G ++VY E
Sbjct: 479 SANGNVIA-----PTYLGRPWKEFSTTVIMETVIGAVVRPSGWMSWVSGVDPPASIVYGE 533
Query: 536 YQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTG 582
Y+NTGPG+ ++RV W G+K + S AEA FT + G+ W+ +TG
Sbjct: 534 YKNTGPGSDVTQRVKWAGYKPVMSDAEAAKFTVATLLHGADWIPATG 580
>At5g04960 pectinesterase
Length = 564
Score = 383 bits (983), Expect = e-106
Identities = 225/576 (39%), Positives = 329/576 (57%), Gaps = 42/576 (7%)
Query: 17 KKKLLISLFTTLLIVASIV-AIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTT 75
KK++ I +++++V +V A+V TT ++++K +S+S +K+ C T
Sbjct: 22 KKRIAIIAISSIVLVCIVVGAVVGTTARDNSKKPPTENNGEPISVS------VKALCDVT 75
Query: 76 LYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTKREKI 135
L+ E CF + S PN + ++ +++ ++ +T + ++ + +
Sbjct: 76 LHKEKCFETLGSAPNASR--SSPEELFKYAVKVTITELSK---VLDGFSNGEHMDNATSA 130
Query: 136 ALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHD 195
A+ C+E I +D+L E + ++ DDL+T +SS T Q TC+D
Sbjct: 131 AMGACVELIGLAVDQLNETMTSSL---------KNFDDLRTWLSSVGTYQETCMDALV-- 179
Query: 196 DADKEVRKVLQEGQI-HVEHMCSNALAMTKNMTD-KDIAEFEQTNMVLGSNKNRKLLEEE 253
+A+K E + + M SNALA+ + D +F + R+LLE
Sbjct: 180 EANKPSLTTFGENHLKNSTEMTSNALAIITWLGKIADTVKF----------RRRRLLETG 229
Query: 254 NG-VGWPEWISAGDRRLLQGSTVK--ADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIK 310
N V + RRLL+ +K A +VVA DGSG ++T+ EA+A K+ K +I
Sbjct: 230 NAKVVVADLPMMEGRRLLESGDLKKKATIVVAKDGSGKYRTIGEALAEVEEKNEKPTIIY 289
Query: 311 IKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARD 370
+K GVY ENV V K K N++ +GDG++ TI++ N +DG+ TF +AT A+ G F+ARD
Sbjct: 290 VKKGVYLENVRVEKTKWNVVMVGDGQSKTIVSAGLNFIDGTPTFETATFAVFGKGFMARD 349
Query: 371 ITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIF 430
+ F NTAGPAKHQAVAL V ADLS FY C + A+QDT+Y H RQF+ +C I GTVDFIF
Sbjct: 350 MGFINTAGPAKHQAVALMVSADLSVFYKCTMDAFQDTMYAHAQRQFYRDCVILGTVDFIF 409
Query: 431 GNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFP 490
GN+AVVFQ C+I RRP GQ+N +TAQGR DPNQNTGI I C I +L ++
Sbjct: 410 GNAAVVFQKCEILPRRPMKGQQNTITAQGRKDPNQNTGISIHNCTIKPLDNLTDIQ---- 465
Query: 491 TYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVT 550
T+LGRPWK++S TV M+S + I+P GW W G+ A +T+ Y EY N+GPGA T RV
Sbjct: 466 TFLGRPWKDFSTTVIMKSFMDKFINPKGWLPWTGDTAPDTIFYAEYLNSGPGASTKNRVK 525
Query: 551 WKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFS 586
W+G K + EA FT FI G++WL +T PF+
Sbjct: 526 WQGLKTSLTKKEANKFTVKPFIDGNNWLPATKVPFN 561
>At3g05620 putative pectinesterase
Length = 543
Score = 382 bits (980), Expect = e-106
Identities = 223/538 (41%), Positives = 311/538 (57%), Gaps = 45/538 (8%)
Query: 66 AILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKL-- 123
+++ AC ELC S I + + N V+ ++ A + +E++
Sbjct: 35 SLVAKACQFIDAHELCVSNIWTHVKESGHGLNPHSVLRAAVK---EAHDKAKLAMERIPT 91
Query: 124 LLRKSLTKREKIALHDCLETIDETLDELKEAQNDLV---------LYPSKKTLYQHADDL 174
++ S+ RE++A+ DC E + ++ EL + ++ L +L
Sbjct: 92 VMMLSIRSREQVAIEDCKELVGFSVTELAWSMLEMNKLHGGGGIDLDDGSHDAAAAGGNL 151
Query: 175 KTLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEF 234
KT +S+A++NQ TCL+GF + +++ ++++ V + SN L M +
Sbjct: 152 KTWLSAAMSNQDTCLEGF--EGTERKYEELIKGSLRQVTQLVSNVLDM-----------Y 198
Query: 235 EQTNMVLGSNKNRKLLEEENGVGWPEWISAGDRRLLQG---STVKADVVVAADGSGNFKT 291
Q N + K E+ + PEW++ D L+ S + + VVA DG G ++T
Sbjct: 199 TQLNAL-----PFKASRNESVIASPEWLTETDESLMMRHDPSVMHPNTVVAIDGKGKYRT 253
Query: 292 VSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGS 351
++EA+ AP S+KRYVI +K GVYKEN+++ KKKTNIM +GDG TIITG RN + G
Sbjct: 254 INEAINEAPNHSTKRYVIYVKKGVYKENIDLKKKKTNIMLVGDGIGQTIITGDRNFMQGL 313
Query: 352 TTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVH 411
TTF +ATVA+ G F+A+DITF+NTAGP QAVALRV +D SAFY C + YQDTLY H
Sbjct: 314 TTFRTATVAVSGRGFIAKDITFRNTAGPQNRQAVALRVDSDQSAFYRCSVEGYQDTLYAH 373
Query: 412 NNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVI 471
+ RQF+ +C I GT+DFIFGN A V QNC I+ R P QK +TAQGR PNQNTG VI
Sbjct: 374 SLRQFYRDCEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPNQNTGFVI 433
Query: 472 QKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTL 531
Q + AT+ PTYLGRPWK YSRTV+M + +S ++ P GW EW GNFAL+TL
Sbjct: 434 QNSYVLATQ---------PTYLGRPWKLYSRTVYMNTYMSQLVQPRGWLEWFGNFALDTL 484
Query: 532 VYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
Y EY N GPG +S RV W G+ ++ A SFT G+FI G WL +TG F+ GL
Sbjct: 485 WYGEYNNIGPGWRSSGRVKWPGYHIMDKRT-ALSFTVGSFIDGRRWLPATGVTFTAGL 541
>At4g33230 pectinesterase - like protein
Length = 609
Score = 380 bits (975), Expect = e-105
Identities = 237/621 (38%), Positives = 344/621 (55%), Gaps = 54/621 (8%)
Query: 2 NKIKQSLAGISNSGNKKKLLISLFTTLLIVASIV--AIVATTTKN---------SNKSKN 50
+KI++ + +K++++ + + L++ A+I+ A T +N +NKSK+
Sbjct: 7 DKIQERVNAERKRKFRKRIILGVVSVLVVAAAIIGGAFAYVTYENKTQEQGKTTNNKSKD 66
Query: 51 NSIASSSLSLSHHSHA-----------ILKSACTTTLYPELCFSAISSEPNITHKITNHK 99
+ S S S S A I+++ C +TLY C + + +E T K T
Sbjct: 67 SPTKSESPSPKPPSSAAQTVKAGQVDKIIQTLCNSTLYKPTCQNTLKNE---TKKDTPQT 123
Query: 100 DVISLSLNITTRAVEHNYFTVEKLLLRKSLTKREKIALHDCLETIDETLDELKEAQ---N 156
D SL + + +++L K+ K +K A+ C +DE +EL + N
Sbjct: 124 DPRSLLKSAIVAVNDDLDQVFKRVLSLKTENKDDKDAIAQCKLLVDEAKEELGTSMKRIN 183
Query: 157 DLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMC 216
D + K + DL + +S+ ++ Q TC+DGF E+RK Q+ +
Sbjct: 184 DSEVNNFAKIV----PDLDSWLSAVMSYQETCVDGFEEGKLKTEIRKNFNSSQV----LT 235
Query: 217 SNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEENGVGWPE----WISAGDRRLLQG 272
SN+LAM K++ D ++ + K R LLE + + W+S +RR+L+
Sbjct: 236 SNSLAMIKSL-DGYLSSVPKV-------KTRLLLEARSSAKETDHITSWLSNKERRMLKA 287
Query: 273 STVKA---DVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNI 329
VKA + VA DGSGNF T++ A+ A P K RY I IK G+Y E+V + KKK N+
Sbjct: 288 VDVKALKPNATVAKDGSGNFTTINAALKAMPAKYQGRYTIYIKHGIYDESVIIDKKKPNV 347
Query: 330 MFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRV 389
+GDG TI+TG+++ TF +AT G F+A+ + F+NTAGP HQAVA+RV
Sbjct: 348 TMVGDGSQKTIVTGNKSHAKKIRTFLTATFVAQGEGFMAQSMGFRNTAGPEGHQAVAIRV 407
Query: 390 GADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNS 449
+D S F NC YQDTLY + +RQ++ +C I GTVDFIFG++A +FQNCDI R+
Sbjct: 408 QSDRSVFLNCRFEGYQDTLYAYTHRQYYRSCVIIGTVDFIFGDAAAIFQNCDIFIRKGLP 467
Query: 450 GQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSS 509
GQKN VTAQGRVD Q TG VI C + +DL+ VK F +YLGRPWK +SRTV M+S+
Sbjct: 468 GQKNTVTAQGRVDKFQTTGFVIHNCTVAPNEDLKPVKAQFKSYLGRPWKPHSRTVVMEST 527
Query: 510 ISDVIDPVGWHEW-NGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTP 568
I DVIDPVGW W +FA++TL Y EY+N GP T+ RV W GF+V+ + EA FT
Sbjct: 528 IEDVIDPVGWLRWQETDFAIDTLSYAEYKNDGPSGATAARVKWPGFRVL-NKEEAMKFTV 586
Query: 569 GNFIGGSSWLGSTGFPFSLGL 589
G F+ G W+ + G P LGL
Sbjct: 587 GPFLQG-EWIQAIGSPVKLGL 606
>At3g10710 putative pectinesterase
Length = 561
Score = 379 bits (972), Expect = e-105
Identities = 236/579 (40%), Positives = 325/579 (55%), Gaps = 47/579 (8%)
Query: 18 KKLLISLFTTLLIVASIV--AIVATTT-KNSNKSKNNSIASSSLSLSHHSHAILKSACTT 74
+K + + +L+I+A IV A+ T K S ++ + S+S+S +K+ C
Sbjct: 23 RKNIAIIAVSLVILAGIVIGAVFGTMAHKKSPETVETNNNGDSISVS------VKAVCDV 76
Query: 75 TLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLTKREK 134
TL+ E CF + S PN + N +++ ++ IT V L ++
Sbjct: 77 TLHKEKCFETLGSAPNASS--LNPEELFRYAVKITIAEVSKAINAFSSSLG----DEKNN 130
Query: 135 IALHDCLETIDETLDELKEAQNDLVLYPSKK--TLYQHADDLKTLISSAITNQVTCLDGF 192
I ++ C E +D T+D L N+ + S T+ + DDL+T +SSA T Q TC++
Sbjct: 131 ITMNACAELLDLTIDNL----NNTLTSSSNGDVTVPELVDDLRTWLSSAGTYQRTCVETL 186
Query: 193 SHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEE 252
+ D + G+ H+++ + +T +A + S K R+ L
Sbjct: 187 APD--------MRPFGESHLKN--------STELTSNALAIITWLGKIADSFKLRRRLLT 230
Query: 253 ENGVGWPEWISAGDRRLLQGSTVK--ADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIK 310
V E RRLLQ + ++ AD+VVA DGSG ++T+ A+ P KS KR +I
Sbjct: 231 TADV---EVDFHAGRRLLQSTDLRKVADIVVAKDGSGKYRTIKRALQDVPEKSEKRTIIY 287
Query: 311 IKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARD 370
+K GVY ENV+V KK N++ +GDG + +I++G NV+DG+ TF +AT A+ G F+ARD
Sbjct: 288 VKKGVYFENVKVEKKMWNVIVVGDGESKSIVSGRLNVIDGTPTFKTATFAVFGKGFMARD 347
Query: 371 ITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIF 430
+ F NTAGP+KHQAVAL V ADL+AFY C + AYQDTLYVH RQF+ C I GTVDFIF
Sbjct: 348 MGFINTAGPSKHQAVALMVSADLTAFYRCTMNAYQDTLYVHAQRQFYRECTIIGTVDFIF 407
Query: 431 GNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFP 490
GNSA V Q+C I RRP GQ+N +TAQGR DPN NTGI I +C I DL V
Sbjct: 408 GNSASVLQSCRILPRRPMKGQQNTITAQGRTDPNMNTGISIHRCNISPLGDLTDVM---- 463
Query: 491 TYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVT 550
T+LGRPWK +S TV M S + ID GW W G+ A +T+ Y EY+NTGPGA T RV
Sbjct: 464 TFLGRPWKNFSTTVIMDSYLHGFIDRKGWLPWTGDSAPDTIFYGEYKNTGPGASTKNRVK 523
Query: 551 WKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
WKG + + S EA FT FI G WL +T PF GL
Sbjct: 524 WKGLRFL-STKEANRFTVKPFIDGGRWLPATKVPFRSGL 561
>At5g51490 pectinesterase
Length = 536
Score = 375 bits (963), Expect = e-104
Identities = 207/463 (44%), Positives = 285/463 (60%), Gaps = 27/463 (5%)
Query: 132 REKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHAD-DLKTLISSAITNQVTCLD 190
+++ L DC++ +T+ +L + + P D D +T +S+A+TN TC
Sbjct: 96 KKQAVLADCIDLYGDTIMQLNRTLHGVS--PKAGAAKSCTDFDAQTWLSTALTNTETCRR 153
Query: 191 GFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLL 250
G S + + ++ +I H+ SN LA+ + + NK
Sbjct: 154 GSSDLNVTDFITPIVSNTKI--SHLISNCLAVNGAL-------------LTAGNKGNTTA 198
Query: 251 EEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLK--SSKRYV 308
++ G+P W+S D+RLL+ V+A++VVA DGSG+F TV A+ A + +S R+V
Sbjct: 199 NQK---GFPTWLSRKDKRLLRA--VRANLVVAKDGSGHFNTVQAAIDVAGRRKVTSGRFV 253
Query: 309 IKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLA 368
I +K G+Y+EN+ V +IM +GDG +TIITG R+V G TT++SAT I G +F+A
Sbjct: 254 IYVKRGIYQENINVRLNNDDIMLVGDGMRSTIITGGRSVQGGYTTYNSATAGIEGLHFIA 313
Query: 369 RDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDF 428
+ ITF+NTAGPAK QAVALR +DLS FY C I YQDTL VH+ RQF+ C+I GTVDF
Sbjct: 314 KGITFRNTAGPAKGQAVALRSSSDLSIFYKCSIEGYQDTLMVHSQRQFYRECYIYGTVDF 373
Query: 429 IFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGN 488
IFGN+A VFQNC I RRP GQ N++TAQGR DP QNTGI I RI DL+ V G
Sbjct: 374 IFGNAAAVFQNCLILPRRPLKGQANVITAQGRADPFQNTGISIHNSRILPAPDLKPVVGT 433
Query: 489 FPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEW--NGNFALNTLVYREYQNTGPGAGTS 546
TY+GRPW ++SRTV +Q+ + +V+ PVGW W F L+TL Y EY+NTGP + T
Sbjct: 434 VKTYMGRPWMKFSRTVVLQTYLDNVVSPVGWSPWIEGSVFGLDTLFYAEYKNTGPASSTR 493
Query: 547 KRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
RV+WKGF V+ A++A +FT G FI G++WL TG PF+ GL
Sbjct: 494 WRVSWKGFHVLGRASDASAFTVGKFIAGTAWLPRTGIPFTSGL 536
>At3g59010 pectinesterase precursor-like protein
Length = 529
Score = 374 bits (961), Expect = e-104
Identities = 237/559 (42%), Positives = 319/559 (56%), Gaps = 53/559 (9%)
Query: 53 IASSSLSLSHHSHAI--LKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLS----- 105
+A++S SL +H I + L + A SS T KI+ H + S S
Sbjct: 1 MATTSFSLPNHKFGIKLMLFLVLNLLSLQTSVFAHSSNSKFT-KISRHPNSDSSSRTKPS 59
Query: 106 -------LNITTRAVEHNYFTVEKLLLRKSLTKREKIAL-----HDCLETIDETLDELKE 153
L+ +++H F L +L+ R L +DCLE +D+TLD L
Sbjct: 60 TSSNKGFLSSVQLSLDHALFA-RSLAFNLTLSHRTSQTLMLDPVNDCLELLDDTLDMLYR 118
Query: 154 AQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVE 213
+V+ K + DD+ T +S+A+TNQ TC S E +EG I ++
Sbjct: 119 ----IVVIKRKDHVN---DDVHTWLSAALTNQETCKQSLS------EKSSFNKEG-IAID 164
Query: 214 HMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEENGVGWPEWISAGDRRLLQGS 273
N + N D +++ ++++ RKLL + + +P W+S+ DR+LL+ S
Sbjct: 165 SFARNLTGLLTNSLDMFVSDKQKSSSSSNLTGGRKLLSDHD---FPTWVSSSDRKLLEAS 221
Query: 274 T--VKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMF 331
++ VVAADGSG +V+EA+A+ K S R VI + AG YKEN+ +P K+ N+M
Sbjct: 222 VEELRPHAVVAADGSGTHMSVAEALASLE-KGSGRSVIHLTAGTYKENLNIPSKQKNVML 280
Query: 332 LGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGA 391
+GDG+ T+I GSR+ G T+ SATVA +G F+ARDITF N+AGP QAVALRVG+
Sbjct: 281 VGDGKGKTVIVGSRSNRGGWNTYQSATVAAMGDGFIARDITFVNSAGPNSEQAVALRVGS 340
Query: 392 DLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQ 451
D S Y C I YQD+LY + RQF+ I+GTVDFIFGNSAVVFQ+C++ +R+ +S Q
Sbjct: 341 DRSVVYRCSIDGYQDSLYTLSKRQFYRETDITGTVDFIFGNSAVVFQSCNLVSRKGSSDQ 400
Query: 452 KNMVTAQGRVDPNQNTGIVIQKCRI-GATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSI 510
N VTAQGR DPNQNTGI I CRI G+TK TYLGRPWK+YSRTV MQS I
Sbjct: 401 -NYVTAQGRSDPNQNTGISIHNCRITGSTK----------TYLGRPWKQYSRTVVMQSFI 449
Query: 511 SDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGN 570
I P GW W+ NFAL TL Y E+ N+GPG+ S RV+W G+ + EAQ FT
Sbjct: 450 DGSIHPSGWSPWSSNFALKTLYYGEFGNSGPGSSVSGRVSWAGYHPALTLTEAQGFTVSG 509
Query: 571 FIGGSSWLGSTGFPFSLGL 589
FI G+SWL STG F GL
Sbjct: 510 FIDGNSWLPSTGVVFDSGL 528
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,700,163
Number of Sequences: 26719
Number of extensions: 539481
Number of successful extensions: 1948
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 87
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 1573
Number of HSP's gapped (non-prelim): 134
length of query: 589
length of database: 11,318,596
effective HSP length: 105
effective length of query: 484
effective length of database: 8,513,101
effective search space: 4120340884
effective search space used: 4120340884
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC137821.5


