

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.2 + phase: 0 /pseudo
(428 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
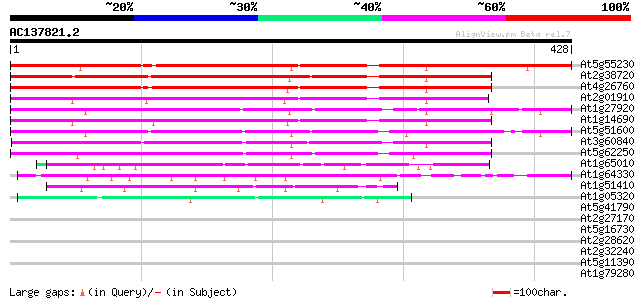
Score E
Sequences producing significant alignments: (bits) Value
At5g55230 unknown protein 283 1e-76
At2g38720 hypothetical protein 281 6e-76
At4g26760 unknown protein 268 4e-72
At2g01910 unknown protein 221 5e-58
At1g27920 hypothetical protein 219 2e-57
At1g14690 hypothetical protein 209 2e-54
At5g51600 putative protein 203 2e-52
At3g60840 putative protein 182 2e-46
At5g62250 cytokinesis regulating protein - like 176 2e-44
At1g65010 hypothetical protein 51 1e-06
At1g64330 unknown protein 47 3e-05
At1g51410 cinnamyl alcohol dehydrogenase, putative 44 2e-04
At1g05320 unknown protein 44 2e-04
At5g41790 myosin heavy chain-like protein 40 0.002
At2g27170 putative chromosome associated protein 40 0.002
At5g16730 putative protein 39 0.005
At2g28620 putative kinesin-like spindle protein 39 0.005
At2g32240 putative myosin heavy chain 39 0.007
At5g11390 unknown protein 38 0.009
At1g79280 hypothetical protein 38 0.009
>At5g55230 unknown protein
Length = 587
Score = 283 bits (725), Expect = 1e-76
Identities = 184/454 (40%), Positives = 277/454 (60%), Gaps = 38/454 (8%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSF--SRGKGT 58
+LLQIEQE ++Y RKVE K + L + L D +E++++ SLG+ + + GT
Sbjct: 40 LLLQIEQECLDVYKRKVEQAAKSRAELLQTLSDANAELSSLTMSLGDKSLVGIPDKSSGT 99
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLT 118
+K+QLA I P +E L +K+ERV+E +++SQI +I + AG G S V VD+ DL+
Sbjct: 100 IKEQLAAIAPALEQLWQQKEERVREFSDVQSQIQKICGDIAG-GLSNEVPI--VDESDLS 156
Query: 119 MEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNG 178
++KL +S L ELQ EK+ R +KV +ST+ +L AV+ +D + +HPSL++ ++
Sbjct: 157 LKKLDDFQSQLQELQKEKSDRLRKVLEFVSTVHDLCAVLGLDFLSTVTEVHPSLDEDTSV 216
Query: 179 AQQSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDVI 234
+SIS+ETL+RL ++VL LK +K+Q LQK+Q L +LW+ M T DE++ + D V
Sbjct: 217 QSKSISNETLSRLAKTVLTLKDDKKQRLQKLQELATQLIDLWNLMD-TPDEERELFDHVT 275
Query: 235 GLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEE 294
IS+SVDEV++ G+L+ D+++Q +VEV RL L S+M KE+AFK+Q+ELEE
Sbjct: 276 CNISSSVDEVTVPGALARDLIEQAEVEVDRLDQLKASRM--------KEIAFKKQSELEE 327
Query: 295 IHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEW 352
I+ AH++++ ++AR+ + LI GN++ +ELL MD QI KAKE+A SR+DI+DRV +W
Sbjct: 328 IYARAHVEVNPESARERIMSLIDSGNVEPTELLADMDSQISKAKEEAFSRKDILDRVEKW 387
Query: 353 KSAAEEEKWLDEYERFI*SVRRKRG-FLSPRFHLW*RI*LQK----------------*K 395
SA EEE WL++Y R RG L+ + RI + K +
Sbjct: 388 MSACEEESWLEDYNRDQNRYSASRGAHLNLKRAEKARILVSKIPAMVDTLVAKTRAWEEE 447
Query: 396 HGRQ-TKKKVSYMKSLHEYNVQRQLREEEKRKSR 428
H V + L EY + RQ REEEKR+ R
Sbjct: 448 HSMSFAYDGVPLLAMLDEYGMLRQEREEEKRRLR 481
>At2g38720 hypothetical protein
Length = 587
Score = 281 bits (718), Expect = 6e-76
Identities = 167/374 (44%), Positives = 247/374 (65%), Gaps = 18/374 (4%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGK-GTL 59
MLL++EQE +IY++KVE T K + L + L ++EIA++ S+LGE VSF++ K G+L
Sbjct: 33 MLLELEQECLDIYNKKVEKTRKFRAELQRSLAQAEAEIASLMSALGEK-VSFAKKKEGSL 91
Query: 60 KQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLTM 119
K+Q++++ PV+EDL KK R KE+ E +QI++I + AG + + +VD+ DLT
Sbjct: 92 KEQISSVKPVLEDLLMKKDRRRKELSETLNQIAEITSNIAGNDYTVS-SGSEVDESDLTQ 150
Query: 120 EKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNGA 179
KL +L++ L +L+NEK +R QKV S+IS + ELS ++S D S+ LN +H SL + S
Sbjct: 151 RKLDELRADLQDLRNEKAVRLQKVNSYISAVHELSEILSFDFSKALNSVHSSLTEFSKTH 210
Query: 180 QQSISDETLARLNESVLLLKQEKQQSLQKVQFL----EELWDFMGITTDEQKAVCDDVIG 235
+SIS++TLAR E V LK EK + L K+Q L +ELW+ M DE++ D
Sbjct: 211 SKSISNDTLARFTELVKSLKAEKHERLLKLQGLGRSMQELWNLMETPMDERRRF-DHCSS 269
Query: 236 LISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEEI 295
L+S+ D+ G LS DI+++ + EV+RL L SKM KEL FKRQ ELEEI
Sbjct: 270 LLSSLPDDALKKGCLSLDIIREAEDEVRRLNSLKSSKM--------KELVFKRQCELEEI 321
Query: 296 HRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEWK 353
RG HMD+++ AAR+ L +LI G+ D+S++L +D QI KA+E+A SR++I+D+V +W+
Sbjct: 322 CRGNHMDINSDAARKSLVELIESGDGDLSDILASIDGQIEKAREEALSRKEILDKVDKWR 381
Query: 354 SAAEEEKWLDEYER 367
A EEE WLD+YE+
Sbjct: 382 HAKEEETWLDDYEK 395
>At4g26760 unknown protein
Length = 578
Score = 268 bits (685), Expect = 4e-72
Identities = 159/375 (42%), Positives = 243/375 (64%), Gaps = 20/375 (5%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGE-DYVSF-SRGKGT 58
+LLQIE+E +Y +KVE K + L + L D E++N+ ++LGE Y+ + GT
Sbjct: 40 LLLQIEEECLNVYKKKVELAAKSRAELLQTLSDATVELSNLTTALGEKSYIDIPDKTSGT 99
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLT 118
+K+QL+ I P +E L +K+ERV+ +++SQI +I E AG G + G VD+ DL+
Sbjct: 100 IKEQLSAIAPALEQLWQQKEERVRAFSDVQSQIQKICEEIAG-GLNNG--PHVVDETDLS 156
Query: 119 MEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNG 178
+++L + L ELQ EK+ R QKV +ST+ +L AV+ +D + +HPSL++++
Sbjct: 157 LKRLDDFQRKLQELQKEKSDRLQKVLEFVSTVHDLCAVLRLDFLSTVTEVHPSLDEANGV 216
Query: 179 AQQSISDETLARLNESVLLLKQEKQQSLQKVQ----FLEELWDFMGITTDEQKAVCDDVI 234
+SIS+ETLARL ++VL LK++K Q L+K+Q L +LW+ M T+DE++ + D V
Sbjct: 217 QTKSISNETLARLAKTVLTLKEDKMQRLKKLQELATQLTDLWNLMD-TSDEERELFDHVT 275
Query: 235 GLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEE 294
ISASV EV+ G+L+ D+++Q +VEV RL L S+M KE+AFK+Q+ELEE
Sbjct: 276 SNISASVHEVTASGALALDLIEQAEVEVDRLDQLKSSRM--------KEIAFKKQSELEE 327
Query: 295 IHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEW 352
I+ AH+++ + R+ + LI GN + +ELL MD QI KAKE+A SR++I+DRV +W
Sbjct: 328 IYARAHIEIKPEVVRERIMSLIDAGNTEPTELLADMDSQIAKAKEEAFSRKEILDRVEKW 387
Query: 353 KSAAEEEKWLDEYER 367
SA EEE WL++Y R
Sbjct: 388 MSACEEESWLEDYNR 402
>At2g01910 unknown protein
Length = 608
Score = 221 bits (564), Expect = 5e-58
Identities = 132/378 (34%), Positives = 218/378 (56%), Gaps = 22/378 (5%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLG----EDYVSFSRGK 56
ML+++E+E +IY RKV++ K L++ + ++E+A++ ++LG + +G
Sbjct: 42 MLMELERECLQIYQRKVDEAANSKAKLHQSVASIEAEVASLMAALGVLNINSPIKLDKGS 101
Query: 57 GTLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQ--SKGVT-DQDVD 113
+LK++LA + P+VE+LR +K+ER+K+ +IK+QI +I E +G +K + ++
Sbjct: 102 KSLKEKLAAVTPLVEELRIQKEERMKQFSDIKAQIEKISGEISGYSDHLNKAMNISLTLE 161
Query: 114 QCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLN 173
+ DLT+ L + ++HL LQ EK+ R KV +++ + L V+ +D S+ ++ +HPSL+
Sbjct: 162 EQDLTLRNLNEYQTHLRTLQKEKSDRLNKVLGYVNEVHALCGVLGVDFSQTVSAVHPSLH 221
Query: 174 DSSNGAQQSISDETLARLNESVLLLKQEKQQSLQK----VQFLEELWDFMGITTDEQKAV 229
+ +ISD TL L + LK E++ QK V L ELW+ M T E +
Sbjct: 222 RTDQEQSTNISDSTLEGLEHMIQKLKTERKSRFQKLKDVVASLFELWNLMD-TPQEDRTK 280
Query: 230 CDDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQ 289
V ++ +S ++ G LS + ++QV EV L L S+M KEL KR+
Sbjct: 281 FGKVTYVVRSSEANITEPGILSTETIEQVSTEVDSLSKLKASRM--------KELVMKRR 332
Query: 290 NELEEIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIID 347
+ELE++ R H+ D + + T LI G +D SELL ++ QI K K++AQSR+DI+D
Sbjct: 333 SELEDLCRLTHIQPDTSTSAEKSTALIDSGLVDPSELLANIEMQINKIKDEAQSRKDIMD 392
Query: 348 RVAEWKSAAEEEKWLDEY 365
R+ W SA EEE WL+EY
Sbjct: 393 RIDRWLSACEEENWLEEY 410
Score = 29.6 bits (65), Expect = 3.3
Identities = 53/247 (21%), Positives = 102/247 (40%), Gaps = 42/247 (17%)
Query: 108 TDQDVDQCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNG 167
T++D +L E L + ++E N K HQ V S + ++ L A + +LN
Sbjct: 37 TEKDRMLMELERECLQIYQRKVDEAANSKAKLHQSVASIEAEVASLMAALG-----VLNI 91
Query: 168 IHP-SLNDSSNGAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLEELWDFMGITTDEQ 226
P L+ S ++ ++ + L E + + K+E+ + +
Sbjct: 92 NSPIKLDKGSKSLKEKLA--AVTPLVEELRIQKEERMKQFSDI----------------- 132
Query: 227 KAVCDDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAF 286
KA + + G IS D ++ ++S + +E Q L + N ++ + + QKE +
Sbjct: 133 KAQIEKISGEISGYSDHLNKAMNIS------LTLEEQDLTLRNLNEYQTHLRTLQKEKSD 186
Query: 287 KRQNEL---EEIHR-----GAHMDMDAKAARQIL--TDLIGNIDMSE-LLQGMDKQIRKA 335
+ L E+H G A L TD + ++S+ L+G++ I+K
Sbjct: 187 RLNKVLGYVNEVHALCGVLGVDFSQTVSAVHPSLHRTDQEQSTNISDSTLEGLEHMIQKL 246
Query: 336 KEQAQSR 342
K + +SR
Sbjct: 247 KTERKSR 253
>At1g27920 hypothetical protein
Length = 592
Score = 219 bits (558), Expect = 2e-57
Identities = 159/456 (34%), Positives = 253/456 (54%), Gaps = 40/456 (8%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGK--GT 58
+LL IEQE E Y RKV+ + L++ L + ++E+ + LGE V K GT
Sbjct: 74 VLLDIEQECVEAYRRKVDHANVSRSRLHQELAESEAELTHFLLCLGERSVPGRPEKKGGT 133
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQ-DVDQCDL 117
L++QL +I P + ++R +K ERVK+ +K +I +I AE AG + T + +D DL
Sbjct: 134 LREQLDSIAPALREMRLRKDERVKQFRSVKGEIQKISAEIAGRSTYEDSTRKITIDDNDL 193
Query: 118 TMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSN 177
+ +KL + ++ L+ L +EKN R QKV +I I +LSA + + S I+ IHPSLND
Sbjct: 194 SNKKLEEYQNELHRLHDEKNERLQKVDIYICAIRDLSATLGTEASMIITKIHPSLNDLY- 252
Query: 178 GAQQSISDETLARLNESVLLLKQEKQQSLQKVQFL----EELWDFMGITTDEQKAVCDDV 233
G ++ISD+ L +LN +V+ L++EK + L+K+ L LW+ M + ++++ V
Sbjct: 253 GISKNISDDILKKLNGTVVSLEEEKHKRLEKLHHLGRALSNLWNLMDASYEDRQKFFH-V 311
Query: 234 IGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELE 293
I L+S++ +V GS++ DI++Q + EV+RL L S++KE K+QKEL E
Sbjct: 312 IDLLSSAPSDVCAPGSITLDIIQQAEAEVKRLDQLKASRIKELFIKKQKEL--------E 363
Query: 294 EIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAE 351
+ +HM+ + I T+L+ G +D +LL MD++I +AKE+A SR+ II++V
Sbjct: 364 DTCNMSHMETPSTEMGNI-TNLVDSGEVDHVDLLAAMDEKIARAKEEAASRKGIIEKVDR 422
Query: 352 WKSAAEEEKWLDEYE-------------RFI*SVRRKRGFLSPRFHLW*RI*LQK*KHGR 398
W A++EE+WL+EY+ R + R R +S L I L K K
Sbjct: 423 WMLASDEERWLEEYDQDENRYSVSRNAHRNLRRAERARITVSKISGLVESI-LVKAKSWE 481
Query: 399 QTKKK------VSYMKSLHEYNVQRQLREEEKRKSR 428
++K V + L EYN RQ +E EK++ R
Sbjct: 482 VERQKVFLYNEVPLVAMLQEYNKLRQEKEMEKQRLR 517
>At1g14690 hypothetical protein
Length = 707
Score = 209 bits (532), Expect = 2e-54
Identities = 133/409 (32%), Positives = 218/409 (52%), Gaps = 51/409 (12%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLG----EDYVSFSRGK 56
ML+++E+E EIY RKV++ K L++ L ++EIA++ ++LG + G
Sbjct: 25 MLMELEKECLEIYRRKVDEAANSKAQLHQSLVSIEAEIASLLAALGVFNSHSPMKAKEGS 84
Query: 57 GTLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVT---DQDVD 113
+LK++LA + P++EDLR +K ER+K+ ++IK+QI ++ E +G T +D
Sbjct: 85 KSLKEKLAAVRPMLEDLRLQKDERMKQFVDIKAQIEKMSGEISGYSDQLNKTMVGSLALD 144
Query: 114 QCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLN 173
+ DLT+ KL + ++HL LQ EK+ R KV +++ + L V+ +D + ++ +HPSL+
Sbjct: 145 EQDLTLRKLNEYQTHLRSLQKEKSDRLNKVLDYVNEVHTLCGVLGVDFGQTVSEVHPSLH 204
Query: 174 DSSNGAQQSISDETLARLNESVLLLKQEKQQSLQKVQ----------------------- 210
+ + +ISD+TL L+ + LK E+ QKV
Sbjct: 205 RTDHEQSTNISDDTLDGLHHMIHKLKTERSVRFQKVDKHACNAILLFVTDYHFDALAFSP 264
Query: 211 ----------FLEELWDFMGITTDEQKAVCDDVIGLISASVDEVSIHGSLSNDIVKQVDV 260
L ELW+ M T+ E++ V ++ +S +++ LS++ ++QV
Sbjct: 265 LSVQLKDVAGSLFELWNLMD-TSQEERTKFASVSYVVRSSESDITEPNILSSETIEQVSA 323
Query: 261 EVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEEIHRGAHMDMDAKAARQILTDLI--GN 318
EV L S+M KEL KR+ ELE + R AH++ D + + T LI G
Sbjct: 324 EVDCFNKLKASRM--------KELVMKRRTELENLCRLAHIEADTSTSLEKSTALIDSGL 375
Query: 319 IDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEWKSAAEEEKWLDEYER 367
+D SELL ++ I K KE+A SR++IIDR+ W SA EEE WL+EY +
Sbjct: 376 VDPSELLTNIELHINKIKEEAHSRKEIIDRIDRWLSACEEENWLEEYNQ 424
>At5g51600 putative protein
Length = 707
Score = 203 bits (516), Expect = 2e-52
Identities = 144/464 (31%), Positives = 243/464 (52%), Gaps = 57/464 (12%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGK---G 57
MLL++E+E E+Y RKV+ + + L + + D ++++A I S++GE V + G
Sbjct: 40 MLLELERECLEVYRRKVDQANRCRAQLRQAIADAEAQLAAICSAMGERPVHIRQSDQSVG 99
Query: 58 TLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDL 117
+LKQ+L I+P +E+++ +K ER + + + QI I + G G+ ++ +D+ +L
Sbjct: 100 SLKQELGRILPELEEMQKRKVERRNQFIVVMEQIDSITNDIKGQGELVH-SEPLIDETNL 158
Query: 118 TMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSN 177
+M KL +L L LQ EK R + ++ H+ T+ +V+ +D +E++ ++P+L+D
Sbjct: 159 SMRKLEELHCQLQVLQKEKIDRVETIRKHLCTLYSHCSVLGMDFNEVVGQVNPTLSDPEG 218
Query: 178 GAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDV 233
+S+SD T+ +L +V L + K Q +Q++Q L ELW+ M +EQ+ +
Sbjct: 219 --PRSLSDHTIEKLGAAVQKLMEVKIQRMQRLQDLATTMLELWNLMDTPIEEQQEY-QHI 275
Query: 234 IGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELE 293
I+AS E++ SLS D +K V+ EV RL + SKMKE V K++ +ELE
Sbjct: 276 TCNIAASEHEITEANSLSEDFIKYVEAEVVRLDEVKASKMKELVLKKR--------SELE 327
Query: 294 EIHRGAHM--DMDAKAARQILTDLIGNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAE 351
EI R H+ D+ + I+ G +D + +L+ +++ I K KE+A SR++I++RV +
Sbjct: 328 EICRKTHLLPVSDSAIDQTIVAIESGIVDATMVLEHLEQHISKIKEEALSRKEILERVEK 387
Query: 352 WKSAAEEEKWLDEYERFI*SVRRKRGFLSPRFHLW*RI*LQK*KHGRQTKKK-------- 403
W SA +EE WL+EY R RG HL L++ + R K
Sbjct: 388 WLSACDEESWLEEYNRDDNRYNAGRG-----AHLT----LKRAEKARNLVTKLPGMVEAL 438
Query: 404 -------------------VSYMKSLHEYNVQRQLREEEKRKSR 428
+ + L EYN+ RQ REEE R+ R
Sbjct: 439 ASKTIVWEQENGIEFLYDGIRLLSMLEEYNILRQEREEEHRRQR 482
>At3g60840 putative protein
Length = 648
Score = 182 bits (463), Expect = 2e-46
Identities = 124/373 (33%), Positives = 211/373 (56%), Gaps = 21/373 (5%)
Query: 2 LLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLG-EDYVSFSRGKGTLK 60
L IE+E +Y RKVE+ + K L K + ++EIA I SS+G ++ S SR LK
Sbjct: 12 LADIEKECLSVYKRKVEEASRGKANLLKEIAVGRAEIAAIGSSMGGQEIHSNSRLGENLK 71
Query: 61 QQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLTME 120
++L N+ ++ LR +K ER+ E+ Q+ ++ + + + ++ DL+++
Sbjct: 72 EELENVNVQLDGLRKRKAERMIRFNEVIDQLLKLSLQLGN--PTDYLKKFAAEETDLSLQ 129
Query: 121 KLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNGAQ 180
+L +L+S L ELQNEK+ R ++V+ + T++ L +V+ D ++ GIH SL DS+
Sbjct: 130 RLEELRSQLGELQNEKSKRLEEVECLLKTLNSLCSVLGEDFKGMIRGIHSSLVDSNT--- 186
Query: 181 QSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDVIGL 236
+ +S TL +L+ ++ L++ K Q +QKVQ L ELW+ + +EQK + +V
Sbjct: 187 RDVSRSTLDKLDMMIVNLREAKLQRMQKVQDLAVSLLELWNLLDTPAEEQK-IFHNVTCS 245
Query: 237 ISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEEIH 296
I+ + E++ LS +K+V+ EV RL + +K+KE + +++ EL EEI
Sbjct: 246 IALTESEITEANILSVASIKRVEDEVIRLSKIKITKIKEVILRKRLEL--------EEIS 297
Query: 297 RGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEWKS 354
R HM + + + I G D +LL+ +D +I K KE+A SR++I+++V +W S
Sbjct: 298 RKMHMATEVLKSENFSVEAIESGVKDPEQLLEQIDSEIAKVKEEASSRKEILEKVEKWMS 357
Query: 355 AAEEEKWLDEYER 367
A EEE WL+EY R
Sbjct: 358 ACEEESWLEEYNR 370
>At5g62250 cytokinesis regulating protein - like
Length = 549
Score = 176 bits (446), Expect = 2e-44
Identities = 118/383 (30%), Positives = 214/383 (55%), Gaps = 27/383 (7%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYV-------SFS 53
+L++IE+E E+Y+RK+E + K + + + D ++ + +I S + E +
Sbjct: 34 ILIEIEEECREVYNRKIEKVKEEKIRIKQEIADSEARVIDICSVMEEPPILGRHHQSDQQ 93
Query: 54 RGKG-TLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVT-DQD 111
G G +LK +L I+ +E++ +K ER + +++ I +R E G + + D
Sbjct: 94 SGNGRSLKDELVKILQKLEEMEKRKSERKIQFIQVIDDIRCVREEINGESDDETCSSDFS 153
Query: 112 VDQCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPS 171
D+ DL++ KL +L L LQ +K R ++++ +I T+ L +V+ ++ E + IHPS
Sbjct: 154 ADESDLSLRKLEELHRELYTLQEQKRNRVKQIQDNIRTLESLCSVLGLNFRETVTKIHPS 213
Query: 172 LNDSSNGAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQK 227
L D+ +SIS+ETL +L SV + K Q +Q++Q L E W+ M +EQ+
Sbjct: 214 LVDTEGS--RSISNETLDKLASSVQQWHETKIQRMQELQDLVTTMLEFWNLMDTPAEEQQ 271
Query: 228 AVCDDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFK 287
D V I+A+V E++ SLS D++++V E+ RL+ L SKMKE V K++
Sbjct: 272 KFMD-VSCNIAATVSEITKPNSLSIDLLEEVKAELCRLEELKWSKMKELVLKKR------ 324
Query: 288 RQNELEEIHRGAHMDMDAK--AARQILTDL-IGNIDMSELLQGMDKQIRKAKEQAQSRRD 344
+ELEEI R H+ ++ + A ++ + G+++ +L+ ++ + K KE+A SR++
Sbjct: 325 --SELEEICRRTHIVLEEEDIAVENVIKAIESGDVNPENILEQIEYRAGKVKEEALSRKE 382
Query: 345 IIDRVAEWKSAAEEEKWLDEYER 367
I+++ +W +A EEE WL+EY +
Sbjct: 383 ILEKADKWLNACEEENWLEEYNQ 405
>At1g65010 hypothetical protein
Length = 1318
Score = 50.8 bits (120), Expect = 1e-06
Identities = 72/347 (20%), Positives = 148/347 (41%), Gaps = 33/347 (9%)
Query: 29 KVLDDFQSEIANIASSLGEDYVSFSRGKGTLKQQLANIIPVV---EDLRFKKKERVKEIL 85
K+L+D ++EI ++ S++ F K +Q+ +++ V E+ +E V ++
Sbjct: 480 KMLEDARNEIDSLKSTVDSIQNEFENSKAGWEQKELHLMGCVKKSEEENSSSQEEVSRLV 539
Query: 86 EIKSQISQI----RAEKAGCGQSKGVTDQDVDQCDLTMEKLGKLKSHLNELQNEKNIRHQ 141
+ + + + E+A + V + +V T+ + L E +K +
Sbjct: 540 NLLKESEEDACARKEEEASLKNNLKVAEGEVKYLQETLGEAKAESMKLKESLLDKEEDLK 599
Query: 142 KVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNGAQQSISDETLARLNESVLLLKQE 201
V + IS++ E + I E L+ + SL D Q SI+ E +KQ
Sbjct: 600 NVTAEISSLREWEGSVLEKIEE-LSKVKESLVDKETKLQ-SITQEAEELKGREAAHMKQI 657
Query: 202 KQQSLQKVQFLEELWDFMGITTDEQKAVCDDVIGLISASVDEVSIHGSLSNDIVKQVDVE 261
++ S ++E I E + + + G + ++E+S+ D V +
Sbjct: 658 EELSTANASLVDEATKLQSIV-QESEDLKEKEAGYLK-KIEELSVANESLADNVTDLQSI 715
Query: 262 VQRLQVLNPSKMKEFVF-KRQKELAFKRQNELEEIHRGAHMDMDAKAARQILTDLIGNID 320
VQ + L K +E + K+ +EL+ ++ +++ + H+D +A
Sbjct: 716 VQESKDL---KEREVAYLKKIEELSVANESLVDKETKLQHIDQEA--------------- 757
Query: 321 MSELLQGMD-KQIRKAKEQAQSRRDIIDRVAEWKSAAEEEKWLDEYE 366
E L+G + ++K +E ++ +++D VA ++ AEE K L E E
Sbjct: 758 --EELRGREASHLKKIEELSKENENLVDNVANMQNIAEESKDLRERE 802
Score = 41.6 bits (96), Expect = 8e-04
Identities = 66/378 (17%), Positives = 149/378 (38%), Gaps = 52/378 (13%)
Query: 21 IKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGKGTLKQQL----ANIIPVVEDLRFK 76
+K + N + S + N+ ED + + +LK L + + E L
Sbjct: 521 VKKSEEENSSSQEEVSRLVNLLKESEEDACARKEEEASLKNNLKVAEGEVKYLQETLGEA 580
Query: 77 KKERVK---EILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLTMEKLGKLKSHLNELQ 133
K E +K +L+ + + + AE + + +G + +++ E L ++ L +
Sbjct: 581 KAESMKLKESLLDKEEDLKNVTAEISSLREWEGSVLEKIEELSKVKESLVDKETKLQSIT 640
Query: 134 NEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNGAQQSISDETLARLNE 193
E + +H+ I ELS + SL D + Q + + + E
Sbjct: 641 QEAEELKGREAAHMKQIEELSTA------------NASLVDEATKLQSIVQESEDLKEKE 688
Query: 194 SVLLLKQEKQQSLQKVQFLEELWDFMGITTDEQKAVCDDVIGLISASVDEVSIHGSLSND 253
+ L K E + S+ + + D I + + +V L ++E+S+ D
Sbjct: 689 AGYLKKIE-ELSVANESLADNVTDLQSIVQESKDLKEREVAYL--KKIEELSVANESLVD 745
Query: 254 I---VKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEEIHRGAHMDMDAKAARQ 310
++ +D E + L+ S +K+ +EL+ + +N ++ + ++ ++K R+
Sbjct: 746 KETKLQHIDQEAEELRGREASHLKKI-----EELSKENENLVDNVANMQNIAEESKDLRE 800
Query: 311 -----------------ILTDLIGNI-----DMSELLQGMDKQIRKAKEQAQSRRDIIDR 348
L D + N+ + EL + ++KA+E ++ ++D+
Sbjct: 801 REVAYLKKIDELSTANGTLADNVTNLQNISEENKELRERETTLLKKAEELSELNESLVDK 860
Query: 349 VAEWKSAAEEEKWLDEYE 366
++ ++ +E + L E E
Sbjct: 861 ASKLQTVVQENEELRERE 878
Score = 37.4 bits (85), Expect = 0.016
Identities = 75/375 (20%), Positives = 152/375 (40%), Gaps = 55/375 (14%)
Query: 10 SEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGKGTLKQQLANIIPV 69
++++ R V+ T + + LN++ +D + E + K L +
Sbjct: 44 TKVHSRLVKGT-ELQTQLNQIQEDLKKA--------DEQIELLKKDKAKAIDDLKESEKL 94
Query: 70 VEDLRFKKKE------RVKEILEIKSQISQIRAEKAGCG--QSKGVTDQ----------- 110
VE+ K KE R +E E++ + + E+AG Q K VT +
Sbjct: 95 VEEANEKLKEALAAQKRAEESFEVE-KFRAVELEQAGLEAVQKKDVTSKNELESIRSQHA 153
Query: 111 -DVDQCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILN--- 166
D+ T E+L ++K L+ + KN K SH ++++ + + + +EIL
Sbjct: 154 LDISALLSTTEELQRVKHELSMTADAKN----KALSHAEEATKIAEIHA-EKAEILASEL 208
Query: 167 GIHPSLNDSSNGAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLEELWDFMGITTDEQ 226
G +L S + +E +++L + LL+ E L+KV LE + EQ
Sbjct: 209 GRLKALLGSKEEKEAIEGNEIVSKLKSEIELLRGE----LEKVSILES-------SLKEQ 257
Query: 227 KAVCDDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQV-LNPSKMKEFVFKRQKELA 285
+ + + + ++ + S +N V++ +V L+ + S + E
Sbjct: 258 EGLVEQ----LKVDLEAAKMAESCTNSSVEEWKNKVHELEKEVEESNRSKSSASESMESV 313
Query: 286 FKRQNELEEIHRGAHMDMDAKAARQILTDLIGNIDMSELLQGMDKQIRKAKEQAQSRRDI 345
K+ EL + D A+ + L + ++L + +Q+ AKE+A ++
Sbjct: 314 MKQLAELNHVLHETKSDNAAQKEKIELLEKTIEAQRTDL-EEYGRQVCIAKEEASKLENL 372
Query: 346 IDRVAEWKSAAEEEK 360
++ + ++EEK
Sbjct: 373 VESIKSELEISQEEK 387
>At1g64330 unknown protein
Length = 555
Score = 46.6 bits (109), Expect = 3e-05
Identities = 97/465 (20%), Positives = 202/465 (42%), Gaps = 72/465 (15%)
Query: 7 QEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGE-DYVSFSRGKGT-----LK 60
+EY +YH+ + T + KV +++ ++ +SS + D S G+G LK
Sbjct: 68 KEYESLYHQYDDLT---GEIRKKVHGKGENDSSSSSSSDSDSDKKSKRNGRGENEIELLK 124
Query: 61 QQLANIIPVVEDLRFK--KKERVKEILEIKSQ--ISQIRAEKAGCGQSKGVTDQDVDQCD 116
+Q+ + + DL+ K + KE +E + Q + +++ CG + T++ +
Sbjct: 125 KQMEDANLEIADLKMKLATTDEHKEAVESEHQEILKKLKESDEICGNLRVETEKLTSENK 184
Query: 117 LTMEKL---GKLKSHLNE-LQNEKNIRH-------QKVKSHISTISELSAVMSIDIS--- 162
EKL G+ +S LN+ L++ K R K K H ST+ E++ +
Sbjct: 185 ELNEKLEVAGETESDLNQKLEDVKKERDGLEAELASKAKDHESTLEEVNRLQGQKNETEA 244
Query: 163 --EILNGIHPSLNDSSNGAQQSISDE-----TLARLNESVLLLKQEKQQSLQKV------ 209
E P+L + N Q+++ ++ TL++ ++ + L +E++ +++K+
Sbjct: 245 ELEREKQEKPALLNQINDVQKALLEQEAAYNTLSQEHKQINGLFEEREATIKKLTDDYKQ 304
Query: 210 --QFLEELWDFMGITTDEQKAVCDDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQV 267
+ LEE M T + DV SA VD SL N++ ++ D ++
Sbjct: 305 AREMLEEYMSKMEETERRMQETGKDVASRESAIVDLEETVESLRNEVERKGDEIESLMEK 364
Query: 268 LNPSKMKEFVFKRQ----KELAFKRQNELEEIHRGAHMDMDAKAARQILTDLIGNIDMSE 323
++ ++K + ++ +++ +++ EL+ I H++ A +I T E
Sbjct: 365 MSNIEVKLRLSNQKLRVTEQVLTEKEGELKRIE-AKHLEEQALLEEKIATT-------HE 416
Query: 324 LLQGMDKQIRKAKEQAQSRRDIIDRVAEWKSAAEEEKWLDEYERFI*SVRRKRGFLSPRF 383
+G+ K+I + + I++R EE+ YE+ + V + L+ +
Sbjct: 417 TYRGLIKEISERVDST-----ILNRFQSLSEKLEEKH--KSYEKTV--VEATKMLLTAKK 467
Query: 384 HLW*RI*LQK*KHGRQTKKKVSYMKSLHEYNVQRQLREEEKRKSR 428
+ ++ K +++ K E ++ Q+REEEK K +
Sbjct: 468 CV---------VEMKKEKDEMAKEKEEVEKKLEGQVREEEKEKEK 503
>At1g51410 cinnamyl alcohol dehydrogenase, putative
Length = 809
Score = 43.9 bits (102), Expect = 2e-04
Identities = 62/293 (21%), Positives = 131/293 (44%), Gaps = 36/293 (12%)
Query: 29 KVLDDFQSEIANIASSLGEDYVSFS--RGKGTLKQQLANIIPVVEDLRFKKKERVKEILE 86
KV D + ++ ++SS D V + R + + + N++ + +++ KK ER K+ +E
Sbjct: 97 KVADGREIGVSGLSSSSAVDEVEEAKWRFQAEMLRSECNLLRIEKEIALKKMERRKKRME 156
Query: 87 --IKSQISQIRAEKAGCGQSKGVTDQDVDQCDLTMEKLGKLKSHLNELQNEKNIRH---- 140
++S + + + K + + D+ +EKL +LKS + +N +H
Sbjct: 157 RTLRSAVLTLLSGKERISEGTKESKVLEDEISYLVEKLNELKSPKVKDMEARNFKHNFDK 216
Query: 141 ------QKVKSHISTISELSAVMSID-ISEILNGIHPSLN--DSSNGAQQSISDETLARL 191
++++ +SE V I ++E +H N ++NG ++S + + L
Sbjct: 217 QASVLRRELERFDEAVSEEVCVKGIQKMAEASFSVHSDQNALRNNNGNIDTLSSK-MEAL 275
Query: 192 NESVLLLKQEKQQSLQKV----QFLEELWDFMGITTDEQKAVCDDVIGLISASV-DEVSI 246
++ VLL + EK+ V +++++ GI E+K C ++ + DEV
Sbjct: 276 SKGVLLERMEKEYGSSLVAPSSSSVQDMY-CKGIKAHEEKKDCSRHCKVVMRKIADEVRA 334
Query: 247 HG---SLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEEIH 296
S +++ QV E++ LQ ++F R A + +E++ +H
Sbjct: 335 EAEQWSQMQEMLNQVRKEMEELQ-----SCRDFWQNR----ALEADSEIQNLH 378
>At1g05320 unknown protein
Length = 841
Score = 43.9 bits (102), Expect = 2e-04
Identities = 73/336 (21%), Positives = 130/336 (37%), Gaps = 42/336 (12%)
Query: 7 QEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGKGTLKQQLANI 66
+E +Y K+ + L + LD +E +A + + + +G L +
Sbjct: 371 EEKIRVYEGKLAEACGQSLSLQEELDQSSAENELLADTNNQLKIKIQELEGYLDSEKETA 430
Query: 67 IPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLTMEKLGKLK 126
I E L K E I ++KS + I K ++ GV D + + + KL L+
Sbjct: 431 I---EKLNQKDTEAKDLITKLKSHENVIEEHKRQVLEASGVADTRKVEVEEALLKLNTLE 487
Query: 127 SHLNELQNEK------NIR-HQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNGA 179
S + EL+ E NI+ +QK+ + S + A +S+ +E +
Sbjct: 488 STIEELEKENGDLAEVNIKLNQKLANQGSETDDFQAKLSVLEAEKYQQAKELQITIEDLT 547
Query: 180 QQSISDETLARLNESVLLLKQEKQQSLQKVQFLEELWDFMGITTDEQKAVCDDVIGLI-- 237
+Q S+ RL + L++EK Q + Q + + K+ DD++ I
Sbjct: 548 KQLTSERE--RLRSQISSLEEEKNQVNEIYQSTKNELVKLQAQLQVDKSKSDDMVSQIEK 605
Query: 238 -SASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFK----------------- 279
SA V E S+ S + + EV+++ L SK++E K
Sbjct: 606 LSALVAEKSVLESKFEQVEIHLKEEVEKVAELT-SKLQEHKHKASDRDVLEEKAIQLHKE 664
Query: 280 ---------RQKELAFKRQNELEEIHRGAHMDMDAK 306
QKE + +ELE + + ++DAK
Sbjct: 665 LQASHTAISEQKEALSHKHSELEATLKKSQEELDAK 700
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 40.4 bits (93), Expect = 0.002
Identities = 63/297 (21%), Positives = 134/297 (44%), Gaps = 50/297 (16%)
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAE--------KAGCGQSKGVTDQ 110
L+QQ+A++ +L + +++ +EI E SQI+ ++ E ++ + G++++
Sbjct: 877 LRQQVASLDSQRAELEIQLEKKSEEISEYLSQITNLKEEIINKVKVHESILEEINGLSEK 936
Query: 111 DVDQCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHP 170
+ +L +E LGK +S L+E K + + V+ H S +M+ ++E++N +
Sbjct: 937 -IKGRELELETLGKQRSELDEELRTK--KEENVQMHDKINVASSEIMA--LTELINNLKN 991
Query: 171 SLNDSSNGAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLEELWDFMGITTDEQKAVC 230
L+ + Q ET A L KQEK + ++ TD QKA+
Sbjct: 992 ELD-----SLQVQKSETEAELERE----KQEKSELSNQI-------------TDVQKALV 1029
Query: 231 DDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQN 290
+ A+ + + N++ K+ + + ++ V + + + + +R KE+ R +
Sbjct: 1030 EQ-----EAAYNTLEEEHKQINELFKETEATLNKVTV-DYKEAQRLLEERGKEVT-SRDS 1082
Query: 291 EL---EEIHRGAHMDMDAKAAR-QILTDLIGNIDMSELLQG----MDKQIRKAKEQA 339
+ EE +++ K + L + I NI++ L + +Q+ KE+A
Sbjct: 1083 TIGVHEETMESLRNELEMKGDEIETLMEKISNIEVKLRLSNQKLRVTEQVLTEKEEA 1139
Score = 37.4 bits (85), Expect = 0.016
Identities = 75/405 (18%), Positives = 158/405 (38%), Gaps = 72/405 (17%)
Query: 16 KVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGKGTLKQQLANIIPVVEDLRF 75
++E + + L+ L + E I+S E + + T+++ +A + + + R
Sbjct: 162 QLESSKQQVSDLSASLKAAEEENKAISSKNVETMNKLEQTQNTIQELMAELGKLKDSHRE 221
Query: 76 KKKE--RVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLTMEKLGKLKSHLNELQ 133
K+ E + E+ E + S I ++ +Q + + + + +L LN +
Sbjct: 222 KESELSSLVEVHETHQRDSSIHVKELE------------EQVESSKKLVAELNQTLNNAE 269
Query: 134 NEKNIRHQKVKSHISTISELSAVMSIDISE----------------ILNGIHPS------ 171
EK + QK+ + I E + +SE L IH +
Sbjct: 270 EEKKVLSQKIAELSNEIKEAQNTIQELVSESGQLKESHSVKDRDLFSLRDIHETHQRESS 329
Query: 172 -----LNDSSNGAQQSISDETL-----ARLNESVLLLKQEKQQSLQKVQ-FLEELWDFMG 220
L ++Q ISD T+ N+++ E L++ Q ++EL D +G
Sbjct: 330 TRVSELEAQLESSEQRISDLTVDLKDAEEENKAISSKNLEIMDKLEQAQNTIKELMDELG 389
Query: 221 ITTDEQKAVCDDVIGLISA----------SVDEVSIHGSLSNDIVKQVDVEVQRLQ---- 266
D K ++ L+ + S+D + + + + E+Q Q
Sbjct: 390 ELKDRHKEKESELSSLVKSADQQVADMKQSLDNAEEEKKMLSQRILDISNEIQEAQKTIQ 449
Query: 267 --VLNPSKMKEFVFKRQKELAFKRQ----NELEEIHRGAHMDMDAKAARQILTDLIGNID 320
+ ++KE +++EL R ++ E R + ++ K Q + DL +++
Sbjct: 450 EHMSESEQLKESHGVKERELTGLRDIHETHQRESSTRLSELETQLKLLEQRVVDLSASLN 509
Query: 321 MSE----LLQGMDKQIRKAKEQAQSR-RDIIDRVAEWKSAAEEEK 360
+E L M +I +QAQS+ ++++ +AE K +++
Sbjct: 510 AAEEEKKSLSSMILEITDELKQAQSKVQELVTELAESKDTLTQKE 554
>At2g27170 putative chromosome associated protein
Length = 1163
Score = 40.0 bits (92), Expect = 0.002
Identities = 43/180 (23%), Positives = 82/180 (44%), Gaps = 31/180 (17%)
Query: 3 LQIEQEYSEIYHRKVEDTIKHK--DYLNKVLDDFQSEIANIASSL-------GEDYVSFS 53
LQ+EQ EI + + HK +Y K+L D ++ I + SS+ G + V
Sbjct: 679 LQVEQLKQEIANANKQKHAIHKAIEYKEKLLGDIRTRIDQVRSSMSMKEAEMGTELVDHL 738
Query: 54 RGKGTLKQQLANIIPVVEDLRFKK---------KERVKEILE------IKSQISQIRAEK 98
+ ++QL+ + P ++DL+ KK +E K LE +K +I++++A
Sbjct: 739 TPEE--REQLSKLNPEIKDLKEKKFAYQADRIERETRKAELEANIATNLKRRITELQATI 796
Query: 99 AGCGQ-----SKGVTDQDVDQCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISEL 153
A S G +Q++D L++ + K + + +EK + +K+K + + L
Sbjct: 797 ASIDDDSLPSSAGTKEQELDDAKLSVNEAAKELKSVCDSIDEKTKQIKKIKDEKAKLKTL 856
>At5g16730 putative protein
Length = 853
Score = 38.9 bits (89), Expect = 0.005
Identities = 66/337 (19%), Positives = 150/337 (43%), Gaps = 27/337 (8%)
Query: 42 ASSLGEDYVSFSRGKGTLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGC 101
A L E ++ + + L +++ +E K + EI ++K +I + A
Sbjct: 326 AKELEEQLEEANKLERSASVSLESVMKQLEGSNDKLHDTETEITDLKERIVTLETTVAKQ 385
Query: 102 GQSKGVTDQDVDQCDLTMEK----LGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVM 157
+ V++Q + + + K + KLKS L ++ EKN +K + S + LS
Sbjct: 386 KEDLEVSEQRLGSVEEEVSKNEKEVEKLKSELETVKEEKNRALKKEQDATSRVQRLSEEK 445
Query: 158 SIDISEILNGIHPSLNDSSNGAQQSISDETLARLNESVLLLKQE--KQQSLQKVQFLEEL 215
S +S++ + + S A +S++ L ++ LK++ Q + +++L
Sbjct: 446 SKLLSDLESS--KEEEEKSKKAMESLA-SALHEVSSEGRELKEKLLSQGDHEYETQIDDL 502
Query: 216 WDFMGITTDEQKAVCDDV---IGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLN-PS 271
+ T ++ + + D+ I ++ ++V++ H S + D E++ ++N
Sbjct: 503 KLVIKATNEKYENMLDEARHEIDVLVSAVEQTKKHFESS-----KKDWEMKEANLVNYVK 557
Query: 272 KMKEFVFKRQKELAFKRQNELEEIHRGAHMDMDAKAARQILT-DLIGNIDMSELLQGMDK 330
KM+E V KE+ N L+ + + + DA ++ T D + ++ E + + +
Sbjct: 558 KMEEDVASMGKEM-----NRLDNLLKRTEEEADAAWKKEAQTKDSLKEVE--EEIVYLQE 610
Query: 331 QIRKAK-EQAQSRRDIIDRVAEWKSAAEEEKWLDEYE 366
+ +AK E + + +++D+ E+++ E + L E
Sbjct: 611 TLGEAKAESMKLKENLLDKETEFQNVIHENEDLKAKE 647
Score = 30.0 bits (66), Expect = 2.6
Identities = 39/224 (17%), Positives = 93/224 (41%), Gaps = 27/224 (12%)
Query: 1 MLLQIEQEY-SEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGKGTL 59
+L Q + EY ++I K+ ++ Y N +LD+ + EI + S++ + F K
Sbjct: 487 LLSQGDHEYETQIDDLKLVIKATNEKYEN-MLDEARHEIDVLVSAVEQTKKHFESSKKDW 545
Query: 60 KQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCD--- 116
+ + AN++ V+ + KE+ + + + + E + + T + + +
Sbjct: 546 EMKEANLVNYVKKMEEDVASMGKEMNRLDNLLKRTEEEADAAWKKEAQTKDSLKEVEEEI 605
Query: 117 -LTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDS 175
E LG+ K+ +L+ + + ++ I +L A + + +I
Sbjct: 606 VYLQETLGEAKAESMKLKENLLDKETEFQNVIHENEDLKAKEDVSLKKI----------- 654
Query: 176 SNGAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLEELWDFM 219
E L++L E +L K++ ++ ++ E+ +D +
Sbjct: 655 ----------EELSKLLEEAILAKKQPEEENGELSESEKDYDLL 688
>At2g28620 putative kinesin-like spindle protein
Length = 1076
Score = 38.9 bits (89), Expect = 0.005
Identities = 76/372 (20%), Positives = 146/372 (38%), Gaps = 65/372 (17%)
Query: 2 LLQIEQEYSEIYHRKVEDTIKHKDYL-------NKVLDD----FQSEIANIASSLGEDYV 50
LL +E++ HR+ TIK K+YL K L D Q+E+AN AS + +
Sbjct: 501 LLDLEEK-----HRQAVATIKEKEYLISNLLKSEKTLVDRAVELQAELANAASDVSNLFA 555
Query: 51 SFSRGKGTLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQ 110
R K ++ ++I + ++ E + + +SQ Q K + D
Sbjct: 556 KIGR-KDKIEDSNRSLIQDFQSQLLRQLELLNN--SVAGSVSQ---------QEKQLQDM 603
Query: 111 DVDQCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHP 170
+ K ++ L K +K + I ++ +++ + D LN ++
Sbjct: 604 ENVMVSFVSAKTKATETLRGSLAQLK----EKYNTGIKSLDDIAGNLDKDSQSTLNDLNS 659
Query: 171 SLNDSSNGAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLEELWDFMGITTDEQKAVC 230
+ S + T +E+ LL + Q SL + E+L F T +Q+
Sbjct: 660 EVTKHSCALEDMFKGFT----SEAYTLL-EGLQGSLHNQE--EKLSAF----TQQQR--- 705
Query: 231 DDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRL-------QVLNPSKMKEFVFKRQKE 283
L S S+D ++ D K +D +L Q +N K+ F K ++
Sbjct: 706 ----DLHSRSMDSAKSVSTVMLDFFKTLDTHANKLTKLAEDAQNVNEQKLSAFTKKFEES 761
Query: 284 LAFKRQNELEEIHRGAHMDMDAKAARQILTDLIGNIDMSELLQGMDKQIRKAKEQAQSRR 343
+A + + LE++ A + + A ++ L I + ++ QG Q +++ + +
Sbjct: 762 IANEEKQMLEKV---AELLASSNARKKELV----QIAVQDIRQGSSSQTGALQQEMSAMQ 814
Query: 344 DIIDRV-AEWKS 354
D + +W S
Sbjct: 815 DSASSIKVQWNS 826
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 38.5 bits (88), Expect = 0.007
Identities = 59/303 (19%), Positives = 139/303 (45%), Gaps = 28/303 (9%)
Query: 69 VVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLTMEKLG----K 124
V + + + ++ + ++ ++S I ++ A+ G + G + + +L + G +
Sbjct: 968 VADTRKVELEDALSKLKNLESTIEELGAKCQGLEKESGDLAEVNLKLNLELANHGSEANE 1027
Query: 125 LKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGI--HPSLNDSSNGAQQS 182
L++ L+ L+ EK ++++ +TI +L+ ++ + ++ + I H N+ N QS
Sbjct: 1028 LQTKLSALEAEKEQTANELEASKTTIEDLTKQLTSEGEKLQSQISSHTEENNQVNAMFQS 1087
Query: 183 ISDE---TLARLNESVLLLKQEKQQSLQKVQ----------FLEELWDFMGITTDEQKA- 228
+E +A+L E + + + + +++ LE ++ + T E KA
Sbjct: 1088 TKEELQSVIAKLEEQLTVESSKADTLVSEIEKLRAVAAEKSVLESHFEELEKTLSEVKAQ 1147
Query: 229 VCDDVIGLISASVDEVSIHGSLSND--IVKQVDV-EVQRLQVLNPSKMKEFVFKRQKELA 285
+ ++V +ASV + L I + DV Q LQ+ + + QK+
Sbjct: 1148 LKENVENAATASVKVAELTSKLQEHEHIAGERDVLNEQVLQLQKELQAAQSSIDEQKQAH 1207
Query: 286 FKRQNELEEIHRGAHMDMDAKAARQILTDLIGNI-DMSELLQGMDKQIR--KAKEQAQSR 342
++Q+ELE + + +++AK ++ +T+ + D+ + +Q D + + +A +
Sbjct: 1208 SQKQSELESALKKSQEEIEAK--KKAVTEFESMVKDLEQKVQLADAKTKETEAMDVGVKS 1265
Query: 343 RDI 345
RDI
Sbjct: 1266 RDI 1268
>At5g11390 unknown protein
Length = 703
Score = 38.1 bits (87), Expect = 0.009
Identities = 41/201 (20%), Positives = 83/201 (40%), Gaps = 17/201 (8%)
Query: 174 DSSNG-------AQQSISDETLARLNESVLLLKQEKQQSLQKVQFLEELWDFMGITTDEQ 226
DSSN AQ E+L E ++LL E +KV LEE + GI T++
Sbjct: 345 DSSNARLADFLVAQTEGLKESLQEAEEKLILLNTENSTLSEKVSSLEEQLNEYGIQTEDA 404
Query: 227 KAVCDDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAF 286
A +I + +E+ + + ++ + + + L+ E R K
Sbjct: 405 DATSGALITDLERINEELKDKLAKTEARAEETESKCKILEESKKELQDELGNFRDKGFTI 464
Query: 287 KRQNELEEIHRGAHMDMD--------AKAARQILTDLIGNIDMSELLQGMDKQIRKAKEQ 338
+ LE+ R + + ++ +K + +L + DM ++++ + ++ KA+ +
Sbjct: 465 HKLASLEKHLRDSDLQLEHAVAAVEASKEKQNLLYSTVS--DMEDVIEDLKSKVLKAENR 522
Query: 339 AQSRRDIIDRVAEWKSAAEEE 359
A + + V+E + EE
Sbjct: 523 ADITEEKLIMVSESNAEVNEE 543
>At1g79280 hypothetical protein
Length = 2111
Score = 38.1 bits (87), Expect = 0.009
Identities = 80/409 (19%), Positives = 167/409 (40%), Gaps = 76/409 (18%)
Query: 2 LLQIEQEYSEIYHRKVEDTIKHKD---YLNKVLDDFQSEIANIASSLGEDYVSFSRGKGT 58
L +I ++Y E + + K+ L +VL + + + I GE Y
Sbjct: 413 LAKIYEKYQEAVDAMRHEQLGRKEAEMILQRVLSELEEKAGFIQEERGE-YERVVEAYCL 471
Query: 59 LKQQLAN-----------IIPVVEDLRFKKKERV---KEILEIKSQIS----QIRAEKAG 100
+ Q+L + I+ + DLR +++E K+I +++ Q++ + R +
Sbjct: 472 VNQKLQDSVSEQSNMEKFIMELKADLRRRERENTLLQKDISDLQKQVTILLKECRDVQLR 531
Query: 101 CGQSKGVTDQDVDQCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSID 160
CG ++ D D D L + +++S +++ +E ++ + + + +L ++
Sbjct: 532 CGAAR---DDDEDDYPLLSDVEMEMESEADKIISEHLLKFKDINGLVEQNVKLRNLVR-S 587
Query: 161 ISEILNGIHPSLNDSSNGAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLEELWDFMG 220
+SE + L ++ ++ +DE A++ +VL +E+ Q ++ + + +
Sbjct: 588 LSEQIESRETELKETFEVDLKNKTDEASAKV-ATVLKRAEEQGQMIESLHTSVAM--YKR 644
Query: 221 ITTDEQKAVCDDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKR 280
+ +EQK D S S D + G R L+ + E KR
Sbjct: 645 LYEEEQKLHSSD-----SRSSDLSPVPG---------------RKNFLHLLEDSEEATKR 684
Query: 281 QKELAFKRQNELEEIHRGAHMD------------MDAKAARQIL---------------T 313
+E AF+R LEE A + M+A AR+ L +
Sbjct: 685 AQEKAFERIRILEEDFAKARSEVIAIRSERDKLAMEANFAREKLEGIMKESERKREEMNS 744
Query: 314 DLIGNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEWKSAAEEEKWL 362
L NI+ S+L+ +++R++ E + +I +++ S ++EK L
Sbjct: 745 VLARNIEFSQLIIDHQRKLRESSESLHAAEEISRKLSMEVSVLKQEKEL 793
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.135 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,488,973
Number of Sequences: 26719
Number of extensions: 338006
Number of successful extensions: 1787
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 90
Number of HSP's that attempted gapping in prelim test: 1648
Number of HSP's gapped (non-prelim): 177
length of query: 428
length of database: 11,318,596
effective HSP length: 102
effective length of query: 326
effective length of database: 8,593,258
effective search space: 2801402108
effective search space used: 2801402108
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137821.2


