

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137703.11 + phase: 0
(323 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
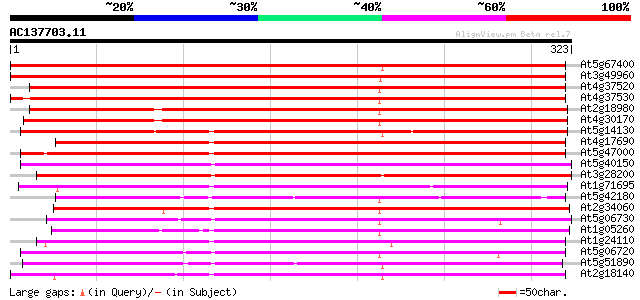
Score E
Sequences producing significant alignments: (bits) Value
At5g67400 peroxidase (emb|CAA66967.1) 448 e-126
At3g49960 peroxidase ATP21a 446 e-126
At4g37520 peroxidase, prxr2 446 e-125
At4g37530 peroxidase - like protein 442 e-124
At2g18980 peroxidase (ATP22a) 417 e-117
At4g30170 peroxidase ATP8a 417 e-117
At5g14130 peroxidase ATP20a (emb|CAA67338.1) 337 5e-93
At4g17690 peroxidase like protein 275 3e-74
At5g47000 peroxidase 262 2e-70
At5g40150 peroxidase ATP26a 253 8e-68
At3g28200 peroxidase like protein 246 9e-66
At1g71695 peroxidase ATP4a 239 1e-63
At5g42180 peroxidase (emb|CAA66960.1) 239 2e-63
At2g34060 putative peroxidase 238 3e-63
At5g06730 peroxidase 235 2e-62
At1g05260 putative peroxidase 232 2e-61
At1g24110 putative peroxidase 229 1e-60
At5g06720 peroxidase (emb|CAA68212.1) 228 3e-60
At5g51890 peroxidase 227 6e-60
At2g18140 putative peroxidase 225 3e-59
>At5g67400 peroxidase (emb|CAA66967.1)
Length = 329
Score = 448 bits (1153), Expect = e-126
Identities = 219/325 (67%), Positives = 256/325 (78%), Gaps = 5/325 (1%)
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
M R+++++V +L+L + + P TT AQL N Y N CPNV+ IV+ VQ+K +QTFVT+PA
Sbjct: 1 MARFSLVVVVTLSLAISMFPDTTTAQLKTNFYGNSCPNVEQIVKKVVQEKIKQTFVTIPA 60
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRLFFHDCFV GCDASV++ S+ NKAEKDHP+N+SLAGDGFD VIKAK ALDA+P C+
Sbjct: 61 TLRLFFHDCFVNGCDASVMIQSTPTNKAEKDHPDNISLAGDGFDVVIKAKKALDAIPSCK 120
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADILALATRDV+ A GPSY VELGRFDGLVS ++ VNG LP P+ + +LN LF
Sbjct: 121 NKVSCADILALATRDVVVAAKGPSYAVELGRFDGLVSTAASVNGNLPGPNNKVTELNKLF 180
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNV 235
A N LTQ DMIALS AHTLGF+HC + NRI VDPTLNK YA +LQ CP+ V
Sbjct: 181 AKNKLTQEDMIALSAAHTLGFAHCGKVFNRIYNFNLTHAVDPTLNKAYAKELQLACPKTV 240
Query: 236 DPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNAN 295
DPRIAINMDPTTPR FDN+Y+KNLQQGKGLFTSDQ+LFTD RS+ TVN +A N FN
Sbjct: 241 DPRIAINMDPTTPRQFDNIYFKNLQQGKGLFTSDQVLFTDGRSKPTVNDWAKNSVAFNKA 300
Query: 296 FITAMTKLGRVGVKNARNGKIRTDC 320
F+TAMTKLGRVGVK RNG IR DC
Sbjct: 301 FVTAMTKLGRVGVKTRRNGNIRRDC 325
>At3g49960 peroxidase ATP21a
Length = 329
Score = 446 bits (1148), Expect = e-126
Identities = 219/325 (67%), Positives = 256/325 (78%), Gaps = 5/325 (1%)
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
M R++++L+ L L + + P TT AQLS Y+ CPNV+ IVR+AVQKK ++TFV VPA
Sbjct: 1 MARFDIVLLIGLCLIISVFPDTTTAQLSRGFYSKTCPNVEQIVRNAVQKKIKKTFVAVPA 60
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRLFFHDCFV GCDASV++ S+ NKAEKDHP+N+SLAGDGFD VI+AK ALD+ P CR
Sbjct: 61 TLRLFFHDCFVNGCDASVMIQSTPKNKAEKDHPDNISLAGDGFDVVIQAKKALDSNPSCR 120
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADIL LATRDV+ AGGPSY VELGRFDGLVS +S V G LP PS N+++LN LF
Sbjct: 121 NKVSCADILTLATRDVVVAAGGPSYEVELGRFDGLVSTASSVEGNLPGPSDNVDKLNALF 180
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQ-----TPVDPTLNKQYAAQLQQMCPRNV 235
N LTQ DMIALS AHTLGF+HC + RI VDPTLNK YA +LQ+ CP+NV
Sbjct: 181 TKNKLTQEDMIALSAAHTLGFAHCGKVFKRIHKFNGINSVDPTLNKAYAIELQKACPKNV 240
Query: 236 DPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNAN 295
DPRIAINMDP TP+TFDN Y+KNLQQGKGLFTSDQ+LFTD RSR TVN++A+N FN
Sbjct: 241 DPRIAINMDPVTPKTFDNTYFKNLQQGKGLFTSDQVLFTDGRSRPTVNAWASNSTAFNRA 300
Query: 296 FITAMTKLGRVGVKNARNGKIRTDC 320
F+ AMTKLGRVGVKN+ NG IR DC
Sbjct: 301 FVIAMTKLGRVGVKNSSNGNIRRDC 325
>At4g37520 peroxidase, prxr2
Length = 329
Score = 446 bits (1146), Expect = e-125
Identities = 218/314 (69%), Positives = 251/314 (79%), Gaps = 5/314 (1%)
Query: 12 LALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFV 71
L L+LCL + AQL N YA CPNV+ IVR+AVQKK QQTF T+PATLRL+FHDCFV
Sbjct: 12 LLLSLCLTLDLSSAQLRRNFYAGSCPNVEQIVRNAVQKKVQQTFTTIPATLRLYFHDCFV 71
Query: 72 QGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILAL 131
GCDASV++AS+ NNKAEKDH ENLSLAGDGFDTVIKAK ALDAVP CRNKVSCADIL +
Sbjct: 72 NGCDASVMIASTNNNKAEKDHEENLSLAGDGFDTVIKAKEALDAVPNCRNKVSCADILTM 131
Query: 132 ATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMI 191
ATRDV+NLAGGP Y VELGR DGL S ++ V G+LP P+ ++N+L +LFA NGL+ DMI
Sbjct: 132 ATRDVVNLAGGPQYDVELGRLDGLSSTAASVGGKLPHPTDDVNKLTSLFAKNGLSLNDMI 191
Query: 192 ALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPT 246
ALSGAHTLGF+HC + NRI T VDPT+NK Y +L+ CPRN+DPR+AINMDPT
Sbjct: 192 ALSGAHTLGFAHCTKVFNRIYTFNKTTKVDPTVNKDYVTELKASCPRNIDPRVAINMDPT 251
Query: 247 TPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRV 306
TPR FDNVYYKNLQQGKGLFTSDQ+LFTD RS+ TV+ +A NG +FN FI +M KLGRV
Sbjct: 252 TPRQFDNVYYKNLQQGKGLFTSDQVLFTDRRSKPTVDLWANNGQLFNQAFINSMIKLGRV 311
Query: 307 GVKNARNGKIRTDC 320
GVK NG IR DC
Sbjct: 312 GVKTGSNGNIRRDC 325
>At4g37530 peroxidase - like protein
Length = 329
Score = 442 bits (1136), Expect = e-124
Identities = 217/325 (66%), Positives = 257/325 (78%), Gaps = 8/325 (2%)
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
M + N++L L L+L L + AQL + YA CPNV+ IVR+AVQKK QQTF T+PA
Sbjct: 4 MNKTNLLL---LILSLFLAINLSSAQLRGDFYAGTCPNVEQIVRNAVQKKIQQTFTTIPA 60
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRL+FHDCFV GCDASV++AS+ NKAEKDH +NLSLAGDGFDTVIKAK A+DAVP CR
Sbjct: 61 TLRLYFHDCFVNGCDASVMIASTNTNKAEKDHEDNLSLAGDGFDTVIKAKEAVDAVPNCR 120
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADIL +ATRDV+NLAGGP Y VELGR DGL S +S V G+LP+P+F+LNQLN LF
Sbjct: 121 NKVSCADILTMATRDVVNLAGGPQYAVELGRRDGLSSSASSVTGKLPKPTFDLNQLNALF 180
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNV 235
A NGL+ DMIALSGAHTLGF+HC + NR+ VDPT+NK Y +L+ CP+N+
Sbjct: 181 AENGLSPNDMIALSGAHTLGFAHCTKVFNRLYNFNKTNNVDPTINKDYVTELKASCPQNI 240
Query: 236 DPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNAN 295
DPR+AINMDP TPR FDNVYYKNLQQGKGLFTSDQ+LFTD+RS+ TV+ +A NG +FN
Sbjct: 241 DPRVAINMDPNTPRQFDNVYYKNLQQGKGLFTSDQVLFTDSRSKPTVDLWANNGQLFNQA 300
Query: 296 FITAMTKLGRVGVKNARNGKIRTDC 320
FI++M KLGRVGVK NG IR DC
Sbjct: 301 FISSMIKLGRVGVKTGSNGNIRRDC 325
>At2g18980 peroxidase (ATP22a)
Length = 323
Score = 417 bits (1073), Expect = e-117
Identities = 210/315 (66%), Positives = 249/315 (78%), Gaps = 9/315 (2%)
Query: 12 LALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFV 71
+AL L + FAQL N Y CPNV++IVR+AV++KFQQTFVT PATLRLFFHDCFV
Sbjct: 10 VALLLIFFSSSVFAQLQTNFYRKSCPNVETIVRNAVRQKFQQTFVTAPATLRLFFHDCFV 69
Query: 72 QGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILAL 131
+GCDAS+L+AS +EKDHP++ SLAGDGFDTV KAK ALD P CRNKVSCADILAL
Sbjct: 70 RGCDASILLASP----SEKDHPDDKSLAGDGFDTVAKAKQALDRDPNCRNKVSCADILAL 125
Query: 132 ATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMI 191
ATRDV+ L GGP+Y VELGR DG +S + V LPQPSF L+QLNT+FA +GL+QTDMI
Sbjct: 126 ATRDVVVLTGGPNYPVELGRRDGRLSTVASVQHSLPQPSFKLDQLNTMFARHGLSQTDMI 185
Query: 192 ALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPT 246
ALSGAHT+GF+HC +FS RI + P+DPTLN +YA QL+QMCP VD RIAINMDPT
Sbjct: 186 ALSGAHTIGFAHCGKFSKRIYNFSPKRPIDPTLNIRYALQLRQMCPIRVDLRIAINMDPT 245
Query: 247 TPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRV 306
+P TFDN Y+KNLQ+G GLFTSDQ+LF+D RSR+TVNSFA++ F FI+A+TKLGRV
Sbjct: 246 SPNTFDNAYFKNLQKGMGLFTSDQVLFSDERSRSTVNSFASSEATFRQAFISAITKLGRV 305
Query: 307 GVKNARNGKIRTDCS 321
GVK G+IR DCS
Sbjct: 306 GVKTGNAGEIRRDCS 320
>At4g30170 peroxidase ATP8a
Length = 325
Score = 417 bits (1071), Expect = e-117
Identities = 206/318 (64%), Positives = 249/318 (77%), Gaps = 9/318 (2%)
Query: 9 VWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHD 68
++S L L+ AQL Y N CPNV++IVR+AV++KFQQTFVT PATLRLFFHD
Sbjct: 9 IFSNFFLLLLLSSCVSAQLRTGFYQNSCPNVETIVRNAVRQKFQQTFVTAPATLRLFFHD 68
Query: 69 CFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADI 128
CFV+GCDAS+++AS +E+DHP+++SLAGDGFDTV+KAK A+D+ P CRNKVSCADI
Sbjct: 69 CFVRGCDASIMIASP----SERDHPDDMSLAGDGFDTVVKAKQAVDSNPNCRNKVSCADI 124
Query: 129 LALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQT 188
LALATR+V+ L GGPSY VELGR DG +S + V +LPQP FNLNQLN +F+ +GL+QT
Sbjct: 125 LALATREVVVLTGGPSYPVELGRRDGRISTKASVQSQLPQPEFNLNQLNGMFSRHGLSQT 184
Query: 189 DMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINM 243
DMIALSGAHT+GF+HC + S RI T +DP++N+ Y QL+QMCP VD RIAINM
Sbjct: 185 DMIALSGAHTIGFAHCGKMSKRIYNFSPTTRIDPSINRGYVVQLKQMCPIGVDVRIAINM 244
Query: 244 DPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKL 303
DPT+PRTFDN Y+KNLQQGKGLFTSDQILFTD RSR+TVNSFA + F FITA+TKL
Sbjct: 245 DPTSPRTFDNAYFKNLQQGKGLFTSDQILFTDQRSRSTVNSFANSEGAFRQAFITAITKL 304
Query: 304 GRVGVKNARNGKIRTDCS 321
GRVGV G+IR DCS
Sbjct: 305 GRVGVLTGNAGEIRRDCS 322
>At5g14130 peroxidase ATP20a (emb|CAA67338.1)
Length = 330
Score = 337 bits (864), Expect = 5e-93
Identities = 179/320 (55%), Positives = 226/320 (69%), Gaps = 9/320 (2%)
Query: 7 ILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFF 66
+++W L + L + + AQLS N+YA+ CP+V+ IV+ AV KF+QT T PATLR+FF
Sbjct: 12 MMMWFLGMLLFSMVAESNAQLSENYYASTCPSVELIVKQAVTTKFKQTVTTAPATLRMFF 71
Query: 67 HDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCA 126
HDCFV+GCDASV +AS N AEKD +N SLAGDGFDTVIKAK A+++ QC VSCA
Sbjct: 72 HDCFVEGCDASVFIASE-NEDAEKDADDNKSLAGDGFDTVIKAKTAVES--QCPGVVSCA 128
Query: 127 DILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLT 186
DILALA RDV+ L GGP + VELGR DGLVS++S V G+LP+P ++ L +FA+NGL+
Sbjct: 129 DILALAARDVVVLVGGPEFKVELGRRDGLVSKASRVTGKLPEPGLDVRGLVQIFASNGLS 188
Query: 187 QTDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDPRIAI 241
TDMIALSGAHT+G SHC+RF+NR+ PVDPT++ YA QL Q C + +P +
Sbjct: 189 LTDMIALSGAHTIGSSHCNRFANRLHNFSTFMPVDPTMDPVYAQQLIQAC-SDPNPDAVV 247
Query: 242 NMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMT 301
++D T+ TFDN YY+NL KGLFTSDQ LF D S+ TV FA N F + F +AM
Sbjct: 248 DIDLTSRDTFDNSYYQNLVARKGLFTSDQALFNDLSSQATVVRFANNAEEFYSAFSSAMR 307
Query: 302 KLGRVGVKNARNGKIRTDCS 321
LGRVGVK G+IR DCS
Sbjct: 308 NLGRVGVKVGNQGEIRRDCS 327
>At4g17690 peroxidase like protein
Length = 326
Score = 275 bits (702), Expect = 3e-74
Identities = 141/295 (47%), Positives = 187/295 (62%), Gaps = 3/295 (1%)
Query: 27 LSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSGNN 86
L+ ++Y CP+ IVR V K Q T TLRLFFHDCF++GCDASVL+A++ N
Sbjct: 26 LTKDYYQKTCPDFNKIVRETVTPKQGQQPTTAAGTLRLFFHDCFMEGCDASVLIATNSFN 85
Query: 87 KAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPSYT 146
KAE+D N SL GD FD V + K AL+ C VSCADILA ATRD++ + GGP Y
Sbjct: 86 KAERDDDLNESLPGDAFDIVTRIKTALEL--SCPGVVSCADILAQATRDLVTMVGGPFYE 143
Query: 147 VELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHCDR 206
V+LGR DG S++ V G LP + ++ + ++F NG T +++ALSG HT+GFSHC
Sbjct: 144 VKLGRKDGFESKAHKVKGNLPLANQSVPDMLSIFKKNGFTLKELVALSGGHTIGFSHCKE 203
Query: 207 FSNRIQTPVDPTLNKQYAAQLQQMCPR-NVDPRIAINMDPTTPRTFDNVYYKNLQQGKGL 265
FSNRI VDP LN ++A L+ +C + +A +DP TP FDN+Y+KNL++G GL
Sbjct: 204 FSNRIFPKVDPELNAKFAGVLKDLCKNFETNKTMAAFLDPVTPGKFDNMYFKNLKRGLGL 263
Query: 266 FTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDC 320
SD ILF D +R V +A N F +F AM KLGRVGVK ++G++R C
Sbjct: 264 LASDHILFKDPSTRPFVELYANNQTAFFEDFARAMEKLGRVGVKGEKDGEVRRRC 318
>At5g47000 peroxidase
Length = 334
Score = 262 bits (669), Expect = 2e-70
Identities = 141/316 (44%), Positives = 197/316 (61%), Gaps = 5/316 (1%)
Query: 7 ILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFF 66
++++ LA+ +I A L ++Y CP+ IVR AV K Q T TLRLFF
Sbjct: 14 VILFCLAVVAPIIS-ADVAILRTDYYQKTCPDFHKIVREAVTTKQVQQPTTAAGTLRLFF 72
Query: 67 HDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCA 126
HDCF++GCDASVL+A++ NKAE+D N SL GD FD V + K AL+ C VSCA
Sbjct: 73 HDCFLEGCDASVLIATNSFNKAERDDDLNDSLPGDAFDIVTRIKTALEL--SCPGVVSCA 130
Query: 127 DILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLT 186
DILA ATRD++ + GGP + V+LGR DG S++ V G +P + + ++ +F NG +
Sbjct: 131 DILAQATRDLVTMVGGPYFDVKLGRKDGFESKAHKVRGNVPMANQTVPDIHGIFKKNGFS 190
Query: 187 QTDMIALSGAHTLGFSHCDRFSNRIQ-TPVDPTLNKQYAAQLQQMCPRN-VDPRIAINMD 244
+M+ALSGAHT+GFSHC FS+R+ + D +N ++AA L+ +C + VD IA D
Sbjct: 191 LREMVALSGAHTIGFSHCKEFSDRLYGSRADKEINPRFAAALKDLCKNHTVDDTIAAFND 250
Query: 245 PTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLG 304
TP FDN+Y+KNL++G GL SD IL D ++ V+ +ATN F +F AM KLG
Sbjct: 251 VMTPGKFDNMYFKNLKRGLGLLASDHILIKDNSTKPFVDLYATNETAFFEDFARAMEKLG 310
Query: 305 RVGVKNARNGKIRTDC 320
VGVK ++G++R C
Sbjct: 311 TVGVKGDKDGEVRRRC 326
>At5g40150 peroxidase ATP26a
Length = 328
Score = 253 bits (647), Expect = 8e-68
Identities = 136/318 (42%), Positives = 191/318 (59%), Gaps = 3/318 (0%)
Query: 7 ILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFF 66
I++ L L+ + + + L+ + Y+ CP I+R + K T T A LRLFF
Sbjct: 12 IILLLLCLSFQSLSFAAESHLTVDFYSKSCPKFLDIIRETITNKQISTPTTAAAALRLFF 71
Query: 67 HDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCA 126
HDCF GCDASVLV+S+ N AE+D NLSL GDGFD VI+AK AL+ C N VSC+
Sbjct: 72 HDCFPNGCDASVLVSSTAFNTAERDSSINLSLPGDGFDVVIRAKTALELA--CPNTVSCS 129
Query: 127 DILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLT 186
DI+A+A RD++ GGP Y + LGR D S+SS V+ LP PS +++L F++ G +
Sbjct: 130 DIIAVAVRDLLVTVGGPYYEISLGRRDSRTSKSSLVSDLLPLPSMQISKLIDQFSSRGFS 189
Query: 187 QTDMIALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNV-DPRIAINMDP 245
+M+ALSGAHT+GFSHC F+NR+ N ++A L++ C + DP I++ D
Sbjct: 190 VQEMVALSGAHTIGFSHCKEFTNRVNPNNSTGYNPRFAVALKKACSNSKNDPTISVFNDV 249
Query: 246 TTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGR 305
TP FDN+Y++N+ +G GL SD LF+D R+R V +A + + F +F AM KL
Sbjct: 250 MTPNKFDNMYFQNIPKGLGLLESDHGLFSDPRTRPFVELYARDQSRFFNDFAGAMQKLSL 309
Query: 306 VGVKNARNGKIRTDCSVL 323
GV R G+IR C +
Sbjct: 310 HGVLTGRRGEIRRRCDAI 327
>At3g28200 peroxidase like protein
Length = 316
Score = 246 bits (629), Expect = 9e-66
Identities = 131/309 (42%), Positives = 188/309 (60%), Gaps = 4/309 (1%)
Query: 16 LCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCD 75
L L +T ++L+ N Y+ CP I+R + K T A +RLFFHDCF GCD
Sbjct: 10 LFLFFFTAQSRLTTNFYSKTCPRFLDIIRDTITNKQITNPTTAAAVIRLFFHDCFPNGCD 69
Query: 76 ASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRD 135
ASVL++S+ N AE+D NLSL GDGFD +++AK AL+ C N VSC+DI+++ATRD
Sbjct: 70 ASVLISSTAFNTAERDSSINLSLPGDGFDVIVRAKTALELA--CPNTVSCSDIISVATRD 127
Query: 136 VINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSG 195
++ GGP Y V LGR D S+SS + LP PS ++++ F + G T +M+ALSG
Sbjct: 128 LLITVGGPYYDVFLGRRDSRTSKSSLLTDLLPLPSTPISKIIQQFESKGFTVQEMVALSG 187
Query: 196 AHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPR-NVDPRIAINMDPTTPRTFDNV 254
AH++GFSHC F R+ + N ++A L++ C DP I++ D TP FDN+
Sbjct: 188 AHSIGFSHCKEFVGRVGRN-NTGYNPRFAVALKKACANYPKDPTISVFNDIMTPNKFDNM 246
Query: 255 YYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNG 314
YY+NL++G GL SD L++D R+R V+ +A N ++F +F AM KL G++ R G
Sbjct: 247 YYQNLKKGLGLLESDHGLYSDPRTRYFVDLYAKNQDLFFKDFAKAMQKLSLFGIQTGRRG 306
Query: 315 KIRTDCSVL 323
+IR C +
Sbjct: 307 EIRRRCDAI 315
>At1g71695 peroxidase ATP4a
Length = 358
Score = 239 bits (610), Expect = 1e-63
Identities = 135/327 (41%), Positives = 187/327 (56%), Gaps = 14/327 (4%)
Query: 6 VILVWSLALTLCLIPYTTFAQ----------LSPNHYANICPNVQSIVRSAVQKKFQQTF 55
+IL+ +A+TL L P + LS N Y CP V++I+R ++K F++
Sbjct: 13 LILISLMAVTLNLFPTVEAKKRSRDAPIVKGLSWNFYQKACPKVENIIRKELKKVFKRDI 72
Query: 56 VTVPATLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDA 115
A LR+ FHDCFVQGC+ASVL+A S + E+ NL+L F + +A +
Sbjct: 73 GLAAAILRIHFHDCFVQGCEASVLLAGSASGPGEQSSIPNLTLRQQAFVVINNLRALVQK 132
Query: 116 VPQCRNKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVN-GRLPQPSFNLN 174
+C VSC+DILALA RD + L+GGP Y V LGR D L S + LP P FN +
Sbjct: 133 --KCGQVVSCSDILALAARDSVVLSGGPDYAVPLGRRDSLAFASQETTLNNLPPPFFNAS 190
Query: 175 QLNTLFANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRN 234
QL FAN L TD++ALSG HT+G +HC F++R+ DPT+N+ +A L++ CP
Sbjct: 191 QLIADFANRNLNITDLVALSGGHTIGIAHCPSFTDRLYPNQDPTMNQFFANSLKRTCPTA 250
Query: 235 VDPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNA 294
+N D +P FDN YY +L +GLFTSDQ LF D R+R V SFA + +F
Sbjct: 251 NSSNTQVN-DIRSPDVFDNKYYVDLMNRQGLFTSDQDLFVDKRTRGIVESFAIDQQLFFD 309
Query: 295 NFITAMTKLGRVGVKNARNGKIRTDCS 321
F AM K+G++ V G+IR++CS
Sbjct: 310 YFTVAMIKMGQMSVLTGTQGEIRSNCS 336
>At5g42180 peroxidase (emb|CAA66960.1)
Length = 317
Score = 239 bits (609), Expect = 2e-63
Identities = 131/300 (43%), Positives = 180/300 (59%), Gaps = 15/300 (5%)
Query: 27 LSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSGNN 86
LSP++Y + CP IV +AV+K A LR+ FHDCFV+GCD SVL+ S G N
Sbjct: 23 LSPHYYDHTCPQADHIVTNAVKKAMSNDQTVPAALLRMHFHDCFVRGCDGSVLLDSKGKN 82
Query: 87 KAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPSYT 146
KAEKD P N+SL F + AK AL+ QC VSCADIL+LA RD + L+GGP++
Sbjct: 83 KAEKDGPPNISL--HAFYVIDNAKKALE--EQCPGIVSCADILSLAARDAVALSGGPTWA 138
Query: 147 VELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHCDR 206
V GR DG +S++ + +LP P+FN++QL F GL+ D++ALSG HTLGF+HC
Sbjct: 139 VPKGRKDGRISKAIETR-QLPAPTFNISQLRQNFGQRGLSMHDLVALSGGHTLGFAHCSS 197
Query: 207 FSNRI-----QTPVDPTLNKQYAAQLQQMCP-RNVDPRIAINMDPTTPRTFDNVYYKNLQ 260
F NR+ Q VDPTLN +AA+L+ +CP N NMD T +FDN+YYK L
Sbjct: 198 FQNRLHKFNTQKEVDPTLNPSFAARLEGVCPAHNTVKNAGSNMDGTV-TSFDNIYYKMLI 256
Query: 261 QGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDC 320
QGK LF+SD+ L ++ V +A + F F+ +M K+ + + ++R +C
Sbjct: 257 QGKSLFSSDESLLAVPSTKKLVAKYANSNEEFERAFVKSMIKMSSI---SGNGNEVRLNC 313
>At2g34060 putative peroxidase
Length = 346
Score = 238 bits (608), Expect = 3e-63
Identities = 126/306 (41%), Positives = 192/306 (62%), Gaps = 11/306 (3%)
Query: 26 QLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSGN 85
+LS ++Y+ CP ++++V S ++F++ ++ PAT+RLFFHDCFV+GCD S+L+ +
Sbjct: 41 ELSADYYSKKCPQLETLVGSVTSQRFKEVPISAPATIRLFFHDCFVEGCDGSILIETKKG 100
Query: 86 NK--AEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGP 143
+K AE++ EN L +GFD++IKAKA +++ C + VSC+DILA+A RD I+LAGGP
Sbjct: 101 SKKLAEREAYENKELREEGFDSIIKAKALVES--HCPSLVSCSDILAIAARDFIHLAGGP 158
Query: 144 SYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSH 203
Y V+ GR+DG S + +V +P+ + ++QL LFA+ GLT +++ LSG+HT+GF+H
Sbjct: 159 YYQVKKGRWDGKRSTAKNVPPNIPRSNSTVDQLIKLFASKGLTVEELVVLSGSHTIGFAH 218
Query: 204 CDRFSNRI-----QTPVDPTLNKQYAAQLQQMCP-RNVDPRIAINMDPTTPRTFDNVYYK 257
C F R+ DP+L+++ +L+ CP + + +D TTP FDN Y+
Sbjct: 219 CKNFLGRLYDYKGTKRPDPSLDQRLLKELRMSCPFSGGSSGVVLPLDATTPFVFDNGYFT 278
Query: 258 NLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNA-RNGKI 316
L GL SDQ LF D R++ A + F F AM K+G +GVK R+G+I
Sbjct: 279 GLGTNMGLLGSDQALFLDPRTKPIALEMARDKQKFLKAFGDAMDKMGSIGVKRGKRHGEI 338
Query: 317 RTDCSV 322
RTDC V
Sbjct: 339 RTDCRV 344
>At5g06730 peroxidase
Length = 358
Score = 235 bits (600), Expect = 2e-62
Identities = 132/309 (42%), Positives = 181/309 (57%), Gaps = 10/309 (3%)
Query: 22 TTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVA 81
T+ AQL+ Y+ CPN +IVRS +Q+ Q + +RL FHDCFV GCD S+L+
Sbjct: 28 TSSAQLNATFYSGTCPNASAIVRSTIQQALQSDARIGGSLIRLHFHDCFVNGCDGSLLLD 87
Query: 82 SSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAG 141
+ + ++EK+ P N + + GF+ V K AL+ C VSC+DILALA+ ++LAG
Sbjct: 88 DTSSIQSEKNAPANAN-STRGFNVVDSIKTALENA--CPGIVSCSDILALASEASVSLAG 144
Query: 142 GPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGF 201
GPS+TV LGR DGL + S N LP P LN + + F GL TD+++LSGAHT G
Sbjct: 145 GPSWTVLLGRRDGLTANLSGANSSLPSPFEGLNNITSKFVAVGLKTTDVVSLSGAHTFGR 204
Query: 202 SHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYY 256
C F+NR+ DPTLN + LQQ+CP+N N+D +TP FDN Y+
Sbjct: 205 GQCVTFNNRLFNFNGTGNPDPTLNSTLLSSLQQLCPQNGSNTGITNLDLSTPDAFDNNYF 264
Query: 257 KNLQQGKGLFTSDQILFTDTRSRNT--VNSFATNGNVFNANFITAMTKLGRVGVKNARNG 314
NLQ GL SDQ LF++T S VNSFA+N +F F+ +M K+G + +G
Sbjct: 265 TNLQSNNGLLQSDQELFSNTGSATVPIVNSFASNQTLFFEAFVQSMIKMGNISPLTGSSG 324
Query: 315 KIRTDCSVL 323
+IR DC V+
Sbjct: 325 EIRQDCKVV 333
>At1g05260 putative peroxidase
Length = 326
Score = 232 bits (591), Expect = 2e-61
Identities = 128/305 (41%), Positives = 180/305 (58%), Gaps = 12/305 (3%)
Query: 25 AQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSG 84
AQL N YAN CPN + IV+ V A +R+ FHDCFV+GCD SVL+ S+
Sbjct: 24 AQLQMNFYANSCPNAEKIVQDFVSNHVSNAPSLAAALIRMHFHDCFVRGCDGSVLINSTS 83
Query: 85 NNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPS 144
N AE+D NL++ G GF IK+ L+A QC VSCADI+ALA+RD + GGP+
Sbjct: 84 GN-AERDATPNLTVRGFGFIDAIKS--VLEA--QCPGIVSCADIIALASRDAVVFTGGPN 138
Query: 145 YTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHC 204
++V GR DG +S +++ +P P+ N+ L TLFAN GL D++ LSGAHT+G SHC
Sbjct: 139 WSVPTGRRDGRISNAAEALANIPPPTSNITNLQTLFANQGLDLKDLVLLSGAHTIGVSHC 198
Query: 205 DRFSNRI-----QTPVDPTLNKQYAAQLQ-QMCPRNVDPRIAINMDPTTPRTFDNVYYKN 258
F+NR+ + DP L+ +YAA L+ + CP D + + MDP + +TFD YY+
Sbjct: 199 SSFTNRLYNFTGRGGQDPALDSEYAANLKSRKCPSLNDNKTIVEMDPGSRKTFDLSYYQL 258
Query: 259 LQQGKGLFTSDQILFTDTRSRNTVNSFATNG-NVFNANFITAMTKLGRVGVKNARNGKIR 317
+ + +GLF SD L T+ + + +N T F + F +M K+GR+ VK G +R
Sbjct: 259 VLKRRGLFQSDSALTTNPTTLSNINRILTGSVGSFFSEFAKSMEKMGRINVKTGSAGVVR 318
Query: 318 TDCSV 322
CSV
Sbjct: 319 RQCSV 323
>At1g24110 putative peroxidase
Length = 326
Score = 229 bits (585), Expect = 1e-60
Identities = 127/315 (40%), Positives = 182/315 (57%), Gaps = 12/315 (3%)
Query: 16 LCLI-----PYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCF 70
LCL P A LS ++Y CP + + V K T TLRLFFHDC
Sbjct: 6 LCLFILVSSPCLLQANLSSDYYTKTCPEFEETLVQIVTDKQIAAPTTAVGTLRLFFHDCM 65
Query: 71 VQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILA 130
V GCDAS+LVAS+ +E+D N SL GD FD + + K A++ +C N VSC+DIL
Sbjct: 66 VDGCDASILVASTPRKTSERDADINRSLPGDAFDVITRIKTAVEL--KCPNIVSCSDILV 123
Query: 131 LATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDM 190
ATR +I++ GGP V+ GR D LVS + V G+L +P+ ++ + ++F ++GLT +M
Sbjct: 124 GATRSLISMVGGPRVNVKFGRKDSLVSDMNRVEGKLARPNMTMDHIISIFESSGLTVQEM 183
Query: 191 IALSGAHTLGFSHCDRFSNRIQTPVDPT----LNKQYAAQLQQMCPRNV-DPRIAINMDP 245
+AL GAHT+GFSHC F++RI D +N +YAA+L+++C D +++ D
Sbjct: 184 VALVGAHTIGFSHCKEFASRIFNKSDQNGPVEMNPKYAAELRKLCANYTNDEQMSAFNDV 243
Query: 246 TTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGR 305
TP FDN+YYKNL+ G GL SD + D R+R+ V+ +A + F F AM K+
Sbjct: 244 FTPGKFDNMYYKNLKHGYGLLQSDHAIAFDNRTRSLVDLYAEDETAFFDAFAKAMEKVSE 303
Query: 306 VGVKNARNGKIRTDC 320
VK + G++R C
Sbjct: 304 KNVKTGKLGEVRRRC 318
>At5g06720 peroxidase (emb|CAA68212.1)
Length = 335
Score = 228 bits (581), Expect = 3e-60
Identities = 130/321 (40%), Positives = 184/321 (56%), Gaps = 10/321 (3%)
Query: 7 ILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFF 66
+ + SL + + I T+ AQL+ Y+ CPN +IVRS +Q+ Q + +RL F
Sbjct: 12 LFIISLIVIVSSIFGTSSAQLNATFYSGTCPNASAIVRSTIQQALQSDTRIGASLIRLHF 71
Query: 67 HDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCA 126
HDCFV GCDAS+L+ +G+ ++EK+ N++ A GF+ V K AL+ C VSC+
Sbjct: 72 HDCFVNGCDASILLDDTGSIQSEKNAGPNVNSAR-GFNVVDNIKTALENA--CPGVVSCS 128
Query: 127 DILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLT 186
D+LALA+ ++LAGGPS+TV LGR D L + + N +P P +L+ + F+ GL
Sbjct: 129 DVLALASEASVSLAGGPSWTVLLGRRDSLTANLAGANSSIPSPIESLSNITFKFSAVGLN 188
Query: 187 QTDMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVDPRIAI 241
D++ALSGAHT G + C F+NR+ DPTLN + LQQ+CP+N
Sbjct: 189 TNDLVALSGAHTFGRARCGVFNNRLFNFSGTGNPDPTLNSTLLSTLQQLCPQNGSASTIT 248
Query: 242 NMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRN--TVNSFATNGNVFNANFITA 299
N+D +TP FDN Y+ NLQ GL SDQ LF+ T S V SFA+N +F F +
Sbjct: 249 NLDLSTPDAFDNNYFANLQSNDGLLQSDQELFSTTGSSTIAIVTSFASNQTLFFQAFAQS 308
Query: 300 MTKLGRVGVKNARNGKIRTDC 320
M +G + NG+IR DC
Sbjct: 309 MINMGNISPLTGSNGEIRLDC 329
>At5g51890 peroxidase
Length = 322
Score = 227 bits (579), Expect = 6e-60
Identities = 124/317 (39%), Positives = 186/317 (58%), Gaps = 11/317 (3%)
Query: 8 LVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFH 67
L++++ + I + A L ++Y CP + I+ V+ LR+FFH
Sbjct: 7 LIFAMIFAVLAIVKPSEAALDAHYYDQSCPAAEKIILETVRNATLYDPKVPARLLRMFFH 66
Query: 68 DCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCAD 127
DCF++GCDAS+L+ S+ +N+AEKD P N+S+ F + AK L+ C VSCAD
Sbjct: 67 DCFIRGCDASILLDSTRSNQAEKDGPPNISVRS--FYVIEDAKRKLEKA--CPRTVSCAD 122
Query: 128 ILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQ 187
++A+A RDV+ L+GGP ++V GR DG +SR+++ LP P+FN++QL FA GL+
Sbjct: 123 VIAIAARDVVTLSGGPYWSVLKGRKDGTISRANETRN-LPPPTFNVSQLIQSFAARGLSV 181
Query: 188 TDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDP-RIAI 241
DM+ LSG HT+GFSHC F +R+Q +DP++N +A L++ CPR + + A
Sbjct: 182 KDMVTLSGGHTIGFSHCSSFESRLQNFSKFHDIDPSMNYAFAQTLKKKCPRTSNRGKNAG 241
Query: 242 NMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMT 301
+ +T FDNVYYK + GKG+F SDQ L D+R++ V +FA + F F +M
Sbjct: 242 TVLDSTSSVFDNVYYKQILSGKGVFGSDQALLGDSRTKWIVETFAQDQKAFFREFAASMV 301
Query: 302 KLGRVGVKNARNGKIRT 318
KLG GVK ++ T
Sbjct: 302 KLGNFGVKETGQVRVNT 318
>At2g18140 putative peroxidase
Length = 337
Score = 225 bits (573), Expect = 3e-59
Identities = 131/331 (39%), Positives = 186/331 (55%), Gaps = 14/331 (4%)
Query: 1 MGRYNVILVWSLALTLCLIPYTTF-----AQLSPNHYANICPNVQSIVRSAVQKKFQQTF 55
+G + ++L + ALTLC+ + L P+ Y + CP + IVRS V K F++
Sbjct: 4 IGSFLILLSLTYALTLCICDNASNFGGNKRNLFPDFYRSSCPRAEEIVRSVVAKAFERET 63
Query: 56 VTVPATLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDA 115
+ +RL FHDCFVQGCD S+L+ +SG+ EK+ N S + GF+ V + KAAL+
Sbjct: 64 RMAASLMRLHFHDCFVQGCDGSLLLDTSGSIVTEKNSNPN-SRSARGFEVVDEIKAALEN 122
Query: 116 VPQCRNKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQ 175
+C N VSCAD L LA RD L GGPS+TV LGR D + + N LP+P +
Sbjct: 123 --ECPNTVSCADALTLAARDSSVLTGGPSWTVPLGRRDSATASRAKPNKDLPEPDNLFDT 180
Query: 176 LNTLFANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQM 230
+ F+N GL TD++ALSG+HT+GFS C F R+ D TL K YAA L+Q
Sbjct: 181 IFLRFSNEGLNLTDLVALSGSHTIGFSRCTSFRQRLYNQSGSGSPDTTLEKSYAAILRQR 240
Query: 231 CPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILF-TDTRSRNTVNSFATNG 289
CPR+ + +D + FDN Y+KNL + GL SDQ+LF ++ +SR V +A +
Sbjct: 241 CPRSGGDQNLSELDINSAGRFDNSYFKNLIENMGLLNSDQVLFSSNEQSRELVKKYAEDQ 300
Query: 290 NVFNANFITAMTKLGRVGVKNARNGKIRTDC 320
F F +M K+G++ +G+IR C
Sbjct: 301 EEFFEQFAESMIKMGKISPLTGSSGEIRKKC 331
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,894,920
Number of Sequences: 26719
Number of extensions: 276648
Number of successful extensions: 1021
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 83
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 680
Number of HSP's gapped (non-prelim): 99
length of query: 323
length of database: 11,318,596
effective HSP length: 99
effective length of query: 224
effective length of database: 8,673,415
effective search space: 1942844960
effective search space used: 1942844960
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137703.11


