

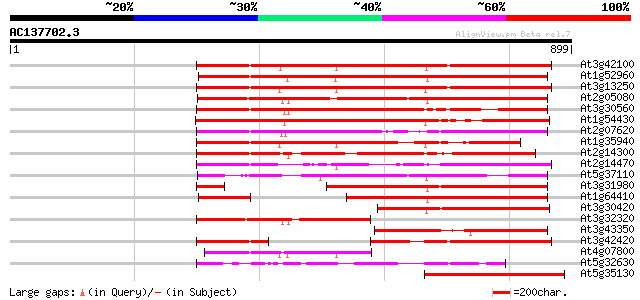
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137702.3 - phase: 2 /pseudo
(899 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g42100 putative protein 564 e-161
At1g52960 hypothetical protein 558 e-159
At3g13250 hypothetical protein 554 e-158
At2g05080 putative helicase 513 e-145
At3g30560 hypothetical protein 487 e-137
At1g54430 hypothetical protein 484 e-137
At2g07620 putative helicase 462 e-130
At1g35940 hypothetical protein 438 e-123
At2g14300 pseudogene; similar to MURA transposase of maize Muta... 431 e-121
At2g14470 pseudogene 383 e-106
At5g37110 putative helicase 376 e-104
At3g31980 hypothetical protein 297 2e-80
At1g64410 unknown protein 293 3e-79
At3g30420 hypothetical protein 250 3e-66
At3g32320 hypothetical protein 245 9e-65
At3g43350 putative protein 244 2e-64
At3g42420 putative protein 229 5e-60
At4g07800 hypothetical protein 224 2e-58
At5g32630 putative protein 212 7e-55
At5g35130 putative protein 176 5e-44
>At3g42100 putative protein
Length = 1752
Score = 564 bits (1453), Expect = e-161
Identities = 291/604 (48%), Positives = 403/604 (66%), Gaps = 41/604 (6%)
Query: 300 NQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNT 359
+ IDKIISAEIPDK+++P+LYE++ + MIHGPCG N +SPC++ KC+K +PKK D T
Sbjct: 807 DDIDKIISAEIPDKDKEPELYEVIKNSMIHGPCGAANMNSPCMVEGKCSKQYPKKHQDIT 866
Query: 360 VIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKY 419
+ DGYP+YRRR +I+K G DN +VVPYN+KL L+Y AHINVEWCNQS SIKY
Sbjct: 867 KVGKDGYPIYRRRMTEDYIEKGGFK-CDNGYVVPYNKKLSLRYQAHINVEWCNQSGSIKY 925
Query: 420 LFKYVNKGHDRVT--------------ATFYQGGDACY---DEIKMYYDCRYLSACEAVW 462
LFKY+NKG DRV AT + ++ DEIK ++DCRY+SA EAVW
Sbjct: 926 LFKYINKGADRVVFIVEPVNQDKTTENATSGEPPNSTEKKKDEIKDWFDCRYVSASEAVW 985
Query: 463 RIFSFDINYREPSVERLNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKYP 522
RI+ F + R +V+RL+FH EG++PV + DI DV+++ D+ F+AW N+
Sbjct: 986 RIYKFPLQDRSTAVQRLSFHDEGKQPVYAKPDADIEDVLERISNEDSMFMAWLTLNKNND 1045
Query: 523 ------EARDLTYGEFPLKFTWKASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNY 576
AR+L Y + P FTW ++W R RG S+GRI++V ++YLR LLN
Sbjct: 1046 VGKNGKRARELLYSQIPAYFTWDGKNKQWVKRIRGFSLGRINYVCRKMEVEYYLRVLLNI 1105
Query: 577 VKGPLCYDDIYTVGGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQL 636
VKGP+ YDDI T GV Y SFKEACFA G+LDDD+ ++D +++AS + Y+R F L
Sbjct: 1106 VKGPMSYDDIKTFNGVVYPSFKEACFARGILDDDQVYIDGLHEASQFCFGDYLRNFFAML 1165
Query: 637 LVTNQFAQPEVVWSKTWQNLSDDMLHRQRR----------ASQLKNYTLAEIDRLLRSHG 686
L+++ A+PE VWS+TW L++D+ +++R ++++NYTL EI++++ +G
Sbjct: 1166 LLSDSLARPEHVWSETWHLLAEDIENKKREDFKNPDLKLTLAEIRNYTLQEIEKIMLRNG 1225
Query: 687 KSMKE--DYPTMPRTDISLIHESRNRLIYDELNYN-QQLLEIEHKKLMSTMTAEQKNVYE 743
++KE D+P R I ++ NRL+ DEL YN L+ +H + + EQ+ +Y+
Sbjct: 1226 ATLKEIQDFPQPSREGI----DNSNRLVVDELRYNIDSNLKEKHDEWFQMLNTEQRGIYD 1281
Query: 744 KIISRVDDNLPGIFFLYGYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRT 803
+I V ++L G+FF+YG+GGTGKTFI + L++AVRSR +IVL VASSGIA+LL+ GGRT
Sbjct: 1282 EITGAVFNDLGGVFFIYGFGGTGKTFIWKTLAAAVRSRGQIVLNVASSGIASLLLEGGRT 1341
Query: 804 AHSRFGIPIIVDEISTCGIHPKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDILR 863
AHSRF IP+ DE S C I PKS LA L+ +A LIIWDEAPMM K CFE+LD+S DIL
Sbjct: 1342 AHSRFAIPLNPDEFSVCKITPKSDLANLIKEASLIIWDEAPMMSKFCFESLDKSFYDILN 1401
Query: 864 VQNN 867
++N
Sbjct: 1402 NKDN 1405
>At1g52960 hypothetical protein
Length = 924
Score = 558 bits (1438), Expect = e-159
Identities = 286/590 (48%), Positives = 391/590 (65%), Gaps = 33/590 (5%)
Query: 303 DKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNTVID 362
DKII+AEIPDK + P LY +V MIHGPCG + +SPC+ KC KYFPK + D T +D
Sbjct: 14 DKIITAEIPDKKKKPGLYAVVKDCMIHGPCGVGHPNSPCMENGKCKKYFPKSYSDTTKVD 73
Query: 363 SDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKYLFK 422
+DG+PVYRRRD G++++K G DNR+V+PYN K+ L+Y AHINVE CNQS SIKYLFK
Sbjct: 74 NDGFPVYRRRDTGIYVEKNGFQ-CDNRYVIPYNEKVSLRYQAHINVELCNQSGSIKYLFK 132
Query: 423 YVNKGHDRVTATFYQGGDACY----DEIKMYYDCRYLSACEAVWRIFSFDINYREPSVER 478
YV+KGHDRVT T DE+K Y+DCRY+SACEA+WRIF F I+YR V +
Sbjct: 133 YVHKGHDRVTVTVEPNDQDTAKKEKDEVKDYFDCRYVSACEAMWRIFKFPIHYRTTPVVK 192
Query: 479 LNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQK----------------YP 522
L FH EG++PV ++ E V+ + T+F+AWF+ N+K P
Sbjct: 193 LFFHEEGKQPVYYKPGETTESVMDRLSSEATQFLAWFQLNKKPPSRTIRANAKKLPKAAP 252
Query: 523 EARDLTYGEFPLKFTWKASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNYVKGPLC 582
+ L + E P FTW + ++K+ R+RG +IGRI+FV + +YLR LLN +G
Sbjct: 253 DPTKLLFEEIPNHFTWNSKEKKFMIRERGFAIGRINFVPRTIEDAYYLRILLNIKRGVTS 312
Query: 583 YDDIYTVGGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQF 642
Y D+ TV GV + SF++A FALGLLDDDKE+++ I A FW +A Y+RRLFV +L++
Sbjct: 313 YKDLKTVKGVVHKSFRDAVFALGLLDDDKEYINGIKDAKFWCSAKYVRRLFVIMLLSESL 372
Query: 643 AQPEVVWSKTWQNLSDDMLHRQRRA----------SQLKNYTLAEIDRLLRSHGKSMKED 692
+PE+VW +TW+ LS+D+ R+R+ + + Y L EI RLL +G S+ +
Sbjct: 373 TKPEMVWDETWRILSEDIERRKRKEWKRPDLQLSDEERQQYCLQEIARLLTKNGVSLSK- 431
Query: 693 YPTMPRTDISLIHESRNRLIYDELNYNQQLLEIEHKKLMSTMTAEQKNVYEKIISRVDDN 752
+ MP+ + E N I DE Y++ L +H++ ++ +T+EQK +Y++I+ V +
Sbjct: 432 WKQMPQISDEHV-EKCNHFILDERKYDRAYLIEKHEEWLTMVTSEQKKIYDEIMDAVLHD 490
Query: 753 LPGIFFLYGYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPI 812
G+FF+YG+GGTGKTF+ + LS+A+RS+ +I L VASSGIAALL+ GGRT HSRFGIPI
Sbjct: 491 RGGVFFVYGFGGTGKTFLWKLLSAAIRSKGDISLNVASSGIAALLLDGGRTTHSRFGIPI 550
Query: 813 IVDEISTCGIHPKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDIL 862
+E STC I S L +LV +A LIIWDE PMM KHCFE+LDR+LRDI+
Sbjct: 551 NPNESSTCNISRGSDLGELVKEANLIIWDETPMMSKHCFESLDRTLRDIM 600
>At3g13250 hypothetical protein
Length = 1419
Score = 554 bits (1427), Expect = e-158
Identities = 285/604 (47%), Positives = 406/604 (67%), Gaps = 41/604 (6%)
Query: 300 NQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNT 359
+ IDKIISAEIP+K+++P+LYE++ + M+HGPCG N SSPC++ +C+K +PKK + T
Sbjct: 475 DDIDKIISAEIPNKDKEPELYEVIKNSMMHGPCGSANTSSPCMVDGQCSKLYPKKHQEIT 534
Query: 360 VIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKY 419
+ +DGYP+YRRR +I+K G DNR+VVPYN+KL L+Y AHINVEWCNQ+ SIKY
Sbjct: 535 KVGADGYPIYRRRLTDDYIEKGGVK-CDNRYVVPYNKKLSLRYQAHINVEWCNQNGSIKY 593
Query: 420 LFKYVNKGHDRV---------------TATFYQGG--DACYDEIKMYYDCRYLSACEAVW 462
LFKY+NKG DRV TA + + DEIK ++DCRY+SA EA+W
Sbjct: 594 LFKYINKGPDRVVFIVEPIKEATSSDTTAPVVESDTTEKKKDEIKDWFDCRYVSASEAIW 653
Query: 463 RIFSFDINYREPSVERLNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKYP 522
RIF F I +R V++L+FH +G++P F+ +ADV+++ D++F+AW N+K
Sbjct: 654 RIFKFPIQHRSTPVQKLSFHDKGKQPAYFDAKAKMADVLERVSNEDSQFLAWLTLNRKNA 713
Query: 523 ------EARDLTYGEFPLKFTWKASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNY 576
ARD Y E P FTW +++ R RG S+GRI++V+ +++YLR LLN
Sbjct: 714 VGKNGKRARDCLYAEIPAYFTWDGENKQFKKRTRGFSLGRINYVSRKMEDEYYLRVLLNI 773
Query: 577 VKGPLCYDDIYTVGGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQL 636
V+GP YDDI TV GV Y S+K ACFA G+LDDD+ +++ + +AS + Y+R F +
Sbjct: 774 VRGPQSYDDIKTVNGVVYPSYKLACFARGILDDDQVYINGLIEASQFCFGDYLRNFFSMM 833
Query: 637 LVTNQFAQPEVVWSKTWQNLSDDMLHRQR----------RASQLKNYTLAEIDRLLRSHG 686
L+++ A+PE VWS+TW LS+D+L ++R +Q++NYTL EI++++ +G
Sbjct: 834 LLSDSLARPEHVWSETWHLLSEDILIKKRDEFKNQELTLTEAQIQNYTLQEIEKIMLFNG 893
Query: 687 KSMK--EDYPTMPRTDISLIHESRNRLIYDELNYNQQL-LEIEHKKLMSTMTAEQKNVYE 743
+++ E +P R I ++ NRLI DEL YN Q L+ +H + +T EQ+ +Y+
Sbjct: 894 ATLEDIEHFPKPSREGI----DNSNRLIIDELRYNNQSDLKKKHSDWIQKLTPEQRGIYD 949
Query: 744 KIISRVDDNLPGIFFLYGYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRT 803
+I + V ++L G+FF+YG+GGTGKTFI + L++AVRS+ +I L VASSGIA+LL+ GGRT
Sbjct: 950 QITNAVFNDLGGVFFVYGFGGTGKTFIWKTLAAAVRSKGQICLNVASSGIASLLLEGGRT 1009
Query: 804 AHSRFGIPIIVDEISTCGIHPKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDILR 863
AHSRF IP+ DE S C I PKS LA L+ +A LIIWDEAPMM K CFEALD+S DI++
Sbjct: 1010 AHSRFSIPLNPDEFSVCKIKPKSDLADLIKEASLIIWDEAPMMSKFCFEALDKSFSDIIK 1069
Query: 864 VQNN 867
+N
Sbjct: 1070 RVDN 1073
>At2g05080 putative helicase
Length = 1219
Score = 513 bits (1320), Expect = e-145
Identities = 268/596 (44%), Positives = 385/596 (63%), Gaps = 52/596 (8%)
Query: 302 IDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNTVI 361
IDK ISAEIPDK +DP+L+E++ +M+HGPCG N PC+ KC+K++PK V T+I
Sbjct: 295 IDKAISAEIPDKLKDPELFEVIKEMMVHGPCGVVNPKCPCMENGKCSKFYPKDHVPKTII 354
Query: 362 DSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKYLF 421
D +G+P+YRRR F++KK + DNR+V+PYNR L L+Y AHINVEWCNQS S+KY+F
Sbjct: 355 DKEGFPIYRRRRIDDFVQKK-DFKCDNRYVIPYNRSLSLRYRAHINVEWCNQSGSVKYIF 413
Query: 422 KYVNKGHDRVTATF------------YQGGDACYD--------EIKMYYDCRYLSACEAV 461
KY++KG DRVT Q +A D E++ +++CRY+SACEA
Sbjct: 414 KYIHKGPDRVTVVVGSSLNSKNKEKGKQKVNADTDGSEPKKKNEVEDFFNCRYVSACEAA 473
Query: 462 WRIFSFDINYREPSVERLNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKY 521
WRI + I+YR SV +L+FHL GE+ + F+ E++ V+ K + + IA
Sbjct: 474 WRILKYPIHYRSTSVMKLSFHLPGEQYIYFKGDEEVETVLNKADLDGSIQIA-------- 525
Query: 522 PEARDLTYGEFPLKFTWKASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNYVKGPL 581
R LTY P +FT+ ++K+ R++G +IGRI++V + +YLR LLN V GP
Sbjct: 526 ---RKLTYPNIPTRFTYDPKEKKFNLRKKGFAIGRINYVPRDIEDGYYLRILLNVVPGPR 582
Query: 582 CYDDIYTVGGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQ 641
++++ TV GV Y +K+AC ALGLLD+D+E++D + + SFW + Y+R+LFV +L +
Sbjct: 583 SFEELKTVNGVLYKEWKDACEALGLLDNDQEYIDDLKRTSFWSSGWYLRQLFVIML--DA 640
Query: 642 FAQPEVVWSKTWQNLSDDMLHRQRRA----------------SQLKNYTLAEIDRLLRSH 685
PE VW+ TWQ+LS+D+ + +++ + K Y L EID +LR +
Sbjct: 641 LISPENVWAATWQHLSEDIQNEKKKYFNRPVTCLFTDLILSDEEKKVYALQEIDHILRRN 700
Query: 686 GKSMKEDYPTMPRTDISLIHESRNRLIYDELNYNQQLLEIEHKKLMSTMTAEQKNVYEKI 745
G S+ Y TMP+ ++ N LI DE Y+++ +H + +T EQK+VY+ I
Sbjct: 701 GTSLTY-YKTMPQVPRDPRFDT-NVLILDEKGYDRESETKKHADSIKKLTLEQKSVYDNI 758
Query: 746 ISRVDDNLPGIFFLYGYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAH 805
I V++N+ G+FF+YG+GGTGKTF+ + LS+A+RS+ +IVL VASSGIA+LL+ GGRTAH
Sbjct: 759 IGAVNENVGGVFFVYGFGGTGKTFLWKTLSAALRSKGDIVLNVASSGIASLLLEGGRTAH 818
Query: 806 SRFGIPIIVDEISTCGIHPKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDI 861
SR GIP+ +E +TC + S A LV +A LIIWDEAPMM +HCFE+LDRSL DI
Sbjct: 819 SRSGIPLNPNEFTTCNMKAGSDRANLVKEASLIIWDEAPMMSRHCFESLDRSLSDI 874
>At3g30560 hypothetical protein
Length = 1473
Score = 487 bits (1254), Expect = e-137
Identities = 260/581 (44%), Positives = 363/581 (61%), Gaps = 55/581 (9%)
Query: 300 NQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNT 359
+ +DKII AEIPDK DPKLY+ V+ MIHGPC N +SPC+ KC+KY+PK V+NT
Sbjct: 582 DHVDKIIFAEIPDKENDPKLYQAVSECMIHGPCRLVNPNSPCMENGKCSKYYPKNHVENT 641
Query: 360 VIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKY 419
+D++GYP+YRRRD G FIKK DNR+VVPYN L+ KY AHINVEWCNQS S+KY
Sbjct: 642 FLDNEGYPIYRRRDTGRFIKKNKYP-CDNRYVVPYNDFLLRKYRAHINVEWCNQSVSVKY 700
Query: 420 LFKYVNKGHDRVTATFYQG-----------GDACYD-----EIKMYYDCRYLSACEAVWR 463
LFKYVNKG DRVT + G+ D +++ Y+DCRY+SACEA+WR
Sbjct: 701 LFKYVNKGPDRVTVSLEPHRKEVVSEENNVGETNNDPQEQNQVEDYFDCRYVSACEAMWR 760
Query: 464 IFSFDINYREPSVERLNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKYPE 523
I + I+YR+ V +L FH +G++P+ ++ E V+ + + +T+F AWFE N++ PE
Sbjct: 761 IKGYPIHYRQTLVTKLTFHEKGKQPIYVKEGETAESVLYRVNDDETQFTAWFELNKRDPE 820
Query: 524 ARDLTYGEFPLKFTWKASQRKWTPRQR-GLSIGRIHFVAPGCGEKFYLRTLLNYVKGPLC 582
A L Y + P +TW + + R+ G +GRI+ V P + ++LR L+N ++GP
Sbjct: 821 AAKLLYEQIPNFYTWNGKDKNFRRRKMPGFVVGRINHVPPKIDDAYHLRILINNIRGPKG 880
Query: 583 YDDIYTVGGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQF 642
+DDI T+ GV + ++++AC+ALGLLDDDK++++ I +A+FW Y+R+LFV +L+
Sbjct: 881 FDDIKTIEGVVHKTYRDACYALGLLDDDKDYINGIEEANFWCFHKYVRKLFVIMLIFESL 940
Query: 643 AQPEVVWSKTWQNLSDDMLHRQRRASQLKNYTLAEIDRLLRSHGKSMKED-YPTMPRTDI 701
+ P VVW TW+ LS+D + R +LK I+ + +S++E P + D
Sbjct: 941 SSPAVVWEHTWKILSEDFQRKVR--DKLKCPAAKNIE---TTWNRSLEEKMLPQLKPGD- 994
Query: 702 SLIHESRNRLIYDELNYNQQLLEIEHKKLMSTMTAEQKNVYEKIISRVDDNLPGIFFLYG 761
+ N+LI DE NYN++ L+ H + +T EQK VY+KI+ V +N G
Sbjct: 995 ---EPAFNQLILDERNYNRETLKTIHDDWLKMLTTEQKKVYDKIMDAVLNNKGG------ 1045
Query: 762 YGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDEISTCG 821
+I L VASSGIA+LL+ GGRTAHSRFGIP+ E STC
Sbjct: 1046 ---------------------DICLNVASSGIASLLLEGGRTAHSRFGIPLTPHETSTCN 1084
Query: 822 IHPKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDIL 862
+ S LA+LV AKLIIWDEAPMM K+CFE+LD+SL+DIL
Sbjct: 1085 MERGSDLAELVTAAKLIIWDEAPMMSKYCFESLDKSLKDIL 1125
>At1g54430 hypothetical protein
Length = 1639
Score = 484 bits (1247), Expect = e-137
Identities = 261/588 (44%), Positives = 370/588 (62%), Gaps = 55/588 (9%)
Query: 299 PNQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDN 358
PN IDK ISAEIP K++DP+ + +V M+HGPCG SSPC+ C+K FP++FV++
Sbjct: 727 PNDIDKYISAEIPVKDKDPEGHTLVEQHMMHGPCGKDRPSSPCMEKGICSKKFPREFVNH 786
Query: 359 TVIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIK 418
T ++ G+ +YRRR++ ++ K G++ +DNR+VVP+N +++ KY AHINVEWCN+S +IK
Sbjct: 787 TKMNESGFILYRRRNDQRYVLK-GQTRLDNRFVVPHNLEILKKYKAHINVEWCNKSSAIK 845
Query: 419 YLFKYVNKGHDRVTATFYQGG------------DACYDEIKMYYDCRYLSACEAVWRIFS 466
YLFKY+ KG D+ T +G + +EI Y DCRYLSACEA+WRIF
Sbjct: 846 YLFKYITKGVDKATFIIQKGNSVNGQGSGNGFEEKPRNEINEYLDCRYLSACEAMWRIFM 905
Query: 467 FDINYREPSVERLNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKYPEARD 526
F+I++ P V+RL HL GE+ +FE+ E++ +V + T +FE N+ +AR
Sbjct: 906 FNIHHHNPPVQRLPLHLPGEQSTIFEEEENLENVEYRYGHERTMLTEYFELNKICEDARK 965
Query: 527 LTYGEFPLKFTWKASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNYVKGPLCYDDI 586
L Y + P F W ++ + +T R++ +IGRI + P G+ +YLR LLN VKG +D +
Sbjct: 966 LKYVQVPTMFVWDSTNKMYTRRKQRENIGRIVNILPTAGDLYYLRILLNKVKGATSFDYL 1025
Query: 587 YTVGGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQFAQPE 646
TVGGV + SFK AC GLLD DKE+ DA+++A+ W T+ +R LFV +L+ + ++P
Sbjct: 1026 KTVGGVVHESFKAACHTRGLLDGDKEWHDAMDEAAQWSTSYLLRSLFVLILIYCEVSEPL 1085
Query: 647 VVWSKTWQNLSDDMLHRQR----------RASQLKNYTLAEIDRLLRSHGKSMKEDYPTM 696
+WS W++++DD+ +Q+ +A +L+ YTL EI+ LLR H KS+ DYP M
Sbjct: 1086 KLWSHCWESMADDVFRKQQKVLNFPQLELKAEELEKYTLIEIETLLRQHEKSL-SDYPEM 1144
Query: 697 PRTDISLIHESRNRLIYDELNYNQQLLEIEHKKLMSTMTAEQKNVYEKIISRVDDNLPGI 756
P+ + L + R+IYD++ L S + E K +
Sbjct: 1145 PQPENKL--NEQQRIIYDDV-------------LKSVINKEGK----------------L 1173
Query: 757 FFLYGYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDE 816
FFLYG GGTGKTF+ + + SA+RS + V+ VASS IAALL+PGGRTAHSRF IPI V E
Sbjct: 1174 FFLYGAGGTGKTFLYKTIISALRSNGKNVMPVASSAIAALLLPGGRTAHSRFKIPINVHE 1233
Query: 817 ISTCGIHPKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDILRV 864
S C I S LA ++ K LIIWDEAPM H+H FEA+DR+LRDIL V
Sbjct: 1234 DSICDIKIGSMLANVLSKVDLIIWDEAPMAHRHTFEAVDRTLRDILSV 1281
>At2g07620 putative helicase
Length = 1241
Score = 462 bits (1188), Expect = e-130
Identities = 259/581 (44%), Positives = 350/581 (59%), Gaps = 56/581 (9%)
Query: 300 NQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNT 359
+ ID I+SAEIPDK +DPKLYE+V MIHGPCG NK SPCI+ KC+K+FPKK V+ T
Sbjct: 433 DDIDNILSAEIPDKAKDPKLYEVVNDCMIHGPCGAANKESPCIVDGKCSKFFPKKLVEQT 492
Query: 360 VIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKY 419
+ DGYP+YRRR++ F++K G DN +VVPYNR L L+Y AHINVEWC QS SIKY
Sbjct: 493 TVGKDGYPIYRRRESEHFVEKGGIK-CDNTYVVPYNRMLSLRYRAHINVEWCKQSGSIKY 551
Query: 420 LFKYVNKGHDRVTAT-----------FYQGGD----ACYD---EIKMYYDCRYLSACEAV 461
LFKY+NKG DRV + G A D EIK Y+DCRY+SA EAV
Sbjct: 552 LFKYINKGQDRVAIVVEPKDKTSNMVLFSGSQKLLVAVIDDDKEIKDYFDCRYVSASEAV 611
Query: 462 WRIFSFDINYREPSVERLNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKY 521
WRIF F I YR V +L++HL G++P+ FED ++I ++ +K D FI + + NQ+
Sbjct: 612 WRIFKFPIQYRTTPVMKLSYHLPGKQPLCFEDTQNIDELSEKKANEDFMFIGFLKLNQEC 671
Query: 522 PEARDLTYGEFPLKFTWKASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNYVKGPL 581
AR Y E P FTW ++W R+RG IGR+++ + ++Y++ LL V GP
Sbjct: 672 EFARQFIYTEIPPYFTWDGQNKQWKLRERGFYIGRMNYASIKMEPEYYMKILLGIVCGPK 731
Query: 582 CYDDIYTVGGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQ 641
+DI T V F F LG++ FW + + ++L+
Sbjct: 732 SDEDIRTYKDVVRRKF----FFLGII------FRIYFLCCFWINVLLDQSMCGKMLLV-- 779
Query: 642 FAQPEVVWSKTWQNLSDDMLHRQRRASQLKNYTLAEIDRLLRSHGKSMKEDYPTMPRTDI 701
LSD ++ NY L EI+ +L +G ++ ED+ MP+
Sbjct: 780 --------------LSD---------AEKINYALLEIEDMLLCNGSTL-EDFKHMPKPTK 815
Query: 702 SLIHESRNRLIYDELNYNQQLLEIEHKKLMSTMTAEQKNVYEKIISRVDDNLPGIFFLYG 761
S NR I +E NYN + L+ +H + MT+EQK +Y++I+ V +N GIFF+YG
Sbjct: 816 EGTDHS-NRFITEEKNYNVEKLKEDHDDWFNKMTSEQKEIYDEIMKAVLENSGGIFFVYG 874
Query: 762 YGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDEISTCG 821
+GGTGKTF+ + LS+AVR + I + VASSGIA LL+ GGRTAHSRFGIPI D+ +TC
Sbjct: 875 FGGTGKTFMWKTLSAAVRMKGLISVNVASSGIAFLLLQGGRTAHSRFGIPINPDDFTTCH 934
Query: 822 IHPKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDIL 862
I P S LA ++ +A LIIWDEAPMM ++CFE+LDRSL D++
Sbjct: 935 IVPNSDLANMLKEASLIIWDEAPMMSRYCFESLDRSLNDVI 975
>At1g35940 hypothetical protein
Length = 1678
Score = 438 bits (1127), Expect = e-123
Identities = 240/552 (43%), Positives = 335/552 (60%), Gaps = 86/552 (15%)
Query: 300 NQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNT 359
+ IDK+ISAEIPDK ++P LYE++ + MIHGPCG N +SPC++ +C+K +PKK D T
Sbjct: 817 DDIDKLISAEIPDKEKEPDLYEVIKNSMIHGPCGSANVNSPCMVDGECSKLYPKKHQDIT 876
Query: 360 VIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKY 419
I SDGYP+YRRR +++K G DNR+VVPYN+KL L+YNAHINVEWCNQ+ SIKY
Sbjct: 877 KIGSDGYPIYRRRKTDDYVEKGGIK-CDNRYVVPYNKKLSLRYNAHINVEWCNQNGSIKY 935
Query: 420 LFKYVNKGHDRV----------------TATFYQGG-DACYDEIKMYYDCRYLSACEAVW 462
LFKY+NKG D+V T QG + +EIK ++DCRY+SA EAVW
Sbjct: 936 LFKYINKGPDKVVFIVEPTQQTTAGDSETPQQEQGSAEKKKNEIKDWFDCRYVSASEAVW 995
Query: 463 RIFSFDINYREPSVERLNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKYP 522
RIF + I + V++L+FH+EG++P F+ +I DV+++ D++F+AW N++
Sbjct: 996 RIFKYPIQHISTPVQKLSFHVEGKQPAYFDPKSNIEDVLERVANVDSQFMAWLTLNRRNA 1055
Query: 523 ------EARDLTYGEFPLKFTWKASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNY 576
AR+ Y E P FTW + + R RG SIGRIH+V+ ++++LR LLN
Sbjct: 1056 VGKNGKRARECLYAEIPAYFTWDGENKSFKKRTRGFSIGRIHYVSRKMEDEYFLRVLLNI 1115
Query: 577 VKGPLCYDDIYTVGGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQL 636
V+GP Y +I T GV Y +FKEACFA G+LDDD+ F+D + +A+
Sbjct: 1116 VRGPTSYAEIKTYDGVVYKTFKEACFARGILDDDQVFIDGLVEATH-------------- 1161
Query: 637 LVTNQFAQPEVVWSKTWQNLSDDMLHRQR----------RASQLKNYTLAEIDRLLRSHG 686
V S+TW L++D+L +R +++KNYTL EI++++ S+G
Sbjct: 1162 -----------VRSQTWHLLAEDILKTKRDEFKNPDLTLTETEIKNYTLQEIEKIMLSNG 1210
Query: 687 KSMKEDYPTMPRTDISLIHESRNRLIYDELNYNQQLLEIEHKKLMSTMTAEQKNVYEKII 746
++ ED P+ E K+++ T EQ+ VY I
Sbjct: 1211 ATL-EDIDEFPKPTRD-----------------------EWKQML---TPEQRGVYNAIT 1243
Query: 747 SRVDDNLPGIFFLYGYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHS 806
V +NL G+FF+YG+GGTGKTFI + LS+A+R R +IVL VASSGIA+LL+ GGRTAHS
Sbjct: 1244 EAVFNNLGGVFFVYGFGGTGKTFIWKTLSAAIRCRGQIVLNVASSGIASLLLEGGRTAHS 1303
Query: 807 RFGIPIIVDEIS 818
RFGIP+ DE S
Sbjct: 1304 RFGIPLNHDEFS 1315
>At2g14300 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 1230
Score = 431 bits (1108), Expect = e-121
Identities = 239/549 (43%), Positives = 333/549 (60%), Gaps = 60/549 (10%)
Query: 300 NQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNT 359
+ +DKII AEIPDK + P+LY+ V+ MIHGPC N +SPC+ KC+KY+PK V+NT
Sbjct: 434 DHVDKIIFAEIPDKEKHPELYQAVSECMIHGPCRLVNPNSPCMENGKCSKYYPKNHVENT 493
Query: 360 VIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKY 419
+D GYP+YRRRD+G FI+K DN +VVPYN L+ KY AHINVEWCNQS SIKY
Sbjct: 494 SLDYKGYPIYRRRDSGRFIEKNKYQ-CDNWYVVPYNDVLLRKYRAHINVEWCNQSVSIKY 552
Query: 420 LFKYVNKGHDRVTATFYQGGDACYD-----EIKMYYDCRYLSACEAVWRIFSFDINYREP 474
LFKYVNKG DRVT G+ D +++ Y+DCR + I+YR+
Sbjct: 553 LFKYVNKGPDRVTQN--NVGEINNDPQERNQVQDYFDCR------------GYPIHYRQT 598
Query: 475 SVERLNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKYPEARDLTYGEFPL 534
SV +L FH +G++ V ++ E V+ + + +T+FIAWFE N++ PEA L Y + P
Sbjct: 599 SVTKLTFHEKGKQSVYVKEGETAESVLYRVNNDETQFIAWFELNKRDPEAAKLLYEQIPN 658
Query: 535 KFTWKASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNYVKGPLCYDDIYTVGGVKY 594
+T I+ V P + ++LR L+N ++ P +DDI TV GV +
Sbjct: 659 FYT-------------------INHVPPKIDDAYHLRILINNIRAPKGFDDIKTVEGVVH 699
Query: 595 NSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQFAQPEVVWSKTWQ 654
++++AC+ALGLLDDDKE++ I +A+FW + Y+R+ FV +L++ + P VVW TW+
Sbjct: 700 KTYRDACYALGLLDDDKEYIHGIEEANFWCSPKYVRKSFVIMLISESLSSPVVVWEHTWK 759
Query: 655 NLSDDMLHRQRRASQLKNYTLAEIDRLLRSHGKSMKEDYPTMPRTDISLIHESRNRLIYD 714
L +D + R +L+ L LL D P N LI D
Sbjct: 760 ILFEDFQRKVR--DKLERPDLWRYKMLLEPG------DEPAF------------NPLIID 799
Query: 715 ELNYNQQLLEIEHKKLMSTMTAEQKNVY-EKIISRVDDNLPGIFFLYGYGGTGKTFILRA 773
E NYN++ L+ +H + T+T E K VY ++I+ V ++ G+FFLY +GGTGKTF+ +
Sbjct: 800 ERNYNRESLKKKHDNWLKTLTPEHKKVYHDEIMDDVLNDKGGVFFLYAFGGTGKTFLWKV 859
Query: 774 LSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDEISTCGIHPKSPLAKLVC 833
LS+A+R + + L VASS IA+LL+ GGRTAHSRFGIP+ E STC + S LA+LV
Sbjct: 860 LSAAIRCKGDTCLNVASSSIASLLLEGGRTAHSRFGIPLTPHETSTCNMERGSDLAELVT 919
Query: 834 KAKLIIWDE 842
AKLIIWDE
Sbjct: 920 AAKLIIWDE 928
>At2g14470 pseudogene
Length = 1265
Score = 383 bits (984), Expect = e-106
Identities = 229/574 (39%), Positives = 311/574 (53%), Gaps = 120/574 (20%)
Query: 300 NQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNT 359
+ IDK+ISAEIPDK ++P+LYE++ + MIHGPCG N SPC++ +C+K +PKK D T
Sbjct: 476 DDIDKLISAEIPDKEKEPELYEVIKNSMIHGPCGSANVKSPCMVDGECSKLYPKKHQDIT 535
Query: 360 VIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKY 419
+ SDGYP+YRRR +++K G DNR+V+PYN+K L+YNAHINVEWCNQ+ SIKY
Sbjct: 536 KVGSDGYPIYRRRKIDDYVEKGGIK-CDNRYVMPYNKKFSLRYNAHINVEWCNQNDSIKY 594
Query: 420 LFKYVNKGHDRVTATFYQGGDACYDEIKMYYDCRYLSACEAVWRIFSFDINYREPSVERL 479
LFKY+NKG D+V I EP+ +
Sbjct: 595 LFKYINKGPDKV-------------------------------------IFIVEPTQQAT 617
Query: 480 NFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKY-----PEARDLTYGEFPL 534
+ E P E + KK I+D WF+ + AR+ Y E P
Sbjct: 618 AG--DSETP----QQEQRSAEKKKNEIKD-----WFDCRRNAVGKNGKRARECLYAEIPA 666
Query: 535 KFTWKASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNYVKGPLCYDDIYTVGGVKY 594
FTW + + R RG SIGRIH+V+ + ++LR LLN
Sbjct: 667 YFTWDGENKAFKKRTRGFSIGRIHYVSRKMEDDYFLRVLLNI------------------ 708
Query: 595 NSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQFAQPEVVWSKTWQ 654
FW + + F LL+++ ++P VWS+TW
Sbjct: 709 ------------------------SVLFWRLS---QEFFAMLLLSDSLSRPAHVWSQTWH 741
Query: 655 NLSDDMLHRQRRASQLKNYTLAEIDRLLRSHGKSMKEDYPTMPRTDISLIHESRNRLIYD 714
L++D+L ++R + KN ED P+ I I S NRLI +
Sbjct: 742 ILAEDILKKKR--DEFKN-----------------PEDIDEFPKPTIDGIDNS-NRLIVE 781
Query: 715 ELNYNQQL-LEIEHKKLMSTMTAEQKNVYEKIISRVDDNLPGIFFLYGYGGTGKTFILRA 773
EL YN++ L+ +H++ +T EQ+ VY +I V +NL G+FF+YG+GGTGKTFI +
Sbjct: 782 ELRYNRESNLKEKHEEWKQMLTPEQRGVYNEITEAVFNNLGGVFFVYGFGGTGKTFIWKT 841
Query: 774 LSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDEISTCGIHPKSPLAKLVC 833
LS+ +R R +IVL VASSGIA+LL+ GGRTAHSRFGIP+ DE S C I PKS LA LV
Sbjct: 842 LSATIRYRDQIVLNVASSGIASLLLEGGRTAHSRFGIPLNPDEFSVCKIKPKSDLANLVK 901
Query: 834 KAKLIIWDEAPMMHKHCFEALDRSLRDILRVQNN 867
KA L+IWDEAPMM + CFEALD+S DI++ +N
Sbjct: 902 KASLVIWDEAPMMSRFCFEALDKSFSDIIKNTDN 935
>At5g37110 putative helicase
Length = 1307
Score = 376 bits (966), Expect = e-104
Identities = 219/576 (38%), Positives = 317/576 (55%), Gaps = 107/576 (18%)
Query: 302 IDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNTVI 361
IDK+ISAEIPDK +DP+L+E++ M+HGPCG N PC+ K
Sbjct: 527 IDKVISAEIPDKLKDPELFEVIKESMVHGPCGVVNPKCPCMENGK--------------- 571
Query: 362 DSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKYLF 421
RR D+ F++KK + DNR+V+PYNR L L+Y AHINVEWCNQS S+KY+F
Sbjct: 572 --------RRTDD--FVEKK-DFKCDNRYVIPYNRSLSLRYRAHINVEWCNQSGSVKYIF 620
Query: 422 KYVNKGHDRVTATFYQGGDACYDEIKMYYDCRYLSACEAVWRIFSFDINYREPSVERLNF 481
KY++KG DRVT S + +E ++ N
Sbjct: 621 KYIHKGPDRVTVVVES----------------------------SLNSKNKENGKQKDNA 652
Query: 482 HLEGEEPVVFEDHED------IADVIKKPHIRDTKFIAWFEANQKYPEARDLTYGEFPLK 535
+G E + ED I V+ + + + F+AWFE N+ AR LTY + P +
Sbjct: 653 DTDGSETKKKNEVEDYFNCRRIETVLNRSDLDGSMFLAWFELNKVSKIARKLTYADIPTR 712
Query: 536 FTWKASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNYVKGPLCYDDIYTVGGVKYN 595
FT+ + ++K+ R++G +IGRI++V + +YLR LLN GP C++++ TV V Y
Sbjct: 713 FTYDSKEKKFNLRKKGFAIGRINYVPRDIEDGYYLRILLNVQPGPRCFEELRTVNDVLYK 772
Query: 596 SFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQFAQPEVVWSKTWQN 655
+K+AC ALGLLD+D+E++D + + SFW + Y+R+LFV +L + PE VW+ TWQ+
Sbjct: 773 EWKDACEALGLLDNDQEYIDDLKRTSFWSSGGYLRQLFVIML--DALISPENVWAATWQH 830
Query: 656 LSDDMLHRQRR----------ASQLKNYTLAEIDRLLRSHGKSMKEDYPTMPRTDISLIH 705
LS+D+ + +R+ + K Y L EID +LR +G S+ Y TMP+
Sbjct: 831 LSEDIQNNKRKYFNRPDLILSDEEKKLYALQEIDHILRRNGTSLTY-YKTMPQVPRDPRF 889
Query: 706 ESRNRLIYDELNYNQQLLEIEHKKLMSTMTAEQKNVYEKIISRVDDNLPGIFFLYGYGGT 765
++ N LI DE Y++ L +H K + +T EQK++Y+ II V++N+ + F+YG+GGT
Sbjct: 890 DT-NVLILDEKGYDRDNLTEKHAKWIKMLTPEQKSIYDDIIGAVNENVGVVVFVYGFGGT 948
Query: 766 GKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDEISTCGIHPK 825
GGRTAHSRFGIP+ +E +TC +
Sbjct: 949 ---------------------------------EGGRTAHSRFGIPLNPNEFTTCNMKVG 975
Query: 826 SPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDI 861
S A LV +A LIIWDEAPMM ++CFE+LDRSL DI
Sbjct: 976 SDRANLVKEASLIIWDEAPMMSRYCFESLDRSLSDI 1011
>At3g31980 hypothetical protein
Length = 1099
Score = 297 bits (761), Expect = 2e-80
Identities = 157/365 (43%), Positives = 230/365 (63%), Gaps = 12/365 (3%)
Query: 508 DTKFIAWFEANQKYPEARDLTYGEFPLKFTWKASQRKWTPRQRGLSIGRIHFVAPGCGEK 567
D+ F+A+ + NQ+ AR TY E P FTW ++W R+RG IGR+++ + +
Sbjct: 384 DSMFMAFLKLNQECEFARQFTYTEIPQYFTWDGQNKQWKLRERGFCIGRMNYASIKMDPE 443
Query: 568 FYLRTLLNYVKGPLCYDDIYTVGGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTAS 627
+Y+R LL V GP +DI T V Y ++KEAC A G+L DD+ ++D I + S +
Sbjct: 444 YYMRILLGIVCGPTSDEDIRTYKDVVYETYKEACLARGILTDDQAYIDTIVEGSLYFFGD 503
Query: 628 YMRRLFVQLLVTNQFAQPEVVWSKTWQNLSDDMLHRQRRA----------SQLKNYTLAE 677
++R LF +L+ A+PE VW K + L +D+ ++R+ ++ +NY L E
Sbjct: 504 HLRNLFSMMLLDKCLARPEYVWEKCSRILIEDIETKKRKQYDNPDLVLTDAERRNYALLE 563
Query: 678 IDRLLRSHGKSMKEDYPTMPRTDISLIHESRNRLIYDELNYNQQLLEIEHKKLMSTMTAE 737
I+ +L +G ++ +D+ MP+ S NR I +E NYN + L +H + MT+E
Sbjct: 564 IEDMLLCNGSTL-QDFKDMPKPTKEGTDHS-NRFITEEKNYNIEKLREDHDDWFNKMTSE 621
Query: 738 QKNVYEKIISRVDDNLPGIFFLYGYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALL 797
QK +Y++II V +N GIFF+YG+GGT KTF+ + LS+AVR R I + VASSGIA+LL
Sbjct: 622 QKGIYDEIIKAVLENSGGIFFVYGFGGTSKTFMWKTLSAAVRMRGLISVNVASSGIASLL 681
Query: 798 IPGGRTAHSRFGIPIIVDEISTCGIHPKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRS 857
+ GGRTAHSRFGIPI D+ +TC I P S LA ++ +A LIIWDEAPMM ++CFE+LDRS
Sbjct: 682 LQGGRTAHSRFGIPINPDDFTTCHIVPNSDLANMLKEASLIIWDEAPMMSRYCFESLDRS 741
Query: 858 LRDIL 862
L D++
Sbjct: 742 LNDVI 746
Score = 72.0 bits (175), Expect = 1e-12
Identities = 31/44 (70%), Positives = 36/44 (81%)
Query: 300 NQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCIL 343
+ ID IISAEIPDK++DPKLYE+V MIHGPCG NK SPCI+
Sbjct: 294 DDIDNIISAEIPDKSKDPKLYEVVKDCMIHGPCGAANKESPCIV 337
>At1g64410 unknown protein
Length = 753
Score = 293 bits (750), Expect = 3e-79
Identities = 155/333 (46%), Positives = 223/333 (66%), Gaps = 12/333 (3%)
Query: 540 ASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNYVKGPLCYDDIYTVGGVKYNSFKE 599
+ ++K+ R+RG +IGRI+FV + +YLR LLN +G D+ TV V Y SF++
Sbjct: 320 SKEKKFMIRERGFAIGRINFVPRTIEDAYYLRILLNIKRGVTSSKDLKTVKAVVYKSFRD 379
Query: 600 ACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQFAQPEVVWSKTWQNLSDD 659
A FALGLLDDDKE+++ I A+FW +A Y+RRLFV +L++ +PE+VW +TW+ LS+D
Sbjct: 380 AVFALGLLDDDKEYINEIKDANFWCSAKYVRRLFVIMLLSESLTKPEMVWDETWKILSED 439
Query: 660 MLHRQRRA----------SQLKNYTLAEIDRLLRSHGKSMKEDYPTMPRTDISLIHESRN 709
+ ++R+ + + Y L EI RLL +G S+ + + MP+ + E N
Sbjct: 440 IERKKRKEWKRPDLQLSDEERQQYCLQEIARLLTKNGVSLSK-WKQMPQISDEHV-EKCN 497
Query: 710 RLIYDELNYNQQLLEIEHKKLMSTMTAEQKNVYEKIISRVDDNLPGIFFLYGYGGTGKTF 769
I DE Y++ L +H++ ++ +T EQK +Y++I+ V + G+FF+YG+GGTGKTF
Sbjct: 498 HFILDERKYDRAYLTEKHEEWLTMVTLEQKKIYDEIMDVVLHDRGGVFFVYGFGGTGKTF 557
Query: 770 ILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDEISTCGIHPKSPLA 829
+ + LS+A+RS+ +I L VASSGIAALL+ GGRTAHSRFGIPI +E STC I
Sbjct: 558 LWKLLSAAIRSKGDISLNVASSGIAALLLDGGRTAHSRFGIPINPNESSTCNISRGLDFG 617
Query: 830 KLVCKAKLIIWDEAPMMHKHCFEALDRSLRDIL 862
+LV +A LIIWDEA MM KHCFE+LDR+LRDI+
Sbjct: 618 ELVKEANLIIWDEAHMMSKHCFESLDRTLRDIM 650
Score = 110 bits (274), Expect = 5e-24
Identities = 46/84 (54%), Positives = 62/84 (73%)
Query: 303 DKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNTVID 362
DK+I+AEIPDK + P+LY +V MIHGPCG + +SPC+ KC KYFPK + D T +D
Sbjct: 211 DKVITAEIPDKKKKPELYAVVKDCMIHGPCGVGHPNSPCMENGKCKKYFPKSYSDTTKVD 270
Query: 363 SDGYPVYRRRDNGVFIKKKGESFV 386
+DG+PVYRRRD G++++K G V
Sbjct: 271 NDGFPVYRRRDTGIYVEKNGHDRV 294
>At3g30420 hypothetical protein
Length = 837
Score = 250 bits (638), Expect = 3e-66
Identities = 133/285 (46%), Positives = 184/285 (63%), Gaps = 11/285 (3%)
Query: 590 GGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQFAQPEVVW 649
GGV + SFK AC A GLLD DKE+ DA+++A+ W T+ +R LFV +L+ + ++P +W
Sbjct: 196 GGVVHESFKAACHARGLLDGDKEWHDAMDEAAQWSTSYLLRSLFVLILIYCEVSEPLKLW 255
Query: 650 SKTWQNLSDDMLHRQRR----------ASQLKNYTLAEIDRLLRSHGKSMKEDYPTMPRT 699
S W++++DD+L +Q+R A +L+ YTL EI+ LLR H KS+ DYP MP+
Sbjct: 256 SHCWESMADDVLRKQQRVLNFPQLELKAKELEKYTLIEIETLLRQHEKSLS-DYPEMPQP 314
Query: 700 DISLIHESRNRLIYDELNYNQQLLEIEHKKLMSTMTAEQKNVYEKIISRVDDNLPGIFFL 759
+ S++ E N L+ E N H L S + +Q+ +Y+ ++ V + +FFL
Sbjct: 315 EKSILEEVNNSLLRQEFQINIDKERETHANLFSKLNEQQRIIYDDVLKSVTNKEGKLFFL 374
Query: 760 YGYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDEIST 819
YG GGTGKTF+ + + SA+RS + V+ VASS IAALL+PGGRTAHS F IPI V E
Sbjct: 375 YGDGGTGKTFLYKTIISALRSNGKNVMPVASSAIAALLLPGGRTAHSWFKIPINVHEDFI 434
Query: 820 CGIHPKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDILRV 864
C I S LA ++ K LIIWDEAPM H+H FEA+DR+LRDIL V
Sbjct: 435 CDIKIGSMLANVLSKVDLIIWDEAPMAHRHTFEAVDRTLRDILSV 479
>At3g32320 hypothetical protein
Length = 494
Score = 245 bits (625), Expect = 9e-65
Identities = 127/296 (42%), Positives = 183/296 (60%), Gaps = 28/296 (9%)
Query: 300 NQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNT 359
+ +D II AEIPDK +DP+ Y+ V+ +IHGPCG N +SPC+ KC+KY+PK V+NT
Sbjct: 174 DHVDMIIFAEIPDKEKDPEQYQAVSECIIHGPCGLVNPNSPCMKNGKCSKYYPKNHVENT 233
Query: 360 VIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKY 419
+D++GYP+YRRRD G FI KK + DN +VVPYN L+ KY AHINVEWCNQS S+KY
Sbjct: 234 SLDNEGYPIYRRRDTGRFI-KKNKYQCDNWYVVPYNDVLLRKYKAHINVEWCNQSVSVKY 292
Query: 420 LFKYVNKGHDRVTATF-----------YQGGDACYD-----EIKMYYDCRYLSACEAVWR 463
LFKYVNKG DRVT + G+ D +++ Y+DC R
Sbjct: 293 LFKYVNKGPDRVTVSVEPHRKEVVSEQNNVGETNNDQQERNQVQDYFDC----------R 342
Query: 464 IFSFDINYREPSVERLNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKYPE 523
I + I+YR+ S+ +L FH +G++PV ++ E V+ + + +T+F AWFE N++ PE
Sbjct: 343 IRGYPIHYRQTSITKLTFHEKGKQPVYVKEGETAESVLYRVNNDETQFTAWFELNKRDPE 402
Query: 524 ARDLTYGEFPLKFTWKASQRKWTPRQR-GLSIGRIHFVAPGCGEKFYLRTLLNYVK 578
A L Y + P +TW + + R+ G +GRI+ V P + ++LR L+N ++
Sbjct: 403 AAKLLYEQIPNFYTWNGKDKDFRGRKMPGFVVGRINHVPPKIDDAYHLRILINNMR 458
>At3g43350 putative protein
Length = 830
Score = 244 bits (622), Expect = 2e-64
Identities = 136/282 (48%), Positives = 181/282 (63%), Gaps = 35/282 (12%)
Query: 585 DIYTVGGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQFAQ 644
D+ TV GV + SF++A FALGLLDDDKE+++AI A+FW +A Y+RRLFV +L++ +
Sbjct: 54 DLKTVKGVVHKSFRDAVFALGLLDDDKEYINAIKDANFWCSAKYVRRLFVIMLLSESLTK 113
Query: 645 PEVVWSKTWQNLSDDMLHRQRRASQLKNYTLAEIDRLLRSHGKSMKEDYPTMPRTDISLI 704
PE+VW +TW+ LS D+ H Q + + Y L EI RLL +G S+ +
Sbjct: 114 PEMVWDETWRILSKDIEHLQLSDEERQQYCLQEIARLLTKNGVSLSK------------- 160
Query: 705 HESRNRLIYDELNYNQQLLEIEHKKLMSTMTA----EQKNVYEKIISRVDDNLPGIFFLY 760
NR HK M+TM K +Y++I+ V + G+FF+Y
Sbjct: 161 ---WNRC---------------HKFQMNTMVDYGDFRAKKIYDEIMDVVLHDRGGVFFVY 202
Query: 761 GYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDEISTC 820
G+GGTGKTF+ + LS+AVRS+ +I L VASSGIAAL + GGRTAHSRF IPI +E STC
Sbjct: 203 GFGGTGKTFLWKLLSAAVRSKGDISLNVASSGIAALRLDGGRTAHSRFDIPINPNESSTC 262
Query: 821 GIHPKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDIL 862
I S L +LV +AKLIIWDEAPMM KHCFE+LDR+L+DI+
Sbjct: 263 NISRGSDLGELVKEAKLIIWDEAPMMSKHCFESLDRTLKDIV 304
>At3g42420 putative protein
Length = 1018
Score = 229 bits (584), Expect = 5e-60
Identities = 123/291 (42%), Positives = 185/291 (63%), Gaps = 28/291 (9%)
Query: 579 GPLCYDDIYTVGGVKYNSFKEACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLV 638
GP C+ I T GV Y ++K FA G+LDDD+ ++D++ AS + +++R F LL+
Sbjct: 534 GPTCFAHIKTYNGVVYPTYKTVSFARGILDDDQVYIDSLVDASQFCFGNFLRNFFAMLLL 593
Query: 639 TNQFAQPEVVWSKTWQNLSDDMLHRQRRASQLKNYTLAEIDRLLRSHGKSMKE--DYPTM 696
+ + +++KNYTL EI++++ +G ++ E ++P
Sbjct: 594 SEL----------------------ELTEAEIKNYTLQEIEKIMLFNGGTITEIENFPQP 631
Query: 697 PRTDISLIHESRNRLIYDELNYNQQLLEIEHKKLMSTMTAEQKNVYEKIISRVDDNLPGI 756
R I ++ NRLI DE+ Y++Q L +H + + +T E++ VY++I V +L G+
Sbjct: 632 TRECI----DNSNRLIVDEMRYDRQYLTGKHAEWIDMLTPEKRGVYDEITGAVFKDLGGV 687
Query: 757 FFLYGYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDE 816
FF+YG+GGT KTFI + LS+A+RSR +IVL VAS+GIA+LL+ GGRTAHSRF IP+ DE
Sbjct: 688 FFVYGFGGTRKTFIWKTLSAAIRSRGDIVLNVASNGIASLLLEGGRTAHSRFSIPLTPDE 747
Query: 817 ISTCGIHPKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDILRVQNN 867
++C I PKS LA L+ KA LIIWDEAP+M K CFE+LD+S DI+ ++N
Sbjct: 748 YTSCRIKPKSDLANLIRKASLIIWDEAPVMSKWCFESLDKSFSDIIGNKDN 798
Score = 144 bits (362), Expect = 3e-34
Identities = 63/115 (54%), Positives = 88/115 (75%), Gaps = 1/115 (0%)
Query: 300 NQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNT 359
+ IDK+ISAEIPDK ++P+LYE++ MIHGPCG NK+SPC++ KC+K +PKKF + T
Sbjct: 420 DDIDKMISAEIPDKEKEPELYEVIKKCMIHGPCGAANKNSPCMVDGKCSKNYPKKFEEIT 479
Query: 360 VIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQS 414
+ DG+PVYRR+ + +++K G DNR+VV YN+ L ++Y AHINVEWCNQ+
Sbjct: 480 KVGKDGFPVYRRKQSHNYVEKSGFR-CDNRYVVSYNKILSVRYGAHINVEWCNQN 533
>At4g07800 hypothetical protein
Length = 448
Score = 224 bits (570), Expect = 2e-58
Identities = 124/295 (42%), Positives = 171/295 (57%), Gaps = 31/295 (10%)
Query: 312 DKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNTVIDSDGYPVYRR 371
D + +LYE++ + MIHG CG N +S C++ +C+K +PKK + T + +DGYPVYRR
Sbjct: 159 DIETEQELYELIKNSMIHGLCGSANTNSLCMVDGQCSKLYPKKHQELTKVGADGYPVYRR 218
Query: 372 RDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKYLFKYVNKGHDRV 431
R I+K G DN +VVPYNR LKY AHINVEWCNQ+ SIKYLFKY+NKG DRV
Sbjct: 219 RPIDGCIEKGGVK-CDNMYVVPYNR-FSLKYQAHINVEWCNQNGSIKYLFKYINKGPDRV 276
Query: 432 --------------TATFYQGGDACY------DEIKMYYDCRYLSACEAVWRIFSFDINY 471
T T GD D IK ++D Y+SA EA+WRIF F I +
Sbjct: 277 VFIVELVKEETNSNTTTL---GDETVTTKKKKDGIKDWFDYIYVSASEAIWRIFKFPIQH 333
Query: 472 REPSVERLNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKY------PEAR 525
R V++L+FH+EG++P F+ + DV+++ D++F+ W N+K AR
Sbjct: 334 RSTPVQKLSFHVEGKQPAYFDAKAKMVDVLERVSNEDSQFMVWLTLNKKNVVGKNGKRAR 393
Query: 526 DLTYGEFPLKFTWKASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNYVKGP 580
+ Y E P FTW + + R RG +GRI++V ++ YL LLN V+GP
Sbjct: 394 NCLYAEIPTYFTWDGENKPFKKRTRGFFLGRINYVLRKMEDENYLIVLLNIVRGP 448
>At5g32630 putative protein
Length = 856
Score = 212 bits (540), Expect = 7e-55
Identities = 150/496 (30%), Positives = 237/496 (47%), Gaps = 125/496 (25%)
Query: 300 NQIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTKYFPKKFVDNT 359
+ IDK+ISAEIPDK+++P+ YE++ + MIHGPCG N +SPC+ V++
Sbjct: 304 DDIDKLISAEIPDKDKEPEFYEVIKNSMIHGPCGAANMNSPCM-------------VEDD 350
Query: 360 VIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLMLKYNAHINVEWCNQSRSIKY 419
I+ G+ DN +VVPYN+KL L+Y AHINVE QS IK
Sbjct: 351 YIEKGGFK------------------CDNSYVVPYNQKLSLRYQAHINVEC--QSNRIKQ 390
Query: 420 LFKYVNKGHDRVTATFYQGGDACYDEIKMYYDCRYLSACEAVWRIFSFDINYREPSVERL 479
K G + DEIK ++DC L + + S RL
Sbjct: 391 --KAATLGEPP------NSTEKKKDEIKDWFDCSCLEDLQISTSASIY-------SSSRL 435
Query: 480 NFHLEGEEPVVFEDHEDIADVIKKPHIRDTKFIAWFEANQKYPEARDLTYGEFPLKFTWK 539
+FH E ++P F+ + I DV+++ D+ F+AW N+ ++
Sbjct: 436 SFHCEWKQPAYFDPNAIIEDVLERISNEDSMFMAWLTLNRNNDVGKN------------- 482
Query: 540 ASQRKWTPRQRGLSIGRIHFVAPGCGEKFYLRTLLNYVKGPLCYDDIYTVGGVKYNSFKE 599
G S+GRI++ A +++YL+ LLN V+GP+ +DI T GV Y SFKE
Sbjct: 483 -----------GFSLGRINYGARKMEDEYYLQVLLNIVRGPMSCEDIKTFNGVLYPSFKE 531
Query: 600 ACFALGLLDDDKEFVDAINQASFWGTASYMRRLFVQLLVTNQFAQPEVVWSKTWQNLSDD 659
ACFA G+LDDD+ +D + +A
Sbjct: 532 ACFARGILDDDQVNIDGLLEAKL------------------------------------- 554
Query: 660 MLHRQRRASQLKNYTLAEIDRLLRSHGKSMKEDYPTMPRTDISLIHESRNRLIYDELNYN 719
+ ++++NYTL EI++++ +G ++K+ M + S+ ++ NRL+ DEL YN
Sbjct: 555 ----KLTVAEIRNYTLQEIEKIMLFNGATLKDIQDFMQPSRESI--DNSNRLVVDELRYN 608
Query: 720 -QQLLEIEHKKLMSTMTAEQKNVYEKIISRVDDNLPGIFFLYGYGGTGKTFILRALSSAV 778
L+ +H + + E + +Y++I + ++L GGT KT + + L++A
Sbjct: 609 IDSNLKEKHDEWFQMLNTEHRGIYDEITGAIFNDL---------GGTEKTSMWKTLAAAF 659
Query: 779 RSRKEIVLTVASSGIA 794
R R +IVL VASSG++
Sbjct: 660 RCRGQIVLNVASSGLS 675
>At5g35130 putative protein
Length = 414
Score = 176 bits (446), Expect = 5e-44
Identities = 93/223 (41%), Positives = 141/223 (62%), Gaps = 1/223 (0%)
Query: 666 RASQLKNYTLAEIDRLLRSHGKSMKEDYPTMPRTDISLIHESRNRLIYDELNYNQQLLEI 725
+ S+L+ YTL E+++++ + S+ E + MP+ D+ +I+E N L+ E YN +
Sbjct: 39 KESELQQYTLIEVEKVMHQYEHSLTE-FKDMPKPDLDVINELGNSLLTQESQYNVHKEKK 97
Query: 726 EHKKLMSTMTAEQKNVYEKIISRVDDNLPGIFFLYGYGGTGKTFILRALSSAVRSRKEIV 785
+H KL ST+ +QK VY+ ++ V++ L +FFLYG G GKT++ + S +RS K+IV
Sbjct: 98 DHIKLFSTLNVQQKEVYDAVMESVENGLGKLFFLYGPRGIGKTYLYSTIISMLRSEKKIV 157
Query: 786 LTVASSGIAALLIPGGRTAHSRFGIPIIVDEISTCGIHPKSPLAKLVCKAKLIIWDEAPM 845
L VA SGIA LL+ GRTAHSRF IP+ V+E S I P + LA+L+ K LIIWDEAPM
Sbjct: 158 LPVALSGIATLLLHAGRTAHSRFKIPLDVNEDSIWHIRPGTMLAELIEKIDLIIWDEAPM 217
Query: 846 MHKHCFEALDRSLRDILRVQNNGRTASLLVERLWYLVVTSDKS 888
H++ F+ALD++LRD++ ++ + L +DKS
Sbjct: 218 THRYAFKALDKTLRDLMSMEKTEAKDQHFGGKTVLLATNADKS 260
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.335 0.146 0.476
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,551,815
Number of Sequences: 26719
Number of extensions: 825004
Number of successful extensions: 3093
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 2947
Number of HSP's gapped (non-prelim): 85
length of query: 899
length of database: 11,318,596
effective HSP length: 108
effective length of query: 791
effective length of database: 8,432,944
effective search space: 6670458704
effective search space used: 6670458704
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC137702.3


