

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137670.4 + phase: 0
(324 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
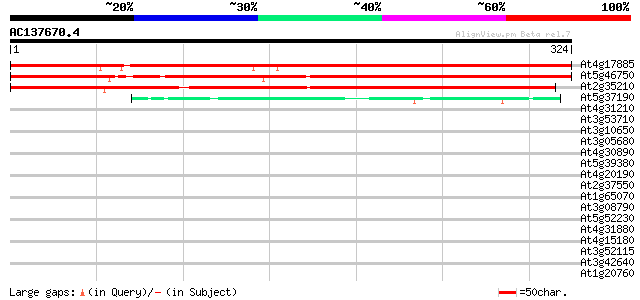
At4g17885 unknown protein 384 e-107
At5g46750 zinc finger protein Glo3-like 350 6e-97
At2g35210 unknown protein 311 2e-85
At5g37190 COP1-interacting protein 4 (CIP4) 43 2e-04
At4g31210 DNA topoisomerase like- protein 37 0.011
At3g53710 ARF GAP-like zinc finger-containing protein ZIGA2 37 0.019
At3g10650 hypothetical protein 37 0.019
At3g05680 unknown protein 36 0.025
At4g30890 ubiquitin-specific protease 24 (UBP24) 36 0.032
At5g39380 putative protein 35 0.055
At4g20190 hypothetical protein 35 0.055
At2g37550 Asp1 (asp1) 35 0.055
At1g65070 35 0.055
At3g08790 hypothetical protein 35 0.072
At5g52230 unknown protein 33 0.21
At4g31880 unknown protein 33 0.21
At4g15180 hypothetical protein 33 0.21
At3g52115 gamma response I protein 33 0.21
At3g42640 plasma membrane H+-ATPase-like protein 33 0.21
At1g20760 unknown protein 33 0.27
>At4g17885 unknown protein
Length = 413
Score = 384 bits (987), Expect = e-107
Identities = 211/336 (62%), Positives = 253/336 (74%), Gaps = 15/336 (4%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALE--KGLPSSP 58
M FGGN+RAQ+FFKQHGWTDGGKIEAKYTSRAA+LYRQIL KEVAK++A E GL SSP
Sbjct: 81 MMFGGNNRAQVFFKQHGWTDGGKIEAKYTSRAADLYRQILAKEVAKAIAEETNSGLLSSP 140
Query: 59 VASQS----SNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPG 114
VA+ SNG V + V +E L K E +TSSP+AS+T + KK IG K+ G
Sbjct: 141 VATSQLPEVSNG---VSSYSVKEELPLSKHEATSATSSPKASNTVVPSTFKKPIGAKRTG 197
Query: 115 KSGGLGARKLNKKPSESFYEQKPEE--PPAPVPSTTNNNVS---ARPSMTSRFEYVDNVQ 169
K+GGLGARKL KP ++ YEQKPEE P P S+TNN S A S SRFEY D++Q
Sbjct: 198 KTGGLGARKLTTKPKDNLYEQKPEEVAPVIPAVSSTNNGESKSSAGSSFASRFEYNDDLQ 257
Query: 170 SSELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAK 229
S G+ NHV+ PKSS+FF+DFGMDS FPKK SN+SK Q+EESDEARKKF+NAK
Sbjct: 258 SGGQSVGGTQVLNHVAPPKSSSFFSDFGMDSSFPKKSSSNSSKSQVEESDEARKKFTNAK 317
Query: 230 SISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFG-DSADSSIDLAASDLINRLSF 288
SISS+Q+FGDQNK D E++ATL KF+ S++ISSADF+G D DS+ID+ ASDLINRLSF
Sbjct: 318 SISSAQYFGDQNKNADLESKATLQKFAGSASISSADFYGHDQDDSNIDITASDLINRLSF 377
Query: 289 QAQQDISSLKNIAGETGKKLSSLASSLMTDLQDRIL 324
QAQQD+SSL NIAGET KKL +LAS + +D+QDR+L
Sbjct: 378 QAQQDLSSLVNIAGETKKKLGTLASGIFSDIQDRML 413
>At5g46750 zinc finger protein Glo3-like
Length = 402
Score = 350 bits (898), Expect = 6e-97
Identities = 194/334 (58%), Positives = 241/334 (72%), Gaps = 19/334 (5%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGLPS---- 56
M FGGN+RAQ+FFKQHGW DGGKIEAKYTSRAA++YRQ L KEVAK+MA E LPS
Sbjct: 78 MMFGGNNRAQVFFKQHGWNDGGKIEAKYTSRAADMYRQTLAKEVAKAMAEETVLPSLSSV 137
Query: 57 ---SPVASQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKP 113
PV S S NGF TSE KE++L + + SSP+AS ++ KK + +K
Sbjct: 138 ATSQPVES-SENGF----TSESPKESSLKQEAAV--VSSPKASQKVVASTFKKPLVSRKS 190
Query: 114 GKSGGLGARKLNKKPSESFYEQKPEEPPAPVP--STTNNNVSARPSMTSRFEYVDNVQSS 171
GK+GGLGARKL K ++ YEQKPEEP +P S TN+ +A S SRFEY D+ QS
Sbjct: 191 GKTGGLGARKLTTKSKDNLYEQKPEEPVPVIPAASPTNDTSAAGSSFASRFEYFDDEQSG 250
Query: 172 ELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAKSI 231
G+ +HV+ PKSSNFF +FGMDS FPKK S++SK Q+EE+DEARKKFSNAKSI
Sbjct: 251 --GQSGTRVLSHVAPPKSSNFFNEFGMDSAFPKKSSSSSSKAQVEETDEARKKFSNAKSI 308
Query: 232 SSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFGDSA-DSSIDLAASDLINRLSFQA 290
SS+QFFG+QN+ D +++ATL KFS S+AISS+D FG DS+ID+ ASDLINR+SFQA
Sbjct: 309 SSAQFFGNQNRDADLDSKATLQKFSGSAAISSSDLFGHGPDDSNIDITASDLINRISFQA 368
Query: 291 QQDISSLKNIAGETGKKLSSLASSLMTDLQDRIL 324
QQD+SS+ N+A ET KL + ASS+ +DLQDR+L
Sbjct: 369 QQDMSSIANLAEETKNKLGTFASSIFSDLQDRML 402
>At2g35210 unknown protein
Length = 395
Score = 311 bits (798), Expect = 2e-85
Identities = 182/322 (56%), Positives = 230/322 (70%), Gaps = 13/322 (4%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKG--LPSSP 58
M +GGN+RAQ+FFKQ+GW+DGGK EAKYTSRAA+LY+QIL KEVAKS A E+ PS P
Sbjct: 78 MIYGGNNRAQVFFKQYGWSDGGKTEAKYTSRAADLYKQILAKEVAKSKAEEELDLPPSPP 137
Query: 59 VASQSSNGFLDVRTSEVLKE-NTLDKAEKLESTS-SPRASHTSASNNLKKSIGGKKPGKS 116
++Q NG ++TSE LKE NTL + EK + SPR S + +KK +G KK GK+
Sbjct: 138 DSTQVPNGLSSIKTSEALKESNTLKQQEKPDVVPVSPRISRS-----VKKPLGAKKTGKT 192
Query: 117 GGLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSR 176
GGLGARKL K S + Y+QKPEE ++ + SAR S +SRF+Y DNVQ+ E D
Sbjct: 193 GGLGARKLTTKSSGTLYDQKPEESVIIQATSPVSAKSARSSFSSRFDYADNVQNRE-DYM 251
Query: 177 GSNTFNHVSVPKSSNFFAD-FGMDSG-FPKKFGSNTSKVQIEESDEARKKFSNAKSISSS 234
+HV+ PKSS FF + M+ G F KK +++SK+QI+E+DEARKKF+NAKSISS+
Sbjct: 252 SPQVVSHVAPPKSSGFFEEELEMNGGRFQKKPITSSSKLQIQETDEARKKFTNAKSISSA 311
Query: 235 QFFGDQNKARDAETRATLSKFSSSSAISSADFFGD-SADSSIDLAASDLINRLSFQAQQD 293
Q+FG+ N + D E +++L KFS SSAISSAD FGD D +DL A DL+NRLS QAQQD
Sbjct: 312 QYFGNDNNSADLEAKSSLKKFSGSSAISSADLFGDGDGDFPLDLTAGDLLNRLSLQAQQD 371
Query: 294 ISSLKNIAGETGKKLSSLASSL 315
ISSLKN+A ET KKL S+ASSL
Sbjct: 372 ISSLKNMAEETKKKLGSVASSL 393
>At5g37190 COP1-interacting protein 4 (CIP4)
Length = 876
Score = 43.1 bits (100), Expect = 2e-04
Identities = 62/253 (24%), Positives = 100/253 (39%), Gaps = 30/253 (11%)
Query: 71 RTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPGKSGGLGARKLNKKPSE 130
R+ +VLKEN D + S S + +N + GG K K + L+ P
Sbjct: 633 RSMQVLKENA-DMGDNFGS--SQKDDIVGGTNKEDQVTGGAKSKKE----KKSLDLHPGG 685
Query: 131 SFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNHVSVPKSS 190
S + +++ S S V S+ +G+ +V KSS
Sbjct: 686 SIDGSMKVKETKGRVQPSSSGTSQLQSRAKNDRNGSKVDLSDASMKGTVNNKKEAVKKSS 745
Query: 191 NFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAKSIS--SSQFFGDQNKARDAET 248
N N SKV + E KK SN+ + + ++ FF D A + E+
Sbjct: 746 NSVT-------------VNNSKVNVNNKKEVVKKISNSVTANKITTNFFKD---AEEDES 789
Query: 249 RATLSKFSSSSAISSADFFGDSADSSIDLAASDLI---NRLSFQAQQDISSLKNIAGETG 305
+ T S S++ + SS+D D S ++L+ NRLS Q + S K+ +
Sbjct: 790 KTTSSDSSNAPSDSSSDNDSDVTSSMYMKQGNNLVGGTNRLSGSLQDILRSSKSY--KAA 847
Query: 306 KKLSSLASSLMTD 318
K +SL+ S+ D
Sbjct: 848 KLTASLSQSVAFD 860
>At4g31210 DNA topoisomerase like- protein
Length = 1179
Score = 37.4 bits (85), Expect = 0.011
Identities = 35/139 (25%), Positives = 54/139 (38%), Gaps = 9/139 (6%)
Query: 57 SPVASQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPGKS 116
S AS +SNG + +K K +K+E SS A + + I KP K
Sbjct: 342 STAASPASNG----NQATTVKSKRRPKNKKVEDKSSSVVPVLEAVSLDESPISVPKP-KH 396
Query: 117 GGLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNN----VSARPSMTSRFEYVDNVQSSE 172
G G RK + E EEP +P PS + + S + S + + V +
Sbjct: 397 SGSGNRKSSSAKKEVAKNHPVEEPKSPAPSNSKSEQQHLKSTKASKAPKQKLVPQHMKNS 456
Query: 173 LDSRGSNTFNHVSVPKSSN 191
++ RG N + P +
Sbjct: 457 IEHRGQNASKPLYPPSGKS 475
>At3g53710 ARF GAP-like zinc finger-containing protein ZIGA2
Length = 459
Score = 36.6 bits (83), Expect = 0.019
Identities = 70/336 (20%), Positives = 120/336 (34%), Gaps = 60/336 (17%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQIL--------------TKEVAK 46
M GGN R FF Q+G I +KY S AA +YR + KE K
Sbjct: 72 MEAGGNERLNKFFAQYGIAKETDIISKYNSNAASVYRDRIQALAEGRPWNDPPVVKEANK 131
Query: 47 SMALEKG---------------------LPSSPVASQSSNGFLDVRTSE---VLKENTLD 82
L +G S +QS+N F E V +++ D
Sbjct: 132 KPPLAQGGYGNNNNNNNGGWDSWDNDDSYKSDMRRNQSANDFRASGNREGAHVKSKSSED 191
Query: 83 --KAEKLESTSSPR--------ASHTSASNNLKKSIGGKKPGKSGGLGARKLNKKPSESF 132
+LE++++ + A + S L S GGK G N + + F
Sbjct: 192 IYTRSQLEASAAGKESFFARRMAENESKPEGLPPSQGGKYVGFGSSSAPPPRNNQQDDVF 251
Query: 133 YEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNHVSVPKSSNF 192
+ + + +A T E+ V+ D + S T N V+ +
Sbjct: 252 SVVSQGFGRLSLVAASAAQSAASVVQTGTKEFTSKVKEGGYDHKVSETVNVVANKTTEIG 311
Query: 193 FADFGMDSGFPKKFGSNTSKVQ--IEESDEARKKFSNAKSISSSQFFGDQNKARDA---- 246
+G+ G T KV+ +E + + S + Q FG+ NKA ++
Sbjct: 312 HRTWGIMKGV---MAMATQKVEEFTKEGSTSWNQQSENEGNGYYQNFGNGNKAANSSVGG 368
Query: 247 ---ETRATLSKFSSSSAISSADFFGDSADSSIDLAA 279
++ +T +++S +S D +G++ + + A
Sbjct: 369 GRPQSSSTSGHYNNSQNSNSWDSWGENENKKTEAVA 404
>At3g10650 hypothetical protein
Length = 1309
Score = 36.6 bits (83), Expect = 0.019
Identities = 55/245 (22%), Positives = 96/245 (38%), Gaps = 20/245 (8%)
Query: 55 PSSPVASQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLK--KSIGGKK 112
P++ + +Q+S D++ L++ + E+ + P S + SN+ +S
Sbjct: 686 PATGLLNQNSGASADIK----LEKTSSTAFGVSEAWAKPTESKKTFSNSASGAESSTSAA 741
Query: 113 PGKSGGL---GARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQ 169
P +G + GA + PS P PP+ ++N+V PS F N
Sbjct: 742 PTLNGSIFSAGANAVTPPPSNGSLTSSPSFPPSISNIPSDNSVGDMPSTVQSFAATHNSS 801
Query: 170 S---SELDSRGSNTFNHVSVPKSSNFFADFGMDSG-FPKKFGSNTSKVQIEESDEARKKF 225
S S SN+ + + P SS FG + F S +S +E++ F
Sbjct: 802 SIFGKLPTSNDSNSQSTSASPLSSTSPFKFGQPAAPFSAPAVSESSGQISKETEVKNATF 861
Query: 226 SNAKSI------SSSQFFGDQNKARDAETRATLS-KFSSSSAISSADFFGDSADSSIDLA 278
N + S+ Q G A+ AE ++ F SSS + + +A +S +
Sbjct: 862 GNTSTFKFGGMASADQSTGIVFGAKSAENKSRPGFVFGSSSVVGGSTLNPSTAAASAPES 921
Query: 279 ASDLI 283
+ LI
Sbjct: 922 SGSLI 926
>At3g05680 unknown protein
Length = 2070
Score = 36.2 bits (82), Expect = 0.025
Identities = 38/136 (27%), Positives = 56/136 (40%), Gaps = 16/136 (11%)
Query: 143 PVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNHVSVPKSSNFFADFGMDSGF 202
P N + P M + V+ +E D+ GS+ F+H+ P +SN +D
Sbjct: 1600 PFLQPDENLMQPAPVMVEQNSPHSIVEETESDANGSSQFSHMGTPVASN------VDENA 1653
Query: 203 PKKFGSNTSKVQIEESDEARKKFSNAKSISSSQFFGDQNKARDAETRATLSK--FSSSSA 260
+F S S + E S SISS + F +Q A +A+ A L S S
Sbjct: 1654 QSEFSSRISVSRPEMS------LIREPSISSDRKFVEQ--ADEAKKMAPLKSAGISESGF 1705
Query: 261 ISSADFFGDSADSSID 276
I + G S +SID
Sbjct: 1706 IPAYHMPGSSGQNSID 1721
>At4g30890 ubiquitin-specific protease 24 (UBP24)
Length = 551
Score = 35.8 bits (81), Expect = 0.032
Identities = 42/167 (25%), Positives = 68/167 (40%), Gaps = 28/167 (16%)
Query: 129 SESFYEQKPEEPP--------APVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNT 180
+ SF+EQKP + P + + N + S+ + E + S GS
Sbjct: 17 TRSFFEQKPTKDPQNSKDKCVGSIQFGSLNLAAENSSVNTNGELKKGEADGTVKSAGSQE 76
Query: 181 FNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQ--------IEESDEARKKFSNAKSIS 232
S P SS+ D D+ P+K N+ +V I+E E+ K +N ++
Sbjct: 77 RLDASRPASSDKNND--SDAKLPRK---NSLRVPEHVVQNGIIKEISESNKSLNNGVAVK 131
Query: 233 SSQFFGDQNKARDAET----RATLSKFSSSSAISSADFFGDSADSSI 275
+ D D E+ +A+ SKF A+ + DF DS+ SI
Sbjct: 132 TDPIGLDNLSMSDGESDPVYKASSSKF---QALDNEDFSSDSSSGSI 175
>At5g39380 putative protein
Length = 507
Score = 35.0 bits (79), Expect = 0.055
Identities = 46/221 (20%), Positives = 83/221 (36%), Gaps = 16/221 (7%)
Query: 42 KEVAKSMALEKGLPSSPVASQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSAS 101
K S K P +P AS S + + S L+++ KA S + + + +
Sbjct: 228 KGTVSSRVASKKAPVTPRASLSPRLSVRLAGSSSLRKSQSLKAASSSSRQNQKPRPVNRT 287
Query: 102 NNLKKSIGGKKPGKSGGLGARKL---NKKPSESFYEQK----PEEPPAPVPSTTNNNVSA 154
+ K + P + L ++ N SE+ Q+ P PP P +T +
Sbjct: 288 DEFNKQLDDY-PVEEKTLHVVEMETTNNVVSENDQNQQGFVEPFLPPLPPTQSTPKDDEC 346
Query: 155 RPSMTSRFEYVDNVQSSELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQ 214
S T +EY +E + N P+++ D ++ +F T
Sbjct: 347 TVSETEEYEYTSGSNEAESEEEEIGLSNGEKKPRAARKEGDSADEAARKLRFRRGTIVDP 406
Query: 215 IEESDEARK-KFSNAKSISSSQFFGDQNKARDAETRATLSK 254
++ARK KF + + ++KA+DA+ R + K
Sbjct: 407 DTVGEKARKLKFRRGRGLG-------EDKAQDAQVRRSFKK 440
>At4g20190 hypothetical protein
Length = 389
Score = 35.0 bits (79), Expect = 0.055
Identities = 31/129 (24%), Positives = 52/129 (40%), Gaps = 7/129 (5%)
Query: 147 TTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKF 206
+++N+ S + + +S++ + SNT N KS N F D S
Sbjct: 164 SSSNSKSLSHKSSGNNSFFSKTESNKSNRSNSNTANS----KSINSFEDGFKCSALCLYL 219
Query: 207 GSNTSKVQIEESDEARKKFSNAKSISSSQFFGDQNKARDA---ETRATLSKFSSSSAISS 263
+ + S + F+ +++SSQ RD RA+L +F S SS
Sbjct: 220 PGFSKGKPVRSSRKGDSSFTRTTTMTSSQSMARTASIRDTAVLSARASLERFECGSWTSS 279
Query: 264 ADFFGDSAD 272
A + D+AD
Sbjct: 280 AMIYDDNAD 288
>At2g37550 Asp1 (asp1)
Length = 456
Score = 35.0 bits (79), Expect = 0.055
Identities = 37/142 (26%), Positives = 54/142 (37%), Gaps = 8/142 (5%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGLPSSPVA 60
M GGN R F Q+G + I +KY S AA +YR + +++A + P+
Sbjct: 72 MDAGGNERLNNFLAQYGISKETDIISKYNSNAASVYRDRI-----QALAEGRQWRDPPIV 126
Query: 61 SQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPGKSGGLG 120
+S G L + + + D S S +S G + SGG G
Sbjct: 127 KESVGGGLMNKKPPLSQGGGRDSGNGGWDNWDNDDSFRSTDMRRNQSAGDFR--SSGGRG 184
Query: 121 ARKLNKKPSESFYEQKPEEPPA 142
A K SE Y + E A
Sbjct: 185 A-PAKSKSSEDIYSRSQLEASA 205
>At1g65070
Length = 857
Score = 35.0 bits (79), Expect = 0.055
Identities = 25/79 (31%), Positives = 36/79 (44%), Gaps = 9/79 (11%)
Query: 52 KGLPSSPVASQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGK 111
K LP S + S+ R V+ +N DK E+ + A TSA +N+K+ IG K
Sbjct: 759 KPLPRSTSSQTSNRSLRSKRQFFVVPDNCTDKHERARQCVA-NAKRTSADSNVKEHIGSK 817
Query: 112 K--------PGKSGGLGAR 122
+ P + G LG R
Sbjct: 818 RNAGGRSGSPARHGHLGKR 836
>At3g08790 hypothetical protein
Length = 607
Score = 34.7 bits (78), Expect = 0.072
Identities = 31/114 (27%), Positives = 46/114 (40%), Gaps = 19/114 (16%)
Query: 55 PSSPVASQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKK-- 112
PSSPVA ++ L ++L +N E TS+P A+H N G
Sbjct: 335 PSSPVAKPDNSIVL----IDMLSDNN---CESSTPTSNPHANHQKVQQNYSNGFGPGHQE 387
Query: 113 -----PGKSGGLGARKLNKKPSESFYEQKPEEPPAPVPST-----TNNNVSARP 156
G S + ++ ++PS Y +P P P++ NNNV A P
Sbjct: 388 QSYYGQGSSAPVWNLQITQQPSSPAYGNQPFSPNFSPPASPHYGGQNNNVLALP 441
>At5g52230 unknown protein
Length = 746
Score = 33.1 bits (74), Expect = 0.21
Identities = 24/102 (23%), Positives = 49/102 (47%), Gaps = 3/102 (2%)
Query: 83 KAEKLESTSSPRASHTSASNNLKKSIGGKKPGKSGGLGARKLNKKPSESFYEQKPEEPPA 142
K ++ S SP + TSA+ +K+ GK+ G+S + KP++ + P +
Sbjct: 495 KQKEEMSPVSPLSCQTSATK-CEKTAAGKRVGRSSPKANLTTSVKPTQISPLRSPNKGKQ 553
Query: 143 PVPSTTNNNVSARPSMTSRFEYVDNVQS--SELDSRGSNTFN 182
P PS + + + R + + + V+ SE+ + +N+F+
Sbjct: 554 PHPSDSGSAIQRRNKLANEYSNSSVVRGTCSEVMEKSTNSFS 595
>At4g31880 unknown protein
Length = 873
Score = 33.1 bits (74), Expect = 0.21
Identities = 25/83 (30%), Positives = 38/83 (45%), Gaps = 2/83 (2%)
Query: 60 ASQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPGKSGGL 119
+S+S V S K ++ K E ++T+S ++ + KS GK KSG
Sbjct: 767 SSRSKKDISSVSKSGKSKASSKKKEEPSKATTSSKSKSGPVKSVPAKSKTGKGKAKSGSA 826
Query: 120 G--ARKLNKKPSESFYEQKPEEP 140
A K + SES E+ P+EP
Sbjct: 827 STPASKAKESASESESEETPKEP 849
Score = 27.7 bits (60), Expect = 8.8
Identities = 30/158 (18%), Positives = 58/158 (35%), Gaps = 10/158 (6%)
Query: 23 KIEAKYTSRAAELYRQILTKEVAKSMALEKGLPSSPVASQSSNGFLDVRTSEVLKENTLD 82
++ + S A + LT E KS + S+ VAS F ++ +V+ D
Sbjct: 212 RLAEQVLSNCASKLKTYLT-EAVKSSGVPLDKYSNIVASICEGTFSALQQDQVVANEKED 270
Query: 83 ---------KAEKLESTSSPRASHTSASNNLKKSIGGKKPGKSGGLGARKLNKKPSESFY 133
+ EK S+P + + K + ++ KK ++
Sbjct: 271 SQGHIKRETEVEKAAEISTPERTDAPKDESGKSGVSNGVAQQNDSSVDTDSMKKQDDTGA 330
Query: 134 EQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSS 171
+ +P++ P + NN +P + + E +N SS
Sbjct: 331 KDEPQQLDNPRNTDLNNTTEEKPDVEHQIEEKENESSS 368
>At4g15180 hypothetical protein
Length = 2351
Score = 33.1 bits (74), Expect = 0.21
Identities = 24/102 (23%), Positives = 48/102 (46%), Gaps = 1/102 (0%)
Query: 219 DEARKKFSNAKSISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFGDSADSSIDLA 278
DE K + + +SS+ + + AE + +S+ S + ++ A +G+ A
Sbjct: 1448 DELMKSWQDGSGLSSATKYNKKLSKTVAEKKY-MSRTSDTFGVNGASDYGEYASDREIKR 1506
Query: 279 ASDLINRLSFQAQQDISSLKNIAGETGKKLSSLASSLMTDLQ 320
+NR SF ++ D SS + G++ S+ AS +D++
Sbjct: 1507 RLSKLNRKSFSSESDTSSELSDNGKSDNYSSASASESESDIR 1548
Score = 28.1 bits (61), Expect = 6.8
Identities = 30/161 (18%), Positives = 57/161 (34%), Gaps = 8/161 (4%)
Query: 75 VLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPGKSGGLGARKLNKKPSESFYE 134
++++ TL + ++ ++ HTS + + +S KP S +K K +
Sbjct: 20 IVEKTTLCGGNESKTAATTENGHTSIATKVPESQPANKPSASSQPVKKKRIVKVIRKVVK 79
Query: 135 QKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNHVSVPKSSNFFA 194
++P++P + PS + +Q E D + V + N
Sbjct: 80 RRPKQPQKQADEQLKDQ---PPSQVVQLPAESQLQIKEQDKKSEFKGGTSGVKEVEN--- 133
Query: 195 DFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAKSISSSQ 235
G DSGF + D + S KS+ S+
Sbjct: 134 --GGDSGFKDEVEEGELGTLKLHEDLENGEISPVKSLQKSE 172
>At3g52115 gamma response I protein
Length = 588
Score = 33.1 bits (74), Expect = 0.21
Identities = 30/110 (27%), Positives = 49/110 (44%), Gaps = 12/110 (10%)
Query: 201 GFPKKFGSNTSKVQIEESDEARKKFSNAKSISSSQFFGD-QNKARDAETRATLSKFSSSS 259
G K+ S S + + EA+ + S + I SQ F + Q+K++ E K S +
Sbjct: 8 GIEAKYISGLSTIMVATIQEAKDRISQIEYIFCSQLFPNFQSKSKAFE------KVYSEA 61
Query: 260 AISSADFFGDSADSSIDLAASDLINRLSFQAQQDISSLKNIAGETGKKLS 309
+++ D + D S + D I L + QQ S + +A E GK S
Sbjct: 62 RLAACDTWKDREKSLL-----DQIEELKVENQQIKSDKEKLAEELGKTAS 106
>At3g42640 plasma membrane H+-ATPase-like protein
Length = 948
Score = 33.1 bits (74), Expect = 0.21
Identities = 37/162 (22%), Positives = 68/162 (41%), Gaps = 23/162 (14%)
Query: 149 NNNVSARPSMTSRFEYVDNVQSSEL---DSRGSNTFNHVSVPKSSNFFADFGMDSGFPKK 205
++ +S + ++T R ++ + ++ D G+ T N +SV KS + FPK
Sbjct: 304 SHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKS--------LIEVFPKN 355
Query: 206 FGSNTSKVQIEESDEARKKFSNAKSISSSQFFGDQNKARDAETRATLSKFSSSSAISSAD 265
S++ V + + +R + +A S GD +AR T F+ ++
Sbjct: 356 MDSDS--VVLMAARASRIENQDAIDASIVGMLGDPKEARAGITEVHFLPFNPVDKRTAIT 413
Query: 266 FFGDSADSSIDLAASDLINRLSFQAQQDISSLKNIAGETGKK 307
+ +S D +R S A + I L N+ GET +K
Sbjct: 414 YIDESGD----------WHRSSKGAPEQIIELCNLQGETKRK 445
>At1g20760 unknown protein
Length = 1019
Score = 32.7 bits (73), Expect = 0.27
Identities = 45/171 (26%), Positives = 69/171 (40%), Gaps = 21/171 (12%)
Query: 114 GKSGGLGARKLNKKPSESFYEQKPEE-------PPAPVPSTTNN-----NVSARPSMTSR 161
G G + + S SF Q+ + P P+ N+ + SAR +
Sbjct: 835 GDDFGGNTARADSPSSRSFGAQRKSQFAFDDSVPSTPLSRFGNSPPRFSDASARDNNFDS 894
Query: 162 FEYVDNVQSSELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEA 221
F D+ +SE + S+ +S S N DFG + F + N+S+
Sbjct: 895 FSRFDSFNTSEAGAGFSSQPERLSRFDSINSSKDFG-GAAFSRFDSINSSR-----DVTG 948
Query: 222 RKKFSNAKSISSSQFFGDQNKARDAETRATLSKFSSSSAIS--SADFFGDS 270
+KFS SI+SS+ FG + +R +T FS S S AD FG +
Sbjct: 949 AEKFSRFDSINSSKDFGGPSLSRFDSMNST-KDFSGSHGYSFDDADPFGST 998
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.306 0.123 0.324
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,817,946
Number of Sequences: 26719
Number of extensions: 291046
Number of successful extensions: 1313
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 96
Number of HSP's that attempted gapping in prelim test: 1257
Number of HSP's gapped (non-prelim): 134
length of query: 324
length of database: 11,318,596
effective HSP length: 99
effective length of query: 225
effective length of database: 8,673,415
effective search space: 1951518375
effective search space used: 1951518375
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137670.4


