

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.8 - phase: 0 /pseudo
(676 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
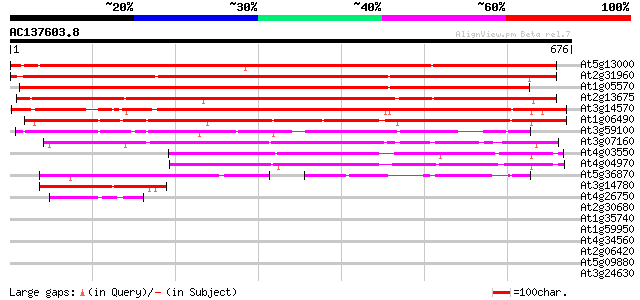
Score E
Sequences producing significant alignments: (bits) Value
At5g13000 callose synthase catalytic subunit -like protein 1073 0.0
At2g31960 glucan synthase like protein 1009 0.0
At1g05570 putative glucan synthase 998 0.0
At2g13675 callose synthase (1,3-beta-glucan synthase) like protein 697 0.0
At3g14570 hypothetical protein 575 e-164
At1g06490 glucan synthase, putative 572 e-163
At3g59100 putative protein 464 e-131
At3g07160 putative glucan synthase 342 4e-94
At4g03550 putative glucan synthase component 335 4e-92
At4g04970 309 3e-84
At5g36870 putative glucan synthase 208 9e-54
At3g14780 unknown protein 111 1e-24
At4g26750 unknown protein 48 2e-05
At2g30680 hypothetical protein 41 0.002
At1g35740 hypothetical protein 41 0.002
At1g59950 hypothetical protein 32 0.89
At4g34560 unknown protein 32 1.2
At2g06420 unknown protein 32 1.5
At5g09880 splicing factor-like protein 30 4.4
At3g24630 hypothetical protein 29 7.5
>At5g13000 callose synthase catalytic subunit -like protein
Length = 1963
Score = 1073 bits (2775), Expect = 0.0
Identities = 539/673 (80%), Positives = 586/673 (86%), Gaps = 18/673 (2%)
Query: 1 MSSRPGPPSSSETQGPPLQRRITRTQTAGNLGEAIFDSEVVPSSLVEIAPILRVANEVEK 60
MS+ G P +Q P QRRI RTQTAGNLGE+ FDSEVVPSSLVEIAPILRVANEVE
Sbjct: 1 MSATRGGPDQGPSQ--PQQRRIIRTQTAGNLGES-FDSEVVPSSLVEIAPILRVANEVES 57
Query: 61 THPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRIKKSDAREM 120
++PRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERE+DPTL GR+KKSDAREM
Sbjct: 58 SNPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLEREHDPTLMGRVKKSDAREM 117
Query: 121 QSFYQHYYKKYIQALQNAADKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDREILETQ 180
QSFYQHYYKKYIQAL NAADKADRAQLTKAYQTANVLFEVLKAVN+TQS+EVDREILE Q
Sbjct: 118 QSFYQHYYKKYIQALHNAADKADRAQLTKAYQTANVLFEVLKAVNLTQSIEVDREILEAQ 177
Query: 181 DKVAEKTEILVPYNILPLDPDSANQAIMRFPEIQAAVFALRNTRGLAWPKDYKKKKDEDI 240
DKVAEKT++ VPYNILPLDPDSANQAIMR+PEIQAAV ALRNTRGL WP+ +KKKKDED+
Sbjct: 178 DKVAEKTQLYVPYNILPLDPDSANQAIMRYPEIQAAVLALRNTRGLPWPEGHKKKKDEDM 237
Query: 241 LDWLGAMFGFQKHNVANQREHLILLLANVHIRQFPKPDQQPK------------LDERAL 288
LDWL MFGFQK NVANQREHLILLLANVHIRQFPKPDQQPK LD++AL
Sbjct: 238 LDWLQEMFGFQKDNVANQREHLILLLANVHIRQFPKPDQQPKFILSFVLIVPSQLDDQAL 297
Query: 289 TEVMKKLFKNYKKWCKYLGRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPEC 348
TEVMKKLFKNYKKWCKYLGRKSSLWLPTIQQE+QQRKLLYM LYLLIWGEAANLRFMPEC
Sbjct: 298 TEVMKKLFKNYKKWCKYLGRKSSLWLPTIQQEMQQRKLLYMALYLLIWGEAANLRFMPEC 357
Query: 349 LCYIYHHMAFELYGMLAGNVSPMTGENIKPAYGGEEEAFLRKVVTPIYNVIAKEAERSKR 408
LCYIYHHMAFELYGMLAGNVSPMTGEN+KPAYGGEE+AFLRKVVTPIY VI EA+RSK+
Sbjct: 358 LCYIYHHMAFELYGMLAGNVSPMTGENVKPAYGGEEDAFLRKVVTPIYEVIQMEAQRSKK 417
Query: 409 GRSKHSQWRNYDDLNEYFWSADCFRLGWPMRADADFFCLPVEHLHFDKLKDNKAD--NRD 466
G+SKHSQWRNYDDLNEYFWS DCFRLGWPMRADADFFCLPV + +K DN RD
Sbjct: 418 GKSKHSQWRNYDDLNEYFWSVDCFRLGWPMRADADFFCLPVAVPNTEKDGDNSKPIVARD 477
Query: 467 RWVGKGNFVEIRSFWHIFRSFDRMWSFFILSLQAMIIVAWNGPGDPTAIFNGDVFKKALS 526
RWVGK NFVEIRSFWH+FRSFDRMWSF+IL LQAMII+AW+G G P+++F DVFKK LS
Sbjct: 478 RWVGKVNFVEIRSFWHVFRSFDRMWSFYILCLQAMIIMAWDG-GQPSSVFGADVFKKVLS 536
Query: 527 VFITAAILKFGQAILDVILNWKAQRSMSMHAKLRYILKVVSGAAWVIVLSVTYAYTWDNP 586
VFITAAI+K GQA+LDVILN+KA +SM++H KLRYILKV S AAWVI+L VTYAY+W +P
Sbjct: 537 VFITAAIMKLGQAVLDVILNFKAHQSMTLHVKLRYILKVFSAAAWVIILPVTYAYSWKDP 596
Query: 587 PGFAQSIQSWFGSNSHSPSMFILAVVVYLSPNMLAAILFLLYIDSNLVVLFPCT*QPRLY 646
P FA++I+SWFGS HSPS+FI+AVV YLSPNMLA L + V T QPRLY
Sbjct: 597 PAFARTIKSWFGSAMHSPSLFIIAVVSYLSPNMLAETNENLLLCCLTDVTIINTLQPRLY 656
Query: 647 VGRGMHESTFSLF 659
VGRGMHES FSLF
Sbjct: 657 VGRGMHESAFSLF 669
>At2g31960 glucan synthase like protein
Length = 1956
Score = 1009 bits (2610), Expect = 0.0
Identities = 501/668 (75%), Positives = 562/668 (84%), Gaps = 20/668 (2%)
Query: 1 MSSRPGPPSSSETQGPPLQRRITRTQTAGNLGEAIFDSEVVPSSLVEIAPILRVANEVEK 60
M+ R GP PP QRRI RTQTAGNLGEA+ DSEVVPSSLVEIAPILRVANEVE
Sbjct: 1 MAQRKGPDP------PPPQRRILRTQTAGNLGEAMLDSEVVPSSLVEIAPILRVANEVEA 54
Query: 61 THPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRIKKSDAREM 120
++PRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLEREN+ TL GR +KSDAREM
Sbjct: 55 SNPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENETTLAGR-QKSDAREM 113
Query: 121 QSFYQHYYKKYIQALQNAADKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDREILETQ 180
QSFYQHYYKKYIQALQNAADKADRAQLTKAYQTA VLFEVLKAVN T+ +EV E
Sbjct: 114 QSFYQHYYKKYIQALQNAADKADRAQLTKAYQTAAVLFEVLKAVNQTEDVEVADE---AH 170
Query: 181 DKVAEKTEILVPYNILPLDPDSANQAIMRFPEIQAAVFALRNTRGLAWPKDYKKKKDEDI 240
KV EK++I VPYNILPLDPDS NQAIMRFPEIQA V ALRNTRGL WP +KKK DED+
Sbjct: 171 TKVEEKSQIYVPYNILPLDPDSQNQAIMRFPEIQATVSALRNTRGLPWPAGHKKKLDEDM 230
Query: 241 LDWLGAMFGFQKHNVANQREHLILLLANVHIRQFPKPDQQPKLDERALTEVMKKLFKNYK 300
LDWL MFGFQK NV+NQREHLILLLANVHIRQFP+P+QQP+LD+RALT VMKKLFKNYK
Sbjct: 231 LDWLQTMFGFQKDNVSNQREHLILLLANVHIRQFPRPEQQPRLDDRALTIVMKKLFKNYK 290
Query: 301 KWCKYLGRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFEL 360
KWCKYLGRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRF+PECLCYIYHHMAFEL
Sbjct: 291 KWCKYLGRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFLPECLCYIYHHMAFEL 350
Query: 361 YGMLAGNVSPMTGENIKPAYGGEEEAFLRKVVTPIYNVIAKEAERSKRGRSKHSQWRNYD 420
YGMLAG+VSPMTGE++KPAYGGE+EAFL+KVVTPIY IAKEA+RS+ G+SKHS+WRNYD
Sbjct: 351 YGMLAGSVSPMTGEHVKPAYGGEDEAFLQKVVTPIYKTIAKEAKRSRGGKSKHSEWRNYD 410
Query: 421 DLNEYFWSADCFRLGWPMRADADFFCLPVEHLHFDKLKDNKADNRDRWVGKGNFVEIRSF 480
DLNEYFWS CFRLGWPMRADADFFC E L D+ +NK DRW+GK NFVEIRSF
Sbjct: 411 DLNEYFWSIRCFRLGWPMRADADFFCQTAEELRLDR-SENKPKTGDRWMGKVNFVEIRSF 469
Query: 481 WHIFRSFDRMWSFFILSLQAMIIVAWNGPGDPTAIFNGDVFKKALSVFITAAILKFGQAI 540
WHIFRSFDRMWSF+ILSLQAMII+AWNG G + IF GDVF K LS+FITAAILK QA+
Sbjct: 470 WHIFRSFDRMWSFYILSLQAMIIIAWNGSGKLSGIFQGDVFLKVLSIFITAAILKLAQAV 529
Query: 541 LDVILNWKAQRSMSMHAKLRYILKVVSGAAWVIVLSVTYAYTWDNPPGFAQSIQSWFGSN 600
LD+ L+WK++ SMS H KLR+I K V+ A WV+++ +TYAY+W P GFA++I++WFG +
Sbjct: 530 LDIALSWKSRHSMSFHVKLRFIFKAVAAAIWVVLMPLTYAYSWKTPSGFAETIKNWFGGH 589
Query: 601 SH-SPSMFILAVVVYLSPNMLAAILF--------LLYIDSNLVVLFPCT*QPRLYVGRGM 651
+ SPS FI+ +++YLSPNML+ +LF L D +V+L QPRLY+GRGM
Sbjct: 590 QNSSPSFFIIVILIYLSPNMLSTLLFAFPFIRRYLERSDYKIVMLMMWWSQPRLYIGRGM 649
Query: 652 HESTFSLF 659
HES SLF
Sbjct: 650 HESALSLF 657
>At1g05570 putative glucan synthase
Length = 1858
Score = 998 bits (2581), Expect = 0.0
Identities = 487/616 (79%), Positives = 548/616 (88%), Gaps = 3/616 (0%)
Query: 12 ETQGPPLQRRITRTQTAGNLGEAIFDSEVVPSSLVEIAPILRVANEVEKTHPRVAYLCRF 71
E PP QRRI RTQT G+LGEA+ DSEVVPSSLVEIAPILRVANEVE ++PRVAYLCRF
Sbjct: 6 EPDPPPPQRRILRTQTVGSLGEAMLDSEVVPSSLVEIAPILRVANEVEASNPRVAYLCRF 65
Query: 72 YAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRIKKSDAREMQSFYQHYYKKY 131
YAFEKAHRLDPTSSGRGVRQFKTALLQRLEREN+ TL GR +KSDAREMQSFYQHYYKKY
Sbjct: 66 YAFEKAHRLDPTSSGRGVRQFKTALLQRLERENETTLAGR-QKSDAREMQSFYQHYYKKY 124
Query: 132 IQALQNAADKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKTEILV 191
IQAL NAADKADRAQLTKAYQTA VLFEVLKAVN T+ +EV EILET +KV EKT+I V
Sbjct: 125 IQALLNAADKADRAQLTKAYQTAAVLFEVLKAVNQTEDVEVADEILETHNKVEEKTQIYV 184
Query: 192 PYNILPLDPDSANQAIMRFPEIQAAVFALRNTRGLAWPKDYKKKKDEDILDWLGAMFGFQ 251
PYNILPLDPDS NQAIMR PEIQAAV ALRNTRGL W +KKK DEDILDWL +MFGFQ
Sbjct: 185 PYNILPLDPDSQNQAIMRLPEIQAAVAALRNTRGLPWTAGHKKKLDEDILDWLQSMFGFQ 244
Query: 252 KHNVANQREHLILLLANVHIRQFPKPDQQPKLDERALTEVMKKLFKNYKKWCKYLGRKSS 311
K NV NQREHLILLLANVHIRQFPKPDQQPKLD+RALT VMKKLF+NYKKWCKYLGRKSS
Sbjct: 245 KDNVLNQREHLILLLANVHIRQFPKPDQQPKLDDRALTIVMKKLFRNYKKWCKYLGRKSS 304
Query: 312 LWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVSPM 371
LWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAG+VSPM
Sbjct: 305 LWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGSVSPM 364
Query: 372 TGENIKPAYGGEEEAFLRKVVTPIYNVIAKEAERSKRGRSKHSQWRNYDDLNEYFWSADC 431
TGE++KPAYGGE+EAFL+KVVTPIY I+KEA+RS+ G+SKHS WRNYDDLNEYFWS C
Sbjct: 365 TGEHVKPAYGGEDEAFLQKVVTPIYQTISKEAKRSRGGKSKHSVWRNYDDLNEYFWSIRC 424
Query: 432 FRLGWPMRADADFFCLPVEHLHFDKLKDNKADNRDRWVGKGNFVEIRSFWHIFRSFDRMW 491
FRLGWPMRADADFFC E L ++ + K+++ DRW+GK NFVEIRSFWHIFRSFDR+W
Sbjct: 425 FRLGWPMRADADFFCQTAEELRLER-SEIKSNSGDRWMGKVNFVEIRSFWHIFRSFDRLW 483
Query: 492 SFFILSLQAMIIVAWNGPGDPTAIFNGDVFKKALSVFITAAILKFGQAILDVILNWKAQR 551
SF+IL LQAMI++AWNG G+ +AIF GDVF K LSVFITAAILK QA+LD+ L+WKA+
Sbjct: 484 SFYILCLQAMIVIAWNGSGELSAIFQGDVFLKVLSVFITAAILKLAQAVLDIALSWKARH 543
Query: 552 SMSMHAKLRYILKVVSGAAWVIVLSVTYAYTWDNPPGFAQSIQSWFGSNSH-SPSMFILA 610
SMS++ KLRY++KV + A WV+V++VTYAY+W N GF+Q+I++WFG +SH SPS+FI+A
Sbjct: 544 SMSLYVKLRYVMKVGAAAVWVVVMAVTYAYSWKNASGFSQTIKNWFGGHSHNSPSLFIVA 603
Query: 611 VVVYLSPNMLAAILFL 626
+++YLSPNML+A+LFL
Sbjct: 604 ILIYLSPNMLSALLFL 619
>At2g13675 callose synthase (1,3-beta-glucan synthase) like protein
Length = 1939
Score = 697 bits (1799), Expect = 0.0
Identities = 369/664 (55%), Positives = 461/664 (68%), Gaps = 21/664 (3%)
Query: 9 SSSETQGPP-LQRRITRTQTAGNLGEAIFDSEVVPSSLVEIAPILRVANEVEKTHPRVAY 67
S+S GP L RR +R+ A + +FD EVVP+SL IAPILRVA E+E PRVAY
Sbjct: 5 STSHDSGPQGLMRRPSRS-AATTVSIEVFDHEVVPASLGTIAPILRVAAEIEHERPRVAY 63
Query: 68 LCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRIKKSDAREMQSFYQHY 127
LCRFYAFEKAHRLDP+S GRGVRQFKT L QRLER+N +L R+KK+D RE++SFYQ Y
Sbjct: 64 LCRFYAFEKAHRLDPSSGGRGVRQFKTLLFQRLERDNASSLASRVKKTDGREVESFYQQY 123
Query: 128 YKKYIQALQNAADKADRAQLTKAYQTANVLFEVLKAVNMTQSME-VDREILETQDKVAEK 186
Y+ Y++AL D+ADRAQL KAYQTA VLFEVL AVN ++ +E V EI+ V EK
Sbjct: 124 YEHYVRALDQG-DQADRAQLGKAYQTAGVLFEVLMAVNKSEKVEAVAPEIIAAARDVQEK 182
Query: 187 TEILVPYNILPLDPDSANQAIMRFPEIQAAVFALRNTRGLAWPKDY----KKKKDEDILD 242
EI PYNILPLD A+Q++M+ E++AAV AL NTRGL WP + KK + D+LD
Sbjct: 183 NEIYAPYNILPLDSAGASQSVMQLEEVKAAVAALGNTRGLNWPSGFEQHRKKTGNLDLLD 242
Query: 243 WLGAMFGFQKHNVANQREHLILLLANVHIRQFPKPDQQPKLDERALTEVMKKLFKNYKKW 302
WL AMFGFQ +NV NQREHL+ L A+ HIR PKP+ KLD+RA+ VM KLFKNYK W
Sbjct: 243 WLRAMFGFQANNVRNQREHLVCLFADNHIRLTPKPEPLNKLDDRAVDTVMSKLFKNYKNW 302
Query: 303 CKYLGRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYG 362
CK+LGRK SL LP Q++QQRK+LYMGLYLLIWGEAAN+RFMPECLCYI+H+MA+EL+G
Sbjct: 303 CKFLGRKHSLRLPQAAQDIQQRKILYMGLYLLIWGEAANIRFMPECLCYIFHNMAYELHG 362
Query: 363 MLAGNVSPMTGENIKPAYGGEEEAFLRKVVTPIYNVIAKEAERSKRGRSKHSQWRNYDDL 422
+LAGNVS +TGENIKP+YGG++EAFLRKV+TPIY V+ EA ++ G++ HS W NYDDL
Sbjct: 363 LLAGNVSIVTGENIKPSYGGDDEAFLRKVITPIYRVVQTEANKNANGKAAHSDWSNYDDL 422
Query: 423 NEYFWSADCFRLGWPMRADADFFCLPVEHLHFDKLKDNKADNRDRWVGKGNFVEIRSFWH 482
NEYFW+ DCF LGWPMR D D F + K KA GK NF E R+FWH
Sbjct: 423 NEYFWTPDCFSLGWPMRDDGDLFKSTRDTTQGKKGSFRKAGR----TGKSNFTETRTFWH 478
Query: 483 IFRSFDRMWSFFILSLQAMIIVAWNGPGDPTAIFNGDVFKKALSVFITAAILKFGQAILD 542
I+ SFDR+W+F++L+LQAMII+A+ + I DV S+FITAA L+F Q++LD
Sbjct: 479 IYHSFDRLWTFYLLALQAMIILAFERV-ELREILRKDVLYALSSIFITAAFLRFLQSVLD 537
Query: 543 VILNWKAQRSMSMHAKLRYILKVVSGAAWVIVLSVTYAYTWDNPPGFAQSIQSWFGSNSH 602
VILN+ LR ILK+V AW +VL + YA + PG + S+
Sbjct: 538 VILNFPGFHRWKFTDVLRNILKIVVSLAWCVVLPLCYAQSVSFAPGKLKQWLSFLPQVKG 597
Query: 603 SPSMFILAVVVYLSPNMLAAILFLLYI--------DSNLVVLFPCT*QPRLYVGRGMHES 654
P ++I+AV +YL PN+LAAI+F+ + D ++ L QPR+YVGRGMHES
Sbjct: 598 VPPLYIMAVALYLLPNVLAAIMFIFPMLRRWIENSDWHIFRLLLWWSQPRIYVGRGMHES 657
Query: 655 TFSL 658
+L
Sbjct: 658 QIAL 661
>At3g14570 hypothetical protein
Length = 1973
Score = 575 bits (1481), Expect = e-164
Identities = 318/711 (44%), Positives = 441/711 (61%), Gaps = 69/711 (9%)
Query: 3 SRP--GPPSSSETQGPPLQRRITRTQTAGNLGEAIFDSEVVPSSLV-EIAPILRVANEVE 59
SRP GP S + R +T + ++ FDSE +P++L EI LR+AN VE
Sbjct: 19 SRPILGPREDSPERATEFTRSLTFRE---HVSSEPFDSERLPATLASEIQRFLRIANLVE 75
Query: 60 KTHPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRIKKSDARE 119
PR+AYLCRF+AFE AH +D S+GRG +F T++ R +KSD RE
Sbjct: 76 SEEPRIAYLCRFHAFEIAHHMDRNSTGRGDEEF--------------TVRRRKEKSDVRE 121
Query: 120 MQSFYQHYYKKYIQALQNAA----DKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDRE 175
++ Y H YK+YI +++ A D + R +L A + A+VL+EVLK V +
Sbjct: 122 LKRVY-HAYKEYI--IRHGAAFNLDNSQREKLINARRIASVLYEVLKTVTSGAGPQA--- 175
Query: 176 ILETQDKVAEKTEILVPYNILPLDPDSANQAIMRFPEIQAAVFALRNTRGLAWPKDYKKK 235
+ ++ + K+E VPYNILPLD +QAIM PEI+AAV +RNTRGL P+++++
Sbjct: 176 -IADRESIRAKSEFYVPYNILPLDKGGVHQAIMHLPEIKAAVAIVRNTRGLPPPEEFQRH 234
Query: 236 KDE-DILDWLGAMFGFQKHNVANQREHLILLLANVHIRQFPKPDQQPKLDERALTEVMKK 294
+ D+ ++L FGFQ NVANQREHLILLL+N IRQ K PK + A+ +MKK
Sbjct: 235 QPFLDLFEFLQYAFGFQNGNVANQREHLILLLSNTIIRQPQKQSSAPKSGDEAVDALMKK 294
Query: 295 LFKNYKKWCKYLGRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYH 354
FKNY WCK+LGRK+++ LP ++QE Q K LY+GLYLLIWGEA+NLRFMPECLCYI+H
Sbjct: 295 FFKNYTNWCKFLGRKNNIRLPYVKQEALQYKTLYIGLYLLIWGEASNLRFMPECLCYIFH 354
Query: 355 HMAFELYGMLAGNVSPMTGENIKPAYGGEEEAFLRKVVTPIYNVIAKEAERSKRGRSKHS 414
HMA+EL+G+L G VS +TGE + PAYGG E+FL VVTPIY V+ KEAE++K G + HS
Sbjct: 355 HMAYELHGVLTGAVSMITGEKVAPAYGGGHESFLADVVTPIYMVVQKEAEKNKNGTADHS 414
Query: 415 QWRNYDDLNEYFWSADCFRLGWPMRADADFFCLPVEH----------LHFDK-------- 456
WRNYDDLNE+FWS +CF +GWPMR + DFFC+ L F K
Sbjct: 415 MWRNYDDLNEFFWSLECFEIGWPMRPEHDFFCVESSETSKPGRWRGMLRFRKQTKKTDEE 474
Query: 457 ---------LKDNKADNRDRWVGKGNFVEIRSFWHIFRSFDRMWSFFILSLQAMIIVAWN 507
L + + RW+GK NFVE RSFW IFRSFDRMWSFF+LSLQA+II+A +
Sbjct: 475 IEDDEELGVLSEEQPKPTSRWLGKTNFVETRSFWQIFRSFDRMWSFFVLSLQALIIMACH 534
Query: 508 GPGDPTAIFNGDVFKKALSVFITAAILKFGQAILDVILNWKAQRSMSMHAKLRYILKVVS 567
G P +FN ++F+ +S+FIT+AILK + ILD+I WKA+ +M ++ K + ++K+
Sbjct: 535 DVGSPLQVFNANIFEDVMSIFITSAILKLIKGILDIIFKWKARNTMPINEKKKRLVKLGF 594
Query: 568 GAAWVIVLSVTYAYTWDNPPGFAQSIQSWFGSNSHSPSMFILAVVVYLSPNMLAAILFLL 627
A W I+L V Y+++ + + ++W G SP +++AV +YL+ + + +LF +
Sbjct: 595 AAMWTIILPVLYSHSRRKYICYFTNYKTWLGEWCFSP--YMVAVTIYLTGSAIELVLFFV 652
Query: 628 -----YIDSNLVVLFPCT---*QPRLYVGRGMHESTFSLFNLDNYKVSIQL 670
YI+++ +F QPRLYVGRGM E+ S F + + + L
Sbjct: 653 PAISKYIETSNHGIFKTLSWWGQPRLYVGRGMQETQVSQFKYTFFWILVLL 703
>At1g06490 glucan synthase, putative
Length = 1933
Score = 572 bits (1474), Expect = e-163
Identities = 318/682 (46%), Positives = 425/682 (61%), Gaps = 45/682 (6%)
Query: 18 LQRRITRTQTA----GNLGEAIFDSEVVPSSLVEIAPILRVANEVEKTHPRVAYLCRFYA 73
+ R++TR T N E DSE+VPSSL IAPILRVAN++++ + RVAYLCRF+A
Sbjct: 24 MSRKMTRAGTMMIEHPNEDERPIDSELVPSSLASIAPILRVANDIDQDNARVAYLCRFHA 83
Query: 74 FEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRIKKSDAREMQSFYQHYYKKYIQ 133
FEKAHR+DPTSSGRGVRQFKT LL +LE E + T + + KSD RE+Q +YQ +Y+ IQ
Sbjct: 84 FEKAHRMDPTSSGRGVRQFKTYLLHKLEEEEEIT-EHMLAKSDPREIQLYYQTFYENNIQ 142
Query: 134 ALQNAADKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKTEILVPY 193
+ K ++ K YQ A VL++VLK V +D + L +V K E Y
Sbjct: 143 ---DGEGKKTPEEMAKLYQIATVLYDVLKTV--VPQARIDDKTLRYAKEVERKKEQYEHY 197
Query: 194 NILPLDPDSANQAIMRFPEIQAAVFALRNTRGLAWPKDYKKKKD------------EDIL 241
NILPL A A+M PEI+AA+ A+ N L P+ + + DIL
Sbjct: 198 NILPLYALGAKTAVMELPEIKAAILAVCNVDNLPRPRFHSASANLDEVDRERGRSFNDIL 257
Query: 242 DWLGAMFGFQKHNVANQREHLILLLANVHIRQFPKPDQQPKLDERALTEVMKKLFKNYKK 301
+WL +FGFQ+ NVANQREHLILLLAN+ +R+ + ++ + ++M+K FKNY
Sbjct: 258 EWLALVFGFQRGNVANQREHLILLLANIDVRKRDL-ENYVEIKPSTVRKLMEKYFKNYNS 316
Query: 302 WCKYLGRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELY 361
WCKYL S L P + QQ LLY+GLYLLIWGEA+N+RFMPECLCYI+H+MA E++
Sbjct: 317 WCKYLRCDSYLRFPA-GCDKQQLSLLYIGLYLLIWGEASNVRFMPECLCYIFHNMANEVH 375
Query: 362 GMLAGNVSPMTGENIKPAYGGEEEAFLRKVVTPIYNVIAKEAERSKRGRSKHSQWRNYDD 421
G+L GNV P+TG+ + A +EEAFLR V+TPIY V+ KE R+K G++ HS+WRNYDD
Sbjct: 376 GILFGNVYPVTGDTYE-AGAPDEEAFLRNVITPIYQVLRKEVRRNKNGKASHSKWRNYDD 434
Query: 422 LNEYFWSADCFRLGWPMRADADFFCLPVEHLHFDKLKDNKADNRD-----RWVGKGNFVE 476
LNEYFW CFRL WPM ADFF +H D++ D + K NFVE
Sbjct: 435 LNEYFWDKRCFRLKWPMNFKADFF------IHTDEISQVPNQRHDQVSHGKRKPKTNFVE 488
Query: 477 IRSFWHIFRSFDRMWSFFILSLQAMIIVAWNGPGDPTAIFNGDVFKKALSVFITAAILKF 536
R+FW+++RSFDRMW F +LSLQ MIIVAW+ G AIF DVF+ L++FIT+A L
Sbjct: 489 ARTFWNLYRSFDRMWMFLVLSLQTMIIVAWHPSGSILAIFTEDVFRNVLTIFITSAFLNL 548
Query: 537 GQAILDVILNWKAQRSMSMHAKLRYILKVVSGAAWVIVLSVTYAYTWDNPPGFAQSIQSW 596
QA LD++L++ A +S+ +RYI K + A W I+L +TY+ + NP G + SW
Sbjct: 549 LQATLDLVLSFGAWKSLKFSQIMRYITKFLMAAMWAIMLPITYSKSVQNPTGLIKFFSSW 608
Query: 597 FGSNSHSPSMFILAVVVYLSPNMLAAILFLL--------YIDSNLVVLFPCT*QPRLYVG 648
GS H S++ A+ +Y+ PN+LAA+ FLL + +V L QP+LY+G
Sbjct: 609 VGSWLHR-SLYDYAIALYVLPNILAAVFFLLPPLRRIMERSNMRIVTLIMWWAQPKLYIG 667
Query: 649 RGMHESTFSLFNLDNYKVSIQL 670
RGMHE F+LF + V + L
Sbjct: 668 RGMHEEMFALFKYTFFWVMLLL 689
>At3g59100 putative protein
Length = 1808
Score = 464 bits (1195), Expect = e-131
Identities = 273/642 (42%), Positives = 373/642 (57%), Gaps = 76/642 (11%)
Query: 8 PSSSETQGPPLQRRITRTQTAGNLGEAIFDSEVVPSSLVEIAPILRVANEVEKTHPRVAY 67
P S + P R T N + DSE+VPSSL IAPILRVANE+EK +PRVAY
Sbjct: 12 PRSLSRRAP--SRATTMMIDRPNEDASAMDSELVPSSLASIAPILRVANEIEKDNPRVAY 69
Query: 68 LCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRIKKSDAREMQSFYQHY 127
LCRF+AFEKAHR+D TSSGRGVRQFKT LL RLE+E + T K ++ K+D RE+Q++YQ++
Sbjct: 70 LCRFHAFEKAHRMDATSSGRGVRQFKTYLLHRLEKEEEET-KPQLAKNDPREIQAYYQNF 128
Query: 128 YKKYIQALQNAADKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKT 187
Y+KYI+ + + + A+L YQ A+VL++VLK V S +VD E ++V K
Sbjct: 129 YEKYIKEGETSRKPEEMARL---YQIASVLYDVLKTV--VPSPKVDYETRRYAEEVERKR 183
Query: 188 EILVPYNILPLDPDSANQAIMRFPEIQAAVFALRNTRGLA---------WPKDYKKKKDE 238
+ YNILPL AI+ PE++AA A+RN R L P + +K + +
Sbjct: 184 DRYEHYNILPLYAVGTKPAIVELPEVKAAFSAVRNVRNLPRRRIHLPSNTPNEMRKARTK 243
Query: 239 --DILDWLGAMFGFQKHNVANQREHLILLLANVHIRQFPKPDQQPKLDERALTEVMKKLF 296
DIL+WL + FGFQ+ NVANQREH+ILLLAN IR+ ++ +L +TE+M K F
Sbjct: 244 LNDILEWLASEFGFQRGNVANQREHIILLLANADIRK-RNDEEYDELKPSTVTELMDKTF 302
Query: 297 KNYKKWCKYLGRKSSLWLPT-----------IQQEVQQRKLLYMGLYLLIWGEAANLRFM 345
K+Y WCKYL S+L + QQ +L+Y+ LYLLIWGEA+N
Sbjct: 303 KSYYSWCKYLHSTSNLKSDVGCFNFILKRFPDDCDKQQLQLIYISLYLLIWGEASN---- 358
Query: 346 PECLCYIYHHMAFELYGMLAGNVSPMTGENIKPAYGGEEEAFLRKVVTPIYNVIAKEAER 405
MA ++YG+L NV ++GE + +EE+FLR V+TPIY VI EA+R
Sbjct: 359 ----------MANDVYGILFSNVEAVSGETYETEEVIDEESFLRTVITPIYQVIRNEAKR 408
Query: 406 SKRGRSKHSQWRNYDDLNEYFWSADCFRLGWPMRADADFFCLPVEHLHFDKLKDNKADNR 465
+K G + HSQWRNYDDLNEYFWS CF++GWP+ ADFF E D+ + +
Sbjct: 409 NKGGTASHSQWRNYDDLNEYFWSKKCFKIGWPLDLKADFFLNSDEITPQDERLNQVTYGK 468
Query: 466 DRWVGKGNFVEIRSFWHIFRSFDRMWSFFILSLQAMIIVAWNGPGDPTAIFNGDVFKKAL 525
+ K NFVE+R+FW++FR FDRMW F +++ QAM+IV W+G G IF+ DVFK L
Sbjct: 469 SK--PKTNFVEVRTFWNLFRDFDRMWIFLVMAFQAMVIVGWHGSGSLGDIFDKDVFKTVL 526
Query: 526 SVFITAAILKFGQAILDVILNWKAQRSMSMHAKLRYILKVVSGAAWVIVLSVTYAYTWDN 585
++FIT+A L Q W ++L + Y+ +
Sbjct: 527 TIFITSAYLTLLQVAF----------------------------MWAVLLPIAYSKSVQR 558
Query: 586 PPGFAQSIQSWFGSNSHSPSMFILAVVVYLSPNMLAAILFLL 627
P G + +W G + S + AV Y+ PN+LAA+LFL+
Sbjct: 559 PTGVVKFFSTWTG-DWKDQSFYTYAVSFYVLPNILAALLFLV 599
>At3g07160 putative glucan synthase
Length = 1931
Score = 342 bits (877), Expect = 4e-94
Identities = 227/654 (34%), Positives = 344/654 (51%), Gaps = 63/654 (9%)
Query: 41 VPSSLV---EIAPILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALL 97
VPSSL +I ILR A+E++ P +A + + + A LDP S GRGV QFKT L+
Sbjct: 36 VPSSLSNNRDIDAILRAADEIQDEDPNIARILCEHGYSLAQNLDPNSEGRGVLQFKTGLM 95
Query: 98 QRLERENDPTLKGRIKKS-DAREMQSFYQHYYKKY-IQALQNA----------ADKADRA 145
++++ G I +S D +Q FY+ Y +K + L+ D+ +R
Sbjct: 96 SVIKQKLAKREVGTIDRSQDILRLQEFYRLYREKNNVDTLKEEEKQLRESGAFTDELERK 155
Query: 146 QLTK--AYQTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKTEILVPYNILPLDPDSA 203
+ + + T VL VL+ + + E+ E+ D A +E + YNI+PLD
Sbjct: 156 TVKRKRVFATLKVLGSVLEQL----AKEIPEELKHVIDSDAAMSEDTIAYNIIPLDAPVT 211
Query: 204 NQAIMRFPEIQAAVFALRNTRGLA-WPKDYK--KKKDEDILDWLGAMFGFQKHNVANQRE 260
A FPE+QAAV AL+ GL P D+ + D+LD+L +FGFQK +V+NQRE
Sbjct: 212 TNATTTFPEVQAAVAALKYFPGLPKLPPDFPIPATRTADMLDFLHYIFGFQKDSVSNQRE 271
Query: 261 HLILLLANVHIRQFPKPDQQPKLDERALTEVMKKLFKNYKKWCKYLGRKSSLWLPTIQQE 320
H++LLLAN R + +PKLD+ A+ +V K +NY KWC YL + + W ++
Sbjct: 272 HIVLLLANEQSRLNIPEETEPKLDDAAVRKVFLKSLENYIKWCDYLCIQPA-W-SNLEAI 329
Query: 321 VQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVSPMTGENIKPAY 380
+KLL++ LY LIWGEAAN+RF+PECLCYI+HHM E+ +L V+ +
Sbjct: 330 NGDKKLLFLSLYFLIWGEAANIRFLPECLCYIFHHMVREMDEILRQQVARPAESCMPVDS 389
Query: 381 GGEEE--AFLRKVVTPIYNVIAKEAERSKRGRSKHSQWRNYDDLNEYFWSADCFRLGWPM 438
G ++ +FL V+ P+Y V++ EA + GR+ HS WRNYDD NEYFWS F LGWP
Sbjct: 390 RGSDDGVSFLDHVIAPLYGVVSAEAFNNDNGRAPHSAWRNYDDFNEYFWSLHSFELGWPW 449
Query: 439 RADADFFCLPVEHLHFDKLKDNKADNRDRWVGKGNFVEIRSFWHIFRSFDRMWSFFILSL 498
R + FF P+ KLK +A +R GK +FVE R+F H++ SF R+W F +
Sbjct: 450 RTSSSFFQKPIPR---KKLKTGRAKHR----GKTSFVEHRTFLHLYHSFHRLWIFLAMMF 502
Query: 499 QAMIIVAWNGPGDPTAIFNGDVFKKALSVFITAAILKFGQAILDVILNWKAQRSMSMHAK 558
QA+ I+A+N + + + LS+ T ++KF +++L+VI+ + A + A
Sbjct: 503 QALAIIAFN----KDDLTSRKTLLQILSLGPTFVVMKFSESVLEVIMMYGAYSTTRRLAV 558
Query: 559 LRYILKVVSGAAWVIVLSVTYAYTWDNPPGFAQSIQSWFGSNSHSP--SMFILAVVVYLS 616
R L+ + W + SV ++ + ++S NS SP ++++ + +Y
Sbjct: 559 SRIFLRFI----WFGLASVFISFLY---------VKSLKAPNSDSPIVQLYLIVIAIYGG 605
Query: 617 PNMLAAILFLLYIDSNL---------VVLFPCT*QPRLYVGRGMHESTFSLFNL 661
+IL + N+ + F Q R YVGRGM+E T NL
Sbjct: 606 VQFFFSILMRIPTCHNIANKCDRWPVIRFFKWMRQERHYVGRGMYERTSDFINL 659
>At4g03550 putative glucan synthase component
Length = 1780
Score = 335 bits (860), Expect = 4e-92
Identities = 190/493 (38%), Positives = 273/493 (54%), Gaps = 45/493 (9%)
Query: 192 PYNILPLDPDSANQAIMRFPEIQAAVFALRNTRGLAWPKDYKKKKDEDILDWLGAMFGFQ 251
PYNI+P++ A+ +RFPE++AA AL+ L P + + D+LDWL FGFQ
Sbjct: 27 PYNIIPVNNLLADHPSLRFPEVRAAAAALKTVGDLRRPPYVQWRSHYDLLDWLALFFGFQ 86
Query: 252 KHNVANQREHLILLLANVHIRQFPKPDQQPKLDERALTEVMKKLFKNYKKWCKYLGRKSS 311
K NV NQREH++L LAN +R P PD LD + +KL NY WC YLG+KS+
Sbjct: 87 KDNVRNQREHMVLHLANAQMRLSPPPDNIDSLDSAVVRRFRRKLLANYSSWCSYLGKKSN 146
Query: 312 LWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVSPM 371
+W+ + +R+LLY+GLYLLIWGEAANLRFMPEC+CYI+H+MA EL +L +
Sbjct: 147 IWISDRNPD-SRRELLYVGLYLLIWGEAANLRFMPECICYIFHNMASELNKILEDCLDEN 205
Query: 372 TGENIKPAYGGEEEAFLRKVVTPIYNVIAKEAERSKRGRSKHSQWRNYDDLNEYFWSADC 431
TG+ P+ G E AFL VV PIY+ I E + SK G H +WRNYDD+NEYFW+ C
Sbjct: 206 TGQPYLPSLSG-ENAFLTGVVKPIYDTIQAEIDESKNGTVAHCKWRNYDDINEYFWTDRC 264
Query: 432 F-RLGWPMRADADFFCLPVEHLHFDKLKDNKADNRDRWVGKGNFVEIRSFWHIFRSFDRM 490
F +L WP+ ++FF +R + VGK FVE R+F++++RSFDR+
Sbjct: 265 FSKLKWPLDLGSNFF-----------------KSRGKSVGKTGFVERRTFFYLYRSFDRL 307
Query: 491 WSFFILSLQAMIIVAWNGPGDPTAIFN--------GDVFKKALSVFITAAILKFGQAILD 542
W L LQA IIVAW D +++ DV + L+VF+T + ++ QA+LD
Sbjct: 308 WVMLALFLQAAIIVAWEEKPDTSSVTRQLWNALKARDVQVRLLTVFLTWSGMRLLQAVLD 367
Query: 543 VILNWKAQRSMSMHAKLRYILKVVSGAAWVIVLSVTYAYTWDNPPGFAQSIQSWFGSNSH 602
+ + R ++KV++ A W++ +V Y W + + W + +
Sbjct: 368 AASQYPLVSRETKRHFFRMLMKVIAAAVWIVAFTVLYTNIWKQ----KRQDRQWSNAATT 423
Query: 603 SPSMFILAVVVYLSPNMLAAILFLL------YIDSNLVVLFPCT--*QPRLYVGRGMHES 654
F+ AV +L P +LA LF++ ++N + F T Q + +VGRG+ E
Sbjct: 424 KIYQFLYAVGAFLVPEILALALFIIPWMRNFLEETNWKIFFALTWWFQGKSFVGRGLREG 483
Query: 655 TFSLFNLDNYKVS 667
+DN K S
Sbjct: 484 L-----VDNIKYS 491
>At4g04970
Length = 1768
Score = 309 bits (792), Expect = 3e-84
Identities = 195/494 (39%), Positives = 261/494 (52%), Gaps = 48/494 (9%)
Query: 193 YNILPLDPDSANQAIMRFPEIQAAVFALRNTRGLAWPKDYKKKKDEDILDWLGAMFGFQK 252
YNI+P+ +R+PE++AA ALR L P D++DWLG +FGFQ
Sbjct: 20 YNIIPIHDFLTEHPSLRYPEVRAAAAALRIVGDLPKPPFADFTPRMDLMDWLGLLFGFQI 79
Query: 253 HNVANQREHLILLLANVHIRQFPKPDQQPKLDERALTEVMKKLFKNYKKWCKYLGRKSSL 312
NV NQRE+L+L LAN +R P P LD L KKL +NY WC +LG + +
Sbjct: 80 DNVRNQRENLVLHLANSQMRLQPPPRHPDGLDPTVLRRFRKKLLRNYTNWCSFLGVRCHV 139
Query: 313 WLPTIQQEVQ-------QRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLA 365
P IQ Q +R+LLY+ LYLLIWGE+ANLRFMPECLCYI+HHMA EL +LA
Sbjct: 140 TSP-IQSRHQTNAVLNLRRELLYVALYLLIWGESANLRFMPECLCYIFHHMAMELNKVLA 198
Query: 366 GNVSPMTGENIKPAYGGEEEAFLRKVVTPIYNVIAKEAERSKRGRSKHSQWRNYDDLNEY 425
G MTG P++ G + AFL+ VV PIY + E E S G HS WRNYDD+NEY
Sbjct: 199 GEFDDMTGMPYWPSFSG-DCAFLKSVVMPIYKTVKTEVESSNNGTKPHSAWRNYDDINEY 257
Query: 426 FWSADCFR-LGWPMRADADFFCLPVEHLHFDKLKDNKADNRDRWVGKGNFVEIRSFWHIF 484
FWS + L WP+ ++FF + VGK FVE RSFW+++
Sbjct: 258 FWSKRALKSLKWPLDYTSNFF---------------DTTPKSSRVGKTGFVEQRSFWNVY 302
Query: 485 RSFDRMWSFFILSLQAMIIVAWNGPGDPTAIFNGDVFKKALSVFITAAILKFGQAILDVI 544
RSFDR+W +L LQA IIVA + P + DV L+VFI+ A L+ Q++LD
Sbjct: 303 RSFDRLWILLLLYLQAAIIVATSDVKFPWQ--DRDVEVALLTVFISWAGLRLLQSVLDAS 360
Query: 545 LNWKAQRSMSMHAKLRYILKVVSGAAWVIVLSVTYAYTWD--NPPGFAQSIQSWFGSNSH 602
+ + +R LK V AW ++ SV YA W N G W + +
Sbjct: 361 TQYSLVSRETYWLFIRLTLKFVVAVAWTVLFSVFYARIWSQKNKDGV------WSRAANE 414
Query: 603 SPSMFILAVVVYLSPNMLAAILFLL------YIDSNLVVLFPCT--*QPRLYVGRGMHES 654
F+ V VY+ P +LA +LF++ + NL V++ T + +VGRGM E
Sbjct: 415 RVVTFLKVVFVYVIPELLALVLFIVPCIRNWVEELNLGVVYFLTWWFYSKTFVGRGMREG 474
Query: 655 TFSLFNLDNYKVSI 668
+DN K ++
Sbjct: 475 L-----VDNVKYTL 483
>At5g36870 putative glucan synthase
Length = 1662
Score = 208 bits (529), Expect = 9e-54
Identities = 134/296 (45%), Positives = 173/296 (58%), Gaps = 24/296 (8%)
Query: 37 DSEVVPSSLVE-IAPILRVANEVEKTHPRVAYLCRF----------------YAFEKAHR 79
DSE+VPSSL E I PILRVA +VE T+PR +L +A +KA+
Sbjct: 24 DSELVPSSLHEDITPILRVAKDVEDTNPRSLFLQDLDIKSVDDSINILSGHSHALDKANE 83
Query: 80 LDPTSSGRGVRQFKTALLQRLERENDPTLKGRIKKSDAREMQSFYQHYYKKYIQALQNAA 139
LDPTSSGR VRQFK +LQ LE+ N+ TLK R K SDA EMQSFYQ Y + I L NA
Sbjct: 84 LDPTSSGRDVRQFKNTILQWLEKNNESTLKARQKSSDAHEMQSFYQQYGDEGINDLLNAG 143
Query: 140 DKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKTEILVPYNILPLD 199
+ +Q TK YQTA VL++VL AV+ +++V +ILE+ +V K +I VPYNILPLD
Sbjct: 144 AGSSSSQRTKIYQTAVVLYDVLDAVHRKANIKVAAKILESHAEVEAKNKIYVPYNILPLD 203
Query: 200 PDSANQAIMRFPEIQAAVFALRNTRGLAWPKDYKKKKDEDILDWLGAMFGFQKHNVANQR 259
PDS N A+MR P+I A + A+R T L W +K DED+LDWL MF FQ
Sbjct: 204 PDSKNHAMMRDPKIVAVLKAIRYTSDLTWQIGHKINDDEDVLDWLKTMFRFQM-----AF 258
Query: 260 EHLILLLANVHIRQF-PKPDQQPKLDERALTEVMKKLFKNYKKWCKYLGR-KSSLW 313
E +L + +++ PK DE LT+V+ ++K + K G K S W
Sbjct: 259 ELFEMLESKGSKKKYKPKNPTYSGKDEDFLTKVVTPVYKTIAEEAKKSGEGKHSEW 314
Score = 192 bits (487), Expect = 7e-49
Identities = 115/273 (42%), Positives = 149/273 (54%), Gaps = 68/273 (24%)
Query: 356 MAFELYGMLAGNVSPMTGENIKPAYGGEEEAFLRKVVTPIYNVIAKEAERSKRGRSKHSQ 415
MAFEL+ ML S + P Y G++E FL KVVTP+Y IA+EA++S G KHS+
Sbjct: 256 MAFELFEMLESKGSKKKYKPKNPTYSGKDEDFLTKVVTPVYKTIAEEAKKS--GEGKHSE 313
Query: 416 WRNYDDLNEYFWSADCF-RLGWPMRADADFFCLPVEHLHFDKLKDNKADNRDRWVGKGNF 474
WRNYDDLNEYFWS +LGWPM+A+ADFFC + L +K
Sbjct: 314 WRNYDDLNEYFWSKQYLDKLGWPMKANADFFCKTSQQLGLNK------------------ 355
Query: 475 VEIRSFWHIFRSFDRMWSFFILSLQAMIIVAWNGPGDPTAIFNGDVFKKALSVFITAAIL 534
+AMII+AWN T+ G VF K LSVFITAA L
Sbjct: 356 -----------------------SEAMIIIAWN----ETSESGGAVFHKVLSVFITAAKL 388
Query: 535 KFGQAILDVILNWKAQRSMSMHAKLRYILKVVSGAAWVIVLSVTYAYTWDNPPGFAQSIQ 594
QA LD+ L+WKA+ SMS H + RYI K V+ A WV+++ +TYAY
Sbjct: 389 NLFQAFLDIALSWKARHSMSTHVRQRYIFKAVAAAVWVLLMPLTYAY------------- 435
Query: 595 SWFGSNSHSPSMFILAVVVYLSPNMLAAILFLL 627
SH+ S+FI+A+++YLSPNML +L L+
Sbjct: 436 ------SHT-SIFIVAILIYLSPNMLPEMLLLI 461
>At3g14780 unknown protein
Length = 347
Score = 111 bits (278), Expect = 1e-24
Identities = 66/169 (39%), Positives = 104/169 (61%), Gaps = 16/169 (9%)
Query: 36 FDSEVVPSSLVE-IAPILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKT 94
+DSE +P +L I LRVAN VE PRVAYLCRFY FE+AHR+D S+GRGVRQFK
Sbjct: 47 YDSEKLPETLASGIQRFLRVANLVESDDPRVAYLCRFYTFEEAHRIDSRSNGRGVRQFKN 106
Query: 95 ALLQRLERENDPTLKGRIKKSDAREMQSFYQHYYKKYI--QALQNAADKADRAQLTKAYQ 152
+LL+RLE++++ T++ R + +D +E++ Y H Y +YI D + + +L A
Sbjct: 107 SLLRRLEKDDEFTIRRRKEINDHKELKRVY-HAYNEYIIRHGASFNLDNSQQEKLINARS 165
Query: 153 TANVLFEVLKAVNMT-------QSMEVDR-----EILETQDKVAEKTEI 189
A+VL+E+L+ + + +S++++R E+L+ + E +I
Sbjct: 166 IASVLYEILRKIESSMGRASTPESIQLNRDEEIGELLDMDTSMEETNKI 214
>At4g26750 unknown protein
Length = 421
Score = 48.1 bits (113), Expect = 2e-05
Identities = 30/116 (25%), Positives = 55/116 (46%), Gaps = 10/116 (8%)
Query: 48 IAPILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPT 107
+ P L+ A+E++K P VAY CR YA E+ ++ + + +L+ +LE++
Sbjct: 11 LLPYLQRADELQKHEPLVAYYCRLYAMERGLKIPQSERTKTTNSILMSLINQLEKDKKSL 70
Query: 108 LKGRIKKSDAREMQSFYQHYYKKYIQALQNAADKADRAQL--TKAYQTANVLFEVL 161
+ D ++ F + K + D+A RA L K + A++ FE+L
Sbjct: 71 ---TLSPDDNMHVEGFALSVFAK-----ADKQDRAGRADLGTAKTFYAASIFFEIL 118
>At2g30680 hypothetical protein
Length = 178
Score = 41.2 bits (95), Expect = 0.002
Identities = 39/134 (29%), Positives = 57/134 (42%), Gaps = 39/134 (29%)
Query: 102 RENDPTLK-GRIKK-SDAREMQSFYQHYYKKYIQ------------------ALQNAADK 141
R ND T + G+ K +D +QSFY YY+K + + +AA +
Sbjct: 33 RRNDETREQGKFKPHTDLPRLQSFYLDYYQKNVMDVIKIIGNQYTILKEIESSAHSAAIR 92
Query: 142 ADRAQLTK-----------------AYQTANVLFEVLKAVNMTQSMEVDREILETQDKVA 184
A A L A TA VLF VL+ +++ +++ E LE V
Sbjct: 93 AIHAALPPHKINGDSRKQADSYIQHACDTAGVLFRVLEL--LSEDVQLPSEFLEADADVK 150
Query: 185 EKTEILVPYNILPL 198
+ EI PYNI+PL
Sbjct: 151 QLAEIFRPYNIIPL 164
>At1g35740 hypothetical protein
Length = 63
Score = 40.8 bits (94), Expect = 0.002
Identities = 24/59 (40%), Positives = 32/59 (53%), Gaps = 2/59 (3%)
Query: 356 MAFELYGMLAGNVSPMTGENIKPAYGGEEEAFLRKVVTPIYNVIA--KEAERSKRGRSK 412
MA+E + NV+ T E KPAY GE + FL+K+VT Y IA KE + R +
Sbjct: 1 MAYERLEIWGNNVNEETRELNKPAYCGERKVFLKKLVTTTYQRIATPKEVQEGSMSRGE 59
>At1g59950 hypothetical protein
Length = 320
Score = 32.3 bits (72), Expect = 0.89
Identities = 17/49 (34%), Positives = 25/49 (50%), Gaps = 2/49 (4%)
Query: 288 LTEVMKKLFKNYKKWCKYLGRKSSLWLPTIQQEVQQRKLLYMGLYLLIW 336
L + +LF K WC L +P IQ+ ++ KL Y+ LYL+ W
Sbjct: 72 LIQSRSELFVTSKLWCA--DAHGGLVVPAIQRSLETLKLDYLDLYLIHW 118
>At4g34560 unknown protein
Length = 221
Score = 32.0 bits (71), Expect = 1.2
Identities = 25/85 (29%), Positives = 41/85 (47%), Gaps = 12/85 (14%)
Query: 473 NFVEIRSFWHIFRSFDRMWSFFILSLQAMIIVAWNGPGDPTAIFNGDVFKKALSVFITAA 532
NF IRSF +FR F+ + ++S + P+ +GD+FK+A ++
Sbjct: 19 NFQAIRSFTSLFRLFELLLLVILISKLSF----------PSVKISGDIFKEAAVFLVSPR 68
Query: 533 ILKF-GQAILDVILNWKAQRSMSMH 556
+ F G AI+ +L K+ R S H
Sbjct: 69 FVFFIGNAIVITLLA-KSGRYSSSH 92
>At2g06420 unknown protein
Length = 249
Score = 31.6 bits (70), Expect = 1.5
Identities = 22/82 (26%), Positives = 36/82 (43%), Gaps = 7/82 (8%)
Query: 231 DYKKK-------KDEDILDWLGAMFGFQKHNVANQREHLILLLANVHIRQFPKPDQQPKL 283
D+KKK K ED+ + L M + + R ++ L+N+ I D+ P++
Sbjct: 119 DFKKKHFKNIVVKHEDVREKLIGMVPLGERSKERLRMMVLYFLSNIIIAPIKTGDKAPQV 178
Query: 284 DERALTEVMKKLFKNYKKWCKY 305
DE L V F +W +Y
Sbjct: 179 DEFCLKAVSDLTFCRNFQWGRY 200
>At5g09880 splicing factor-like protein
Length = 527
Score = 30.0 bits (66), Expect = 4.4
Identities = 16/45 (35%), Positives = 25/45 (55%)
Query: 177 LETQDKVAEKTEILVPYNILPLDPDSANQAIMRFPEIQAAVFALR 221
LE +D VA++ P N + +D +SA +RF ++AA A R
Sbjct: 455 LEIRDDVADECSKYGPVNHIYVDKNSAGFVYLRFQSVEAAAAAQR 499
>At3g24630 hypothetical protein
Length = 724
Score = 29.3 bits (64), Expect = 7.5
Identities = 32/150 (21%), Positives = 65/150 (43%), Gaps = 16/150 (10%)
Query: 39 EVVPSSLVEIAPILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSG--RGVRQFKTAL 96
+V PSSL +LRV+ ++K + + L + ++FE R + + RG + +L
Sbjct: 69 DVPPSSL----QLLRVSKGIQKLNVAIESLSKGFSFEAVSRPEDIAKDLLRGALDLEESL 124
Query: 97 -----LQRLERENDPTLK--GRIKKSDAREMQSFYQHYYKKYIQALQNAADKADRAQLTK 149
+Q + + P ++ GR R M + +K + +N A K +L K
Sbjct: 125 AMLSSIQEDDSKRKPMIRNDGRSDLRFQRSMSDRFGERIEKRMMVQENVASKDCYEELRK 184
Query: 150 AYQTANVLFEVLKAVNMTQSMEVDREILET 179
+ + F V+ T ++E + ++ +
Sbjct: 185 VIRES---FLRQNLVSQTTTIETKKRVVRS 211
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,068,257
Number of Sequences: 26719
Number of extensions: 644440
Number of successful extensions: 1645
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1548
Number of HSP's gapped (non-prelim): 31
length of query: 676
length of database: 11,318,596
effective HSP length: 106
effective length of query: 570
effective length of database: 8,486,382
effective search space: 4837237740
effective search space used: 4837237740
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC137603.8


