

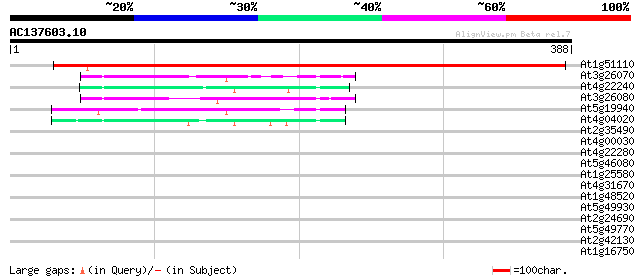
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.10 + phase: 0
(388 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g51110 unknown protein 477 e-135
At3g26070 unknown protein 56 3e-08
At4g22240 fibrillin precursor-like protein 51 1e-06
At3g26080 unknown protein 50 2e-06
At5g19940 unknown protein 43 3e-04
At4g04020 putative fibrillin 42 4e-04
At2g35490 putative fibrillin 34 0.16
At4g00030 unknown protein 33 0.27
At4g22280 putative protein 32 0.59
At5g46080 serine/threonine protein kinase-like protein 30 2.3
At1g25580 unknown protein 30 2.3
At4g31670 unknown protein 30 2.9
At1g48520 unknown protein 30 2.9
At5g49930 putative protein 29 5.0
At2g24690 unknown protein 29 5.0
At5g49770 receptor protein kinase-like 28 6.6
At2g42130 unknown protein 28 6.6
At1g16750 unknown protein 28 8.6
>At1g51110 unknown protein
Length = 409
Score = 477 bits (1227), Expect = e-135
Identities = 239/360 (66%), Positives = 286/360 (79%), Gaps = 6/360 (1%)
Query: 31 NSRFVSAGCSKVEQISIVTEES-----ENSLIQALVGIQGRGRSSSPQQLNAIERAIQVL 85
N R + CS ++ T++S E LI AL+GIQGRG+S+SP+QLN +E A++VL
Sbjct: 48 NCRTLRISCSSSSTVTDQTQQSSFNDAELKLIDALIGIQGRGKSASPKQLNDVESAVKVL 107
Query: 86 EHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQEVYLQ-TNDPRVT 144
E + G+ +PT+S LIEGRW+L+FTTRPGTASPIQRTF GVD F+VFQ+VYL+ TNDPRV+
Sbjct: 108 EGLEGIQNPTDSDLIEGRWRLMFTTRPGTASPIQRTFTGVDVFTVFQDVYLKATNDPRVS 167
Query: 145 NIVSFSDAIGELKVEAAASIGDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGW 204
NIV FSD IGELKVEA ASI DGKR+LFRFDRAAF KFLPFKVPYPVPF+LLGDEAKGW
Sbjct: 168 NIVKFSDFIGELKVEAVASIKDGKRVLFRFDRAAFDLKFLPFKVPYPVPFRLLGDEAKGW 227
Query: 205 LDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKLLTAISSGVGVREAIDKLISLNKNSG 264
LDTTYLS SGNLRISRGNKGTTFVLQK+T PRQKLL IS GV EAID+ ++ N NS
Sbjct: 228 LDTTYLSPSGNLRISRGNKGTTFVLQKETVPRQKLLATISQDKGVAEAIDEFLASNSNSA 287
Query: 265 EEDPELEEGEWQMIWNSQTVTDSWLENAANGLMGKQIVEKNGRIKYVVNILLGLKFSMSG 324
E++ EL EG WQMIW+SQ TDSW+ENAANGLMG+QI+EK+GRIK+ VNI+ +FSM G
Sbjct: 288 EDNYELLEGSWQMIWSSQMYTDSWIENAANGLMGRQIIEKDGRIKFEVNIIPAFRFSMKG 347
Query: 325 IFVKSSPKVYEVTMDDAAIIGGPFGYPLEFGKKFILEILFNDGKVRISRGDNEIIFVHAR 384
F+KS Y++ MDDAAIIGG FGYP++ L+IL+ D K+RISRG + IIFVH R
Sbjct: 348 KFIKSESSTYDLKMDDAAIIGGAFGYPVDITNNIELKILYTDEKMRISRGFDNIIFVHIR 407
>At3g26070 unknown protein
Length = 242
Score = 56.2 bits (134), Expect = 3e-08
Identities = 46/193 (23%), Positives = 92/193 (46%), Gaps = 27/193 (13%)
Query: 50 EESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFT 109
++ + L++A+ ++ RG ++SP I++ + +E + +P S L+ G+W+LI+T
Sbjct: 72 KQLKQELLEAIEPLE-RGATASPDDQLRIDQLARKVEAVNPTKEPLKSDLVNGKWELIYT 130
Query: 110 TRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPRVTNIVS---FSDAIGELKVEAAASIGD 166
T + F+ S+ + + +V N+ + ++ G++K + +
Sbjct: 131 TSASILQAKKPRFLR----SITNYQSINVDTLKVQNMETWPFYNSVTGDIKPLNSKKVA- 185
Query: 167 GKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTT 226
K +F+ F+P K P D A+G L+ TY+ LR+SRG+KG
Sbjct: 186 VKLQVFKI------LGFIPIKAP---------DSARGELEITYVDE--ELRLSRGDKGNL 228
Query: 227 FVLQKQTEPRQKL 239
F+L K +P ++
Sbjct: 229 FIL-KMFDPTYRI 240
>At4g22240 fibrillin precursor-like protein
Length = 310
Score = 50.8 bits (120), Expect = 1e-06
Identities = 56/228 (24%), Positives = 88/228 (38%), Gaps = 45/228 (19%)
Query: 49 TEESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSS-LIEGRWQLI 107
TE + SL+ +L G RG S+S + I I LE PT + L+ G+W L
Sbjct: 83 TERLKRSLVDSLYGTD-RGLSASSETRAEIGDLITQLESKNPTPAPTEALFLLNGKWILA 141
Query: 108 FTTRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPRVTNIVSFSDAIG--ELKVEAAASIG 165
+T+ + R V + + + +++ V N V F+ +G + A I
Sbjct: 142 YTSFVNLFPLLSRGIVPLIKVDEISQT-IDSDNFTVQNSVRFAGPLGTNSISTNAKFEIR 200
Query: 166 DGKRILFRFDRAAFSFKFLPFKVPYP---------------------------------- 191
KR+ +F++ L + P
Sbjct: 201 SPKRVQIKFEQGVIGTPQLTDSIEIPEYVEVLGQKIDLNPIRGLLTSVQDTASSVARTIS 260
Query: 192 ----VPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEP 235
+ F L D A+ WL TTYL ++RISRG+ G+ FVL K+ P
Sbjct: 261 SQPPLKFSLPADNAQSWLLTTYLDK--DIRISRGDGGSVFVLIKEGSP 306
>At3g26080 unknown protein
Length = 234
Score = 50.4 bits (119), Expect = 2e-06
Identities = 47/195 (24%), Positives = 83/195 (42%), Gaps = 30/195 (15%)
Query: 50 EESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFT 109
++ ++ L++A+ ++ RG ++SP I++ + +E + +P S LI G+W+LI+T
Sbjct: 63 KQLKHELVEAIEPLE-RGATASPDDQLLIDQLARKVEAVNPTKEPLKSDLINGKWELIYT 121
Query: 110 TRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPR----VTNIVSFS-DAIGELKVEAAASI 164
T LQ PR +TN + D + ++E
Sbjct: 122 T---------------------SAAILQAKKPRFLRSLTNYQCINMDTLKVQRMETWPFY 160
Query: 165 GDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKG 224
L + + K FK+ +P K A+G L+ TY+ LRISRG KG
Sbjct: 161 NSVTGDLTPLNSKTVAVKLQVFKILGFIPVKAPDGTARGELEITYVDE--ELRISRG-KG 217
Query: 225 TTFVLQKQTEPRQKL 239
+ K +P ++
Sbjct: 218 NLLFILKMFDPTYRI 232
>At5g19940 unknown protein
Length = 239
Score = 43.1 bits (100), Expect = 3e-04
Identities = 49/209 (23%), Positives = 85/209 (40%), Gaps = 18/209 (8%)
Query: 30 SNSRFVSAGCSKVEQISIVTEESENSLIQAL---VGIQGRGRSSSPQQLNAIERAIQVLE 86
S SR + S V ++ + LI L V G + SP+Q + + L+
Sbjct: 42 SRSRRLGLVVSSVSAPNVELRTGPDDLISTLLSKVANSDGGVTLSPEQHKEVAQVAGELQ 101
Query: 87 HIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPRVTNI 146
V +P + LI G W++++ +RP + R+ +G FF + + V N
Sbjct: 102 KYC-VKEPVKNPLIFGDWEVVYCSRPTSPGGGYRSVIGRLFFKTKEMIQAIDAPDIVRNK 160
Query: 147 VS---FSDAIGELKVEAAASIGDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKG 203
VS F G++ + D + + F+ L FK G E++
Sbjct: 161 VSINAFGFLDGDVSLTGKLKALDSEWVQVIFEPPEIKVGSLEFK---------YGFESEV 211
Query: 204 WLDTTYLSHSGNLRISRGNKGTTFVLQKQ 232
L TY+ LR+ G+KG+ FV +++
Sbjct: 212 KLRITYVDE--KLRLGLGSKGSLFVFRRR 238
>At4g04020 putative fibrillin
Length = 318
Score = 42.4 bits (98), Expect = 4e-04
Identities = 62/247 (25%), Positives = 91/247 (36%), Gaps = 52/247 (21%)
Query: 30 SNSRFVSAGCSKVEQISIVTEESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIG 89
++S VS +E + TE + SL +L G RG S S I I LE
Sbjct: 73 ASSSTVSVADKAIESVE-ETERLKRSLADSLYGTD-RGLSVSSDTRAEISELITQLESKN 130
Query: 90 GVSDPTNSS-LIEGRWQLIFTTRPGTASPIQRTF---VGVDFFSVFQEVYLQTNDPRVTN 145
P + L+ G+W L +T+ G + R V VD S + ++ V N
Sbjct: 131 PTPAPNEALFLLNGKWILAYTSFVGLFPLLSRRIEPLVKVDEISQT----IDSDSFTVQN 186
Query: 146 IVSFSDAIG--ELKVEAAASIGDGKRILFRFDRAAF-------------SFKFLPFKVPY 190
V F+ A I KR+ +F++ S + L K+
Sbjct: 187 SVRFAGPFSTTSFSTNAKFEIRSPKRVQIKFEQGVIGTPQLTDSIEIPESVEVLGQKIDL 246
Query: 191 -------------------------PVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGT 225
P+ F L D + WL TTYL +LRISRG+ G+
Sbjct: 247 NPIKGLLTSVQDTASSVARTISNQPPLKFSLPSDNTQSWLLTTYLDK--DLRISRGDGGS 304
Query: 226 TFVLQKQ 232
+VL K+
Sbjct: 305 VYVLIKE 311
>At2g35490 putative fibrillin
Length = 376
Score = 33.9 bits (76), Expect = 0.16
Identities = 21/48 (43%), Positives = 28/48 (57%), Gaps = 9/48 (18%)
Query: 185 PFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQ 232
P K+P+P G+ WL TTYL +LRISRG+ G FVL ++
Sbjct: 331 PLKLPFP------GNRGSSWLLTTYLDK--DLRISRGD-GGLFVLARE 369
>At4g00030 unknown protein
Length = 212
Score = 33.1 bits (74), Expect = 0.27
Identities = 38/142 (26%), Positives = 70/142 (48%), Gaps = 21/142 (14%)
Query: 252 AIDKLISLNKNSGEEDPELEEGEWQMIWNSQTVTDSWLENAANGLMGK------QIVEKN 305
AI+ + + ++S D L W+++W ++ +E A GL G Q+++ N
Sbjct: 66 AIESMTVIGRSSITTDDSLS-ATWRLLWTTEKEQLFIIEKA--GLFGTTAGDVLQVIDVN 122
Query: 306 GRIKYVVNILL----GLKFSMSGIFVKSSPKVYEVTMDDAAIIGGPFGYPLE-FGKKFIL 360
RI + N++ G+ F S I + +SP+ + A + G + PL FGK +
Sbjct: 123 KRI--LNNVITFPPDGVFFVRSDIDI-ASPQRVNFRFNSAVLRGKNWELPLPPFGKGWF- 178
Query: 361 EILFNDGKVRIS---RGDNEII 379
E ++ DG++R++ RGD I+
Sbjct: 179 ENVYMDGEIRVAKDIRGDYLIV 200
>At4g22280 putative protein
Length = 467
Score = 32.0 bits (71), Expect = 0.59
Identities = 15/62 (24%), Positives = 30/62 (48%)
Query: 79 ERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQEVYLQT 138
ER I++ + + +++ T++S++ RW+ +F RP Q G F V + T
Sbjct: 61 ERFIRIWQRMDRINEATSTSVLSMRWRYLFAFRPNLCLDDQEVGGGDSFIDFVDRVLVVT 120
Query: 139 ND 140
+
Sbjct: 121 GN 122
>At5g46080 serine/threonine protein kinase-like protein
Length = 332
Score = 30.0 bits (66), Expect = 2.3
Identities = 27/102 (26%), Positives = 45/102 (43%), Gaps = 6/102 (5%)
Query: 128 FSVFQEVYLQTNDPRVTNIVSFSDAIGELKVEAAASIGDGKRILFRFDRAAFSFKFLPFK 187
FS + ++ P V +I+ FS+A GE ++ +G+GK + + S + F
Sbjct: 110 FSTVIKSLSSSHHPNVVSILGFSEAPGE-RIVVTEFVGEGKSLSDHLHGGSNSATAVEF- 167
Query: 188 VPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVL 229
+ FK+ A+G YL N RI G ++ VL
Sbjct: 168 -GWKTRFKIAAGAARG---LEYLHEIANPRIVHGRFTSSNVL 205
>At1g25580 unknown protein
Length = 449
Score = 30.0 bits (66), Expect = 2.3
Identities = 17/46 (36%), Positives = 26/46 (55%), Gaps = 2/46 (4%)
Query: 260 NKNSGEEDPELEEGEWQMIWNSQTVTDSWLENAANGLMGKQIVEKN 305
N+N GEEDP + Q I NSQ + ++ + + L+G Q E+N
Sbjct: 335 NENQGEEDPTWFDSGSQFILNSQQLVEAL--SLCDDLLGSQDREEN 378
>At4g31670 unknown protein
Length = 631
Score = 29.6 bits (65), Expect = 2.9
Identities = 14/31 (45%), Positives = 20/31 (64%)
Query: 26 SPNPSNSRFVSAGCSKVEQISIVTEESENSL 56
SP+PS S S CS+VE+I + ES +S+
Sbjct: 544 SPSPSPSVLASECCSEVERIDTLDSESNSSI 574
>At1g48520 unknown protein
Length = 550
Score = 29.6 bits (65), Expect = 2.9
Identities = 20/65 (30%), Positives = 30/65 (45%), Gaps = 5/65 (7%)
Query: 296 LMGKQIVEKNGRIKYVVNILLGLK--FSMSGIFVKSSPKVYEVTMDDAAIIGGPF---GY 350
++ ++VE R+ +N L LK F F PK Y+++ D I G +
Sbjct: 117 VLNSKVVEFGVRLGLALNCDLSLKSKFDRKQYFYPDLPKGYQISQFDIPIASGGYVDVDI 176
Query: 351 PLEFG 355
PLEFG
Sbjct: 177 PLEFG 181
>At5g49930 putative protein
Length = 1080
Score = 28.9 bits (63), Expect = 5.0
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 8/45 (17%)
Query: 266 EDPELEEGEWQMIWNSQTVTDSWLENAANG--------LMGKQIV 302
ED +L++ E Q++ + V + WLE+ NG LM KQI+
Sbjct: 251 EDKKLDDNEIQLLVQAVIVFEDWLEDIINGQKVPEGYILMQKQIL 295
>At2g24690 unknown protein
Length = 682
Score = 28.9 bits (63), Expect = 5.0
Identities = 23/92 (25%), Positives = 39/92 (42%), Gaps = 11/92 (11%)
Query: 191 PVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNK----------GTTFVLQKQTEPRQKLL 240
P LLG + W+ + L G + + +G K G + L+ E R +L
Sbjct: 216 PGEITLLGTDGAKWMASLLLEKKGRMSLGKGWKDFAKANGLKTGDSITLEPIWEDRTPVL 275
Query: 241 T-AISSGVGVREAIDKLISLNKNSGEEDPELE 271
+ SSG G E + +S+ +SG ++E
Sbjct: 276 SIKSSSGKGQSEFSKESLSIKPSSGNMTKKVE 307
>At5g49770 receptor protein kinase-like
Length = 946
Score = 28.5 bits (62), Expect = 6.6
Identities = 22/70 (31%), Positives = 33/70 (46%), Gaps = 3/70 (4%)
Query: 205 LDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKLLTAISSGVGVREAIDKLISLNKNSG 264
LDTT + +SGNL +G + V + EP +S V E+I +L+ LN N+
Sbjct: 862 LDTTIIQNSGNL---KGFEKYVDVALQCVEPEGVNRPTMSEVVQELESILRLVGLNPNAD 918
Query: 265 EEDPELEEGE 274
E G+
Sbjct: 919 SATYEEASGD 928
>At2g42130 unknown protein
Length = 270
Score = 28.5 bits (62), Expect = 6.6
Identities = 19/58 (32%), Positives = 30/58 (50%), Gaps = 6/58 (10%)
Query: 52 SENSLIQALVGIQG-----RGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRW 104
++ SL+ AL G G++++ Q+++ ER I LE + PT S EGRW
Sbjct: 71 AKESLLLALKDAGGFEALVTGKTTNMQRIDVNER-ITSLERLNPTPRPTTSPCFEGRW 127
>At1g16750 unknown protein
Length = 529
Score = 28.1 bits (61), Expect = 8.6
Identities = 13/38 (34%), Positives = 20/38 (52%)
Query: 43 EQISIVTEESENSLIQALVGIQGRGRSSSPQQLNAIER 80
E + EE++ IQA VG+ RG+ PQ L+ +
Sbjct: 433 ENVGEELEEAQKDYIQASVGVSPRGKLIVPQMLHCFAK 470
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,725,113
Number of Sequences: 26719
Number of extensions: 375346
Number of successful extensions: 965
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 944
Number of HSP's gapped (non-prelim): 26
length of query: 388
length of database: 11,318,596
effective HSP length: 101
effective length of query: 287
effective length of database: 8,619,977
effective search space: 2473933399
effective search space used: 2473933399
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137603.10


