

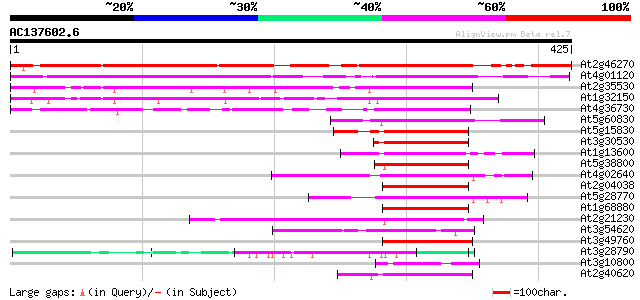
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137602.6 + phase: 0
(425 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g46270 G-box binding bZIP transcription factor GBF3 / AtbZip55 393 e-109
At4g01120 G-box binding bZip transcription factor GBF2 / AtbZip54 311 3e-85
At2g35530 bZIP transcription factor AtbZip16 202 4e-52
At1g32150 putative G-Box binding protein 197 9e-51
At4g36730 G-box binding bZip transcription factor GBF1 / AtbZip41 177 1e-44
At5g60830 bZip transcription factor AtbZip70 64 2e-10
At5g15830 bZIP transcription factor AtbZip3 62 5e-10
At3g30530 bZip transcription factor AtbZip42 59 4e-09
At1g13600 bZip transcription factor AtbZip58 59 4e-09
At5g38800 bZIP transcription factor AtbZip43 57 1e-08
At4g02640 bZIP protein BZO2H1 56 3e-08
At2g04038 bZip transcription factor AtbZip48 55 7e-08
At5g28770 bZip transcription factor BZO2H3 / AtbZip63 55 1e-07
At1g68880 bZip transcription factor AtbZip8 52 6e-07
At2g21230 bZIP transcription factor AtbZip30 50 2e-06
At3g54620 bZIP transcription factor-like protein 50 2e-06
At3g49760 bZIP transcription factor AtbZip5 50 3e-06
At3g28790 unknown protein 49 4e-06
At3g10800 bZIP transcription factor AtbZip28 49 4e-06
At2g40620 bZIP transcription factor AtbZip18 49 4e-06
>At2g46270 G-box binding bZIP transcription factor GBF3 / AtbZip55
Length = 382
Score = 393 bits (1009), Expect = e-109
Identities = 231/430 (53%), Positives = 281/430 (64%), Gaps = 53/430 (12%)
Query: 1 MGNSDEEKS--TKTEKPSSPVTVDQTNQTNVHVYPDWAAMQAYYGPRVAMPPYYNSPVA- 57
MGNS EE TK++KPSSP +QTNVHVYPDWAAMQAYYGPRVAMPPYYNS +A
Sbjct: 1 MGNSSEEPKPPTKSDKPSSP----PVDQTNVHVYPDWAAMQAYYGPRVAMPPYYNSAMAA 56
Query: 58 SGHTPHPYMWGPPQPMMPPYGHPYAAMYPHGG-VYTHPAVPIGPHPHSQGISSSPATGTP 116
SGH P PYMW P Q MM PYG PYAA+YPHGG VY HP +P+G P Q GT
Sbjct: 57 SGHPPPPYMWNP-QHMMSPYGAPYAAVYPHGGGVYAHPGIPMGSLPQGQKDPPLTTPGTL 115
Query: 117 LSIETPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEGSSDGS 176
LSI+TP KS+GNTD GLMKKLK FDGLAMS+GNG+ E+ R S T+GS+DGS
Sbjct: 116 LSIDTPTKSTGNTDNGLMKKLKEFDGLAMSLGNGNPENGAD-EHKRSRNSSETDGSTDGS 174
Query: 177 DGNTSGANQTRRKRSREGTPTTDGEGKTNTQGSQISKEIAASDKMMAVAPA-GVTGQLVG 235
DGNT+GA++ + KRSREGTPT DG K++ + +V+P+ G TG +
Sbjct: 175 DGNTTGADEPKLKRSREGTPTKDG------------KQLVQASSFHSVSPSSGDTGVKLI 222
Query: 236 PVASSAMTTALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESA 295
+ + + S V + +NP + Q A++PPE W+QNERELKRERRKQSNRESA
Sbjct: 223 QGSGAIL--------SPGVSANSNPFMS-QSLAMVPPETWLQNERELKRERRKQSNRESA 273
Query: 296 RRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFKIAKLGQ 355
RRSRLRKQAE EELARKVE+L AE+ +LRSE+N+L E S++LR NA L +K K +
Sbjct: 274 RRSRLRKQAETEELARKVEALTAENMALRSELNQLNEKSDKLRGANATLLDKLKCS---- 329
Query: 356 PKEIILTNIDSQRTTPVSTENLLSRVNNNSGSNDRTVEDENGYCDNKPNSGAKLHQLLDA 415
+ ++ P N+LSRV NSG+ D+ DN NS +KLHQLLD
Sbjct: 330 ---------EPEKRVPA---NMLSRV-KNSGAGDK----NKNQGDNDSNSTSKLHQLLDT 372
Query: 416 SPRADAVAAG 425
PRA AVAAG
Sbjct: 373 KPRAKAVAAG 382
>At4g01120 G-box binding bZip transcription factor GBF2 / AtbZip54
Length = 360
Score = 311 bits (798), Expect = 3e-85
Identities = 192/428 (44%), Positives = 247/428 (56%), Gaps = 73/428 (17%)
Query: 1 MGNSDEEKSTK-TEKPSSPVTVDQTNQTNVHVYP-DWAAMQAYYGPRVAMPPYYNSPVAS 58
MG+++E T ++KPS +Q+N VHVY DWAAMQAYYGPRV +P YYNS +A
Sbjct: 1 MGSNEEGNPTNNSDKPSQAAAPEQSN---VHVYHHDWAAMQAYYGPRVGIPQYYNSNLAP 57
Query: 59 GHTPHPYMWGPPQPMMPPYGHPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLS 118
GH P PYMW P PMM PYG PY P GGVY HP V +G P S+ TPL+
Sbjct: 58 GHAPPPYMWASPSPMMAPYGAPYPPFCPPGGVYAHPGVQMGSQPQGPVSQSASGVTTPLT 117
Query: 119 IETPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAE-PGAESRQSQSVNTEGSSDGSD 177
I+ P S+GN+D G MKKLK FDGLAMSI N SAE +E R SQS +GSS+GSD
Sbjct: 118 IDAPANSAGNSDHGFMKKLKEFDGLAMSISNNKVGSAEHSSSEHRSSQSSENDGSSNGSD 177
Query: 178 GNTSGANQTRRKRSREGTPTTDGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGPV 237
GNT+G Q+RRKR ++ +P+T GE ++ + E D M
Sbjct: 178 GNTTGGEQSRRKRRQQRSPST-GERPSSQNSLPLRGENEKPDVTM--------------- 221
Query: 238 ASSAMTTALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESARR 297
+ M TA+ +NS+ ++ PQP W NE+E+KRE+RKQSNRESARR
Sbjct: 222 GTPVMPTAMSFQNSAGMN------GVPQP--------W--NEKEVKREKRKQSNRESARR 265
Query: 298 SRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFKIAKLGQPK 357
SRLRKQAE E+L+ KV++L AE+ SLRS++ +L SE+LR+EN A+ ++ K G+
Sbjct: 266 SRLRKQAETEQLSVKVDALVAENMSLRSKLGQLNNESEKLRLENEAILDQLKAQATGK-- 323
Query: 358 EIILTNIDSQRTTPVSTENLLSRVN-NNSGSNDRTVEDENGYCDNKPNSGAKLHQLLDAS 416
TENL+SRV+ NNS S +TV+ HQLL+AS
Sbjct: 324 ----------------TENLISRVDKNNSVSGSKTVQ----------------HQLLNAS 351
Query: 417 PRADAVAA 424
P D VAA
Sbjct: 352 PITDPVAA 359
>At2g35530 bZIP transcription factor AtbZip16
Length = 409
Score = 202 bits (513), Expect = 4e-52
Identities = 144/382 (37%), Positives = 199/382 (51%), Gaps = 45/382 (11%)
Query: 1 MGNSDEEKSTKTEKPSS---PVTVDQTNQTNVHVY-PDWAAMQAYYGPRVAMPPYYNSPV 56
M S +EK KT PSS P + + ++ + + PDW+ QAY MPP + V
Sbjct: 6 MEKSSKEKEPKTPPPSSTAPPSSQEPSSAVSAGMATPDWSGFQAYS----PMPPPHGY-V 60
Query: 57 ASGHTPHPYMWGPPQPMMPPYG---HPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSPAT 113
AS PHPYMWG Q MMPPYG HPY AMYP GG+Y HP++P G +P+S SP
Sbjct: 61 ASSPQPHPYMWGV-QHMMPPYGTPPHPYVAMYPPGGMYAHPSMPPGSYPYSPYAMPSPNG 119
Query: 114 GTPLSIETPPKSSGNTDQGLMKK---LKGFDGLAMSIGNGHAESAEPGAES--------R 162
T +S T + G+ Q +K+ +K G S+ ++ EPG S
Sbjct: 120 MTEVSGNTTGGTDGDAKQSEVKEKLPIKRSRGSLGSLNMITGKNNEPGKNSGASANGAYS 179
Query: 163 QSQSVNTEGSSDGSDGNT-----SGANQTRRKRSREGTPTTDG--EGKTNTQGSQISKEI 215
+S ++GSS+GSDGN+ SG + + + E + +G G T +S+ +
Sbjct: 180 KSGESASDGSSEGSDGNSQNDSGSGLDGKDAEAASENGGSANGPQNGSAGTPILPVSQTV 239
Query: 216 AASDKMMAVAPAGVTGQLVGPVASSAMTTALELRNSSSVHSKTNPTSTPQPSAVLP---- 271
A P T +G A T+A +H K STP P V P
Sbjct: 240 PIMPMTAAGVPGPPTNLNIGMDYWGAPTSA----GIPGMHGKV---STPVPGVVAPGSRD 292
Query: 272 ---PEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEIN 328
+ W+Q++RELKR+RRKQSNRESARRSRLRKQAE +ELA++ E LN E+ +LR+EIN
Sbjct: 293 GGHSQPWLQDDRELKRQRRKQSNRESARRSRLRKQAECDELAQRAEVLNEENTNLRAEIN 352
Query: 329 RLAENSERLRMENAALKEKFKI 350
+L E L EN +LK++ +
Sbjct: 353 KLKSQCEELTTENTSLKDQLSL 374
>At1g32150 putative G-Box binding protein
Length = 389
Score = 197 bits (501), Expect = 9e-51
Identities = 150/403 (37%), Positives = 209/403 (51%), Gaps = 52/403 (12%)
Query: 1 MGNSDEEKSTKTEKP---------SSPVTVDQTNQTN-----VHVYPDWAAMQAYYGPRV 46
MG+S+ EKS K ++P S+P TV ++ V V DW+ QAY
Sbjct: 1 MGSSEMEKSGKEKEPKTTPPSTSSSAPATVVSQEPSSAVSAGVAVTQDWSGFQAYS---- 56
Query: 47 AMPPYYNSPVASGHTPHPYMWGPPQPMMPPYG---HPYAAMYPHGGVYTHPAVPIGPHPH 103
MPP+ VAS PHPYMWG Q MMPPYG HPY MYP GG+Y HP++P G +P+
Sbjct: 57 PMPPH--GYVASSPQPHPYMWGV-QHMMPPYGTPPHPYVTMYPPGGMYAHPSLPPGSYPY 113
Query: 104 SQGISSSP-----ATGTPLS-IETPPKSSGNTDQGLMKKLKGFDG-LAMSIGNGHAESAE 156
S SP A+G S IE K S ++ +K+ KG G L M IG +
Sbjct: 114 SPYAMPSPNGMAEASGNTGSVIEGDGKPSDGKEKLPIKRSKGSLGSLNMIIGKNNEAGKN 173
Query: 157 PGAESR----QSQSVNTEGSSDGSDGNTSGANQTRRKRSREGTPTTDGEGKTNTQGSQIS 212
GA + +S ++GSSDGSD N+ + +R ++G ++ G + G +
Sbjct: 174 SGASANGACSKSAESGSDGSSDGSDANSQNDSGSRHN-GKDGETASESGGSAH--GPPRN 230
Query: 213 KEIAASDKMMAVAPAGVTGQLVGPVASSAMTTALELRNSSSVHSKTNPTSTPQPSAVLP- 271
++ +A+ P TG + GP + L S S P V+
Sbjct: 231 GSNLPVNQTVAIMPVSATG-VPGPPTN--------LNIGMDYWSGHGNVSGAVPGVVVDG 281
Query: 272 --PEAWIQ--NERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEI 327
+ W+Q +ERE+KR+RRKQSNRESARRSRLRKQAE +ELA++ E LN E++SLR+EI
Sbjct: 282 SQSQPWLQVSDEREIKRQRRKQSNRESARRSRLRKQAECDELAQRAEVLNGENSSLRAEI 341
Query: 328 NRLAENSERLRMENAALKEKFKIAKLGQPKEIILTNIDSQRTT 370
N+L E L EN++LK KF A + ++ + QR+T
Sbjct: 342 NKLKSQYEELLAENSSLKNKFSSAPSLEGGDLDKNEQEPQRST 384
>At4g36730 G-box binding bZip transcription factor GBF1 / AtbZip41
Length = 315
Score = 177 bits (448), Expect = 1e-44
Identities = 129/353 (36%), Positives = 179/353 (50%), Gaps = 67/353 (18%)
Query: 1 MGNSDEEKSTKTEKPSSPVTVDQTNQTNVHVYPDWA-AMQAYYGPRVAMPPYYNSPVASG 59
MG S+++ KT KP+S + YPDW +MQAYYG P++ SPV S
Sbjct: 1 MGTSEDKMPFKTTKPTS-----SAQEVPPTPYPDWQNSMQAYYGGGGTPNPFFPSPVGSP 55
Query: 60 HTPHPYMWGPPQPMMPPYGHP--YAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPL 117
+PHPYMWG MMPPYG P Y AMYP G VY HP++P+ P+ S P P
Sbjct: 56 -SPHPYMWGAQHHMMPPYGTPVPYPAMYPPGAVYAHPSMPMPPN-------SGPTNKEPA 107
Query: 118 SIETP-PKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEGSSDGS 176
+ KS GN+ KK +G D GN A S +SV T GSSD +
Sbjct: 108 KDQASGKKSKGNSK----KKAEGGDKALSGSGNDGA--------SHSDESV-TAGSSDEN 154
Query: 177 DGNTSGANQTRRKRSREGTPTTDGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGP 236
D N + Q ++ G D ++ T EI S M VAP
Sbjct: 155 DENANQQEQGSIRKPSFGQMLADASSQSTTG------EIQGSVPMKPVAPG--------- 199
Query: 237 VASSAMTTALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESAR 296
+ + ++L +S A +P +++ERELKR++RKQSNRESAR
Sbjct: 200 ---TNLNIGMDLWSSQ---------------AGVP----VKDERELKRQKRKQSNRESAR 237
Query: 297 RSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFK 349
RSRLRKQAE E+L ++VESL+ E+ SLR E+ RL+ ++L+ EN +++++ +
Sbjct: 238 RSRLRKQAECEQLQQRVESLSNENQSLRDELQRLSSECDKLKSENNSIQDELQ 290
>At5g60830 bZip transcription factor AtbZip70
Length = 188
Score = 63.9 bits (154), Expect = 2e-10
Identities = 53/165 (32%), Positives = 76/165 (45%), Gaps = 25/165 (15%)
Query: 244 TALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNER---ELKRERRKQSNRESARRSRL 300
T + S+V ++ N S P+A E NE E +R RR SNRESARRSR+
Sbjct: 41 TDFNVHLQSNVSTRINNQSHLDPNA----ENIFHNEGLAPEERRARRMVSNRESARRSRM 96
Query: 301 RKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFKIAKLGQPKEII 360
RK+ + EEL ++VE L + L ++ L E++ ++ EN+ LKEK L
Sbjct: 97 RKKKQIEELQQQVEQLMMLNHHLSEKVINLLESNHQILQENSQLKEKVSSFHL------- 149
Query: 361 LTNIDSQRTTPVSTENLLSRVNNNSGSNDRTVEDENGYCDNKPNS 405
+ + LL N S NDR V G N+P +
Sbjct: 150 -----------LMADVLLPMRNAESNINDRNVNYLRGEPSNRPTN 183
>At5g15830 bZIP transcription factor AtbZip3
Length = 186
Score = 62.4 bits (150), Expect = 5e-10
Identities = 38/102 (37%), Positives = 64/102 (62%), Gaps = 11/102 (10%)
Query: 246 LELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESARRSRLRKQAE 305
L L++ S +S T+ +T E ++ NER ++RR SNRESARRSR+RKQ
Sbjct: 48 LNLQSPVSNNSTTSDDATE--------EIFVINER---KQRRMVSNRESARRSRMRKQRH 96
Query: 306 AEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEK 347
+EL +V L +E+ L ++N++++N++ + EN++LKE+
Sbjct: 97 LDELLSQVAWLRSENHQLLDKLNQVSDNNDLVIQENSSLKEE 138
>At3g30530 bZip transcription factor AtbZip42
Length = 173
Score = 59.3 bits (142), Expect = 4e-09
Identities = 34/72 (47%), Positives = 48/72 (66%), Gaps = 3/72 (4%)
Query: 276 IQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSE 335
I NER ++RR SNRESARRSR+RKQ +EL +V L E+ L ++N L+E+ +
Sbjct: 77 IINER---KQRRMISNRESARRSRMRKQRHLDELWSQVMWLRIENHQLLDKLNNLSESHD 133
Query: 336 RLRMENAALKEK 347
++ ENA LKE+
Sbjct: 134 KVLQENAQLKEE 145
>At1g13600 bZip transcription factor AtbZip58
Length = 196
Score = 59.3 bits (142), Expect = 4e-09
Identities = 49/147 (33%), Positives = 78/147 (52%), Gaps = 13/147 (8%)
Query: 251 SSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELA 310
SSS + + TS S ++ + +ER ++RR SNRESARRSR+RKQ +EL
Sbjct: 57 SSSSNGQDLMTSNNSTSDEDHQQSMVIDER---KQRRMISNRESARRSRMRKQRHLDELW 113
Query: 311 RKVESLNAESASLRSEINRLAENSERLRMENAALKEKFKIAKLGQPKEIILTNIDSQRTT 370
+V L ++ L ++NR++E+ E ENA LKE + + L Q +++ I S
Sbjct: 114 SQVIRLRTDNHCLMDKLNRVSESHELALKENAKLKE--ETSDLRQ----LISEIKSHN-- 165
Query: 371 PVSTENLLSRVNNNSGSNDRTVEDENG 397
+N R +S SN R+ +++G
Sbjct: 166 --EDDNSFLRELEDSISNSRSDSNQSG 190
>At5g38800 bZIP transcription factor AtbZip43
Length = 165
Score = 57.4 bits (137), Expect = 1e-08
Identities = 31/74 (41%), Positives = 50/74 (66%), Gaps = 3/74 (4%)
Query: 277 QNEREL---KRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAEN 333
+N +E+ ++++RK SNRESARRSR+RKQ + +EL +V L E+ L ++N + E+
Sbjct: 63 ENHKEIINERKQKRKISNRESARRSRMRKQRQVDELWSQVMWLRDENHQLLRKLNCVLES 122
Query: 334 SERLRMENAALKEK 347
E++ EN LKE+
Sbjct: 123 QEKVIEENVQLKEE 136
>At4g02640 bZIP protein BZO2H1
Length = 417
Score = 56.2 bits (134), Expect = 3e-08
Identities = 50/201 (24%), Positives = 83/201 (40%), Gaps = 17/201 (8%)
Query: 199 DGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGPVASSAMTTALELRNSSSVHSKT 258
D TQ + +M GVT L V + ++ SS +S
Sbjct: 145 DSTSSPETQLQPVQSSPLTQGSLMTPGELGVTSSLPAEVKKTGVSMKQVTSGSSREYSDD 204
Query: 259 NPTSTP-QPSAVLPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLN 317
+ + L PE ++K+ RR SNRESARRSR RKQ + +L +V L
Sbjct: 205 EDLDEENETTGSLKPE-------DVKKSRRMLSNRESARRSRRRKQEQTSDLETQVNDLK 257
Query: 318 AESASLRSEINRLAENSERLRMENAALKEKFKI--AKLGQPKEIILTNIDSQRTTPVSTE 375
E +SL +++ + + + N LK + AK+ +E + +R T ++
Sbjct: 258 GEHSSLLKQLSNMNHKYDEAAVGNRILKADIETLRAKVKMAEETV------KRVTGMN-P 310
Query: 376 NLLSRVNNNSGSNDRTVEDEN 396
LL R + ++ +N + N
Sbjct: 311 MLLGRSSGHNNNNRMPITGNN 331
>At2g04038 bZip transcription factor AtbZip48
Length = 166
Score = 55.1 bits (131), Expect = 7e-08
Identities = 29/65 (44%), Positives = 43/65 (65%)
Query: 283 KRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENA 342
+++RR SNRESARRSR+RKQ +EL +V L E+ L ++NR++E + EN+
Sbjct: 74 RKQRRMLSNRESARRSRMRKQRHLDELWSQVIRLRNENNCLIDKLNRVSETQNCVLKENS 133
Query: 343 ALKEK 347
LKE+
Sbjct: 134 KLKEE 138
>At5g28770 bZip transcription factor BZO2H3 / AtbZip63
Length = 307
Score = 54.7 bits (130), Expect = 1e-07
Identities = 51/174 (29%), Positives = 75/174 (42%), Gaps = 25/174 (14%)
Query: 227 AGVTGQLVGPVASSAMTTALELR-NSSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRE 285
A + P+ SSA+T+ EL + +TN N +KR
Sbjct: 106 ASLASSKATPMMSSAITSGSELSGDEEEADGETN-----------------MNPTNVKRV 148
Query: 286 RRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALK 345
+R SNRESARRSR RKQA EL +V L E++ L + + + +EN LK
Sbjct: 149 KRMLSNRESARRSRRRKQAHLSELETQVSQLRVENSKLMKGLTDVTQTFNDASVENRVLK 208
Query: 346 EKFKI--AKLGQPKEII--LTNIDSQ-RTTP--VSTENLLSRVNNNSGSNDRTV 392
+ AK+ +E + LT + P VST +L S +N+ + V
Sbjct: 209 ANIETLRAKVKMAEETVKRLTGFNPMFHNMPQIVSTVSLPSETSNSPDTTSSQV 262
>At1g68880 bZip transcription factor AtbZip8
Length = 138
Score = 52.0 bits (123), Expect = 6e-07
Identities = 29/65 (44%), Positives = 42/65 (64%)
Query: 283 KRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENA 342
++ RRK SNRESARRSR+RKQ EEL + L ++ SL E+++ E E++ EN
Sbjct: 47 RKRRRKVSNRESARRSRMRKQRHMEELWSMLVQLINKNKSLVDELSQARECYEKVIEENM 106
Query: 343 ALKEK 347
L+E+
Sbjct: 107 KLREE 111
>At2g21230 bZIP transcription factor AtbZip30
Length = 519
Score = 50.4 bits (119), Expect = 2e-06
Identities = 56/229 (24%), Positives = 95/229 (41%), Gaps = 11/229 (4%)
Query: 137 LKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEG-SSDGSDGNTSGANQTRRKRSREGT 195
L F G GN + E E SR S + T G SS S+G++S + + S +
Sbjct: 223 LNSFGGEDGKNGNENVEEME---SSRGSGTKKTNGGSSSDSEGDSSASGNVKVALSSSSS 279
Query: 196 PTTDGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGPVASSAMTTALELRNSSSVH 255
G + + ++ M G L P +SSA + +S
Sbjct: 280 GVKRRAGGDIAPTGRHYRSVSMDSCFMGKLNFGDESSLKLPPSSSAKVSPTNSGEGNSSA 339
Query: 256 SKTNPTSTPQPSAVLPPEAWIQNEREL-----KRERRKQSNRESARRSRLRKQAEAEELA 310
++ +A + A + E+ KR +R +NR SA RS+ RK EL
Sbjct: 340 YSVEFGNSEFTAAEMKKIAADEKLAEIVMADPKRVKRILANRVSAARSKERKTRYMAELE 399
Query: 311 RKVESLNAESASLRSEINRLAENSERLRMENAALKEKFKIAKLGQPKEI 359
KV++L E+ +L +++ L +S L +N+ L KF++ + Q ++
Sbjct: 400 HKVQTLQTEATTLSAQLTHLQRDSMGLTNQNSEL--KFRLQAMEQQAQL 446
Score = 36.6 bits (83), Expect = 0.027
Identities = 38/151 (25%), Positives = 51/151 (33%), Gaps = 23/151 (15%)
Query: 60 HTPHPYMWGPPQPMMP--PYGHPYAAMYP-HGGVYTHPA--------VPIGPHPHSQGIS 108
H HP+ PP P+ P PY A + P H + P+ P+ P S +S
Sbjct: 44 HFRHPFTGAPPPPIPPISPYSQIPATLQPRHSRSMSQPSSFFSFDSLPPLNPSAPSVSVS 103
Query: 109 SSPATGTPLSIETPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVN 168
TG S PP + S G E+ P R+S S
Sbjct: 104 VEEKTGAGFSPSLPPSPFTMCHS------------SSSRNAGDGENLPPRKSHRRSNSDV 151
Query: 169 TEGSSDGSDGNTSGANQTRRKRSREGTPTTD 199
T G S N + +RS G T+D
Sbjct: 152 TFGFSSMMSQNQKSPPLSSLERSISGEDTSD 182
>At3g54620 bZIP transcription factor-like protein
Length = 403
Score = 50.1 bits (118), Expect = 2e-06
Identities = 46/163 (28%), Positives = 73/163 (44%), Gaps = 19/163 (11%)
Query: 200 GEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGPVASSAMTTALELRNSSSVHSKTN 259
G K + S + A ++A G + P S+ + R +S + S+ +
Sbjct: 154 GTVKPEDSSASASNQKQAQGSIVAQTSPGASSVRFSPTTSTQKKPDVPARQTS-ISSRDD 212
Query: 260 PTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAE 319
L +A + ++KR RR SNRESARRSR RKQ + E +V L AE
Sbjct: 213 SDDDD-----LDGDADNGDPTDVKRARRMLSNRESARRSRRRKQEQMNEFDTQVGQLRAE 267
Query: 320 SASLRSEINRLAENSER----------LRMENAALKEKFKIAK 352
++L INRL++ + + LR + L+ K K+A+
Sbjct: 268 HSTL---INRLSDMNHKYDAAAVDNRILRADIETLRTKVKMAE 307
>At3g49760 bZIP transcription factor AtbZip5
Length = 156
Score = 49.7 bits (117), Expect = 3e-06
Identities = 23/68 (33%), Positives = 45/68 (65%)
Query: 283 KRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENA 342
++++RK SNRESA+RSR +KQ EE++ ++ L ++ L++++ + + +R +MEN
Sbjct: 72 RKKKRKLSNRESAKRSREKKQKHLEEMSIQLNQLKIQNQELKNQLRYVLYHCQRTKMEND 131
Query: 343 ALKEKFKI 350
L + +I
Sbjct: 132 RLLMEHRI 139
>At3g28790 unknown protein
Length = 608
Score = 49.3 bits (116), Expect = 4e-06
Identities = 79/349 (22%), Positives = 132/349 (37%), Gaps = 53/349 (15%)
Query: 3 NSDEEKSTKTEKPSSPVTVDQTNQTNVHVYPDWAAMQAYYGPRVAMPPYYNSPVASGHTP 62
N+ + ++ SS T + + ++ + Y D + P + P ++P S TP
Sbjct: 234 NTGSKTEAGSKSSSSAKTKEVSGGSSGNTYKDTTGSSSGASPSGSPTPTPSTPTPSTPTP 293
Query: 63 HPYMWGPPQPMMPPYGHPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLSIETP 122
P P P P P P S+PA TP + +T
Sbjct: 294 -----STPTPSTP--------------------TPSTPTP------STPAPSTPAAGKTS 322
Query: 123 PKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEGSSDGSDGNTSG 182
K S + MKK S +ESA G+ S+ ++ +GSS + +T+G
Sbjct: 323 EKGS---ESASMKK--------ESNSKSESESAASGSVSKTKET--NKGSSGDTYKDTTG 369
Query: 183 ANQTRRKRSREG--TPTTDGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGPVASS 240
+ S G TP+T +GK +++GS S AS A A A S
Sbjct: 370 TSSGSPSGSPSGSPTPSTSTDGKASSKGS-ASASAGASASASAGASASAEESAASQKKES 428
Query: 241 AMTTALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNERELK--RERRKQSNRESARRS 298
++ +++SV TS+ S + E ELK E+ K S SA+ S
Sbjct: 429 NSKSSSSSSSTTSVKEVETQTSSEVNSFISNLEKKYTGNSELKVFFEKLKTSMSASAKLS 488
Query: 299 RLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEK 347
+ A+EL + S ++ A ++ SE + A+ +++
Sbjct: 489 ----TSNAKELVTGMRSAASKIAEAMMFVSSRFSKSEETKTSMASCQQE 533
Score = 43.9 bits (102), Expect = 2e-04
Identities = 62/289 (21%), Positives = 101/289 (34%), Gaps = 46/289 (15%)
Query: 108 SSSPATGTPLSIETPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSV 167
S ++G+ S + +SS NT G + A SI E G++S S S
Sbjct: 192 SIGSSSGSDGSSSSDNESSSNTKSQGTSSKSGSESTAGSIETNTGSKTEAGSKS--SSSA 249
Query: 168 NTEGSSDGSDGNT--------SGANQTRRKRSREGTPT---------------------- 197
T+ S GS GNT SGA+ + TPT
Sbjct: 250 KTKEVSGGSSGNTYKDTTGSSSGASPSGSPTPTPSTPTPSTPTPSTPTPSTPTPSTPTPS 309
Query: 198 -----TDGEGKTNTQGSQIS--KEIAASDKMMAVAPAGVTGQLVGPVASSAMTTALELRN 250
T GKT+ +GS+ + K+ + S A +G + S+ T +
Sbjct: 310 TPAPSTPAAGKTSEKGSESASMKKESNSKSESESAASGSVSKTKETNKGSSGDTYKDTTG 369
Query: 251 SSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRERRKQSN-------RESARRSRLRKQ 303
+SS +P+ +P PS +A + ++ ESA +
Sbjct: 370 TSSGSPSGSPSGSPTPSTSTDGKASSKGSASASAGASASASAGASASAEESAASQKKESN 429
Query: 304 AEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFKIAK 352
+++ + S+ SE+N N E+ N+ LK F+ K
Sbjct: 430 SKSSSSSSSTTSVKEVETQTSSEVNSFISNLEKKYTGNSELKVFFEKLK 478
Score = 42.0 bits (97), Expect = 6e-04
Identities = 46/166 (27%), Positives = 69/166 (40%), Gaps = 30/166 (18%)
Query: 171 GSSDGSDGNTSGANQ------TRRKRSREGTPTTDGEGKTNT-----QGSQISKEIAASD 219
GSS GSDG++S N+ ++ S+ G+ +T G +TNT GS+ S +A
Sbjct: 194 GSSSGSDGSSSSDNESSSNTKSQGTSSKSGSESTAGSIETNTGSKTEAGSKSSS--SAKT 251
Query: 220 KMMAVAPAG-----VTGQLVGPVASSAMTTALELRNSSSVHSKTNPTSTPQPSAVLP--- 271
K ++ +G TG G S + T S+ T STP PS P
Sbjct: 252 KEVSGGSSGNTYKDTTGSSSGASPSGSPTPTPSTPTPSTPTPSTPTPSTPTPSTPTPSTP 311
Query: 272 ----PEAWIQNER-----ELKRERRKQSNRESARRSRLRKQAEAEE 308
P A +E+ +K+E +S ESA + K E +
Sbjct: 312 APSTPAAGKTSEKGSESASMKKESNSKSESESAASGSVSKTKETNK 357
>At3g10800 bZIP transcription factor AtbZip28
Length = 675
Score = 49.3 bits (116), Expect = 4e-06
Identities = 32/79 (40%), Positives = 47/79 (58%), Gaps = 10/79 (12%)
Query: 278 NERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERL 337
++R+L R+ R NRESA+ SRLRK+ + EEL RKV+S+NA A L +I +
Sbjct: 190 DKRKLIRQIR---NRESAQLSRLRKKQQTEELERKVKSMNATIAELNGKI-------AYV 239
Query: 338 RMENAALKEKFKIAKLGQP 356
EN AL+++ +A P
Sbjct: 240 MAENVALRQQMAVASGAPP 258
>At2g40620 bZIP transcription factor AtbZip18
Length = 367
Score = 49.3 bits (116), Expect = 4e-06
Identities = 33/105 (31%), Positives = 55/105 (51%), Gaps = 6/105 (5%)
Query: 249 RNSSSVHSKTNPTSTPQPSAVLPP---EAWIQNERELKRERRKQSNRESARRSRLRKQAE 305
R+S SV + S A+ P E W+ + KR +R +NR+SA RS+ RK
Sbjct: 116 RHSLSVDGSSTLESIEAKKAMAPDKLAELWVVDP---KRAKRIIANRQSAARSKERKARY 172
Query: 306 AEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFKI 350
EL RKV++L E+ +L ++++ ++ L EN LK + ++
Sbjct: 173 ILELERKVQTLQTEATTLSAQLSLFQRDTTGLSSENTELKLRLQV 217
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.305 0.123 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,178,236
Number of Sequences: 26719
Number of extensions: 483629
Number of successful extensions: 2741
Number of sequences better than 10.0: 396
Number of HSP's better than 10.0 without gapping: 147
Number of HSP's successfully gapped in prelim test: 253
Number of HSP's that attempted gapping in prelim test: 1936
Number of HSP's gapped (non-prelim): 797
length of query: 425
length of database: 11,318,596
effective HSP length: 102
effective length of query: 323
effective length of database: 8,593,258
effective search space: 2775622334
effective search space used: 2775622334
T: 11
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137602.6


