

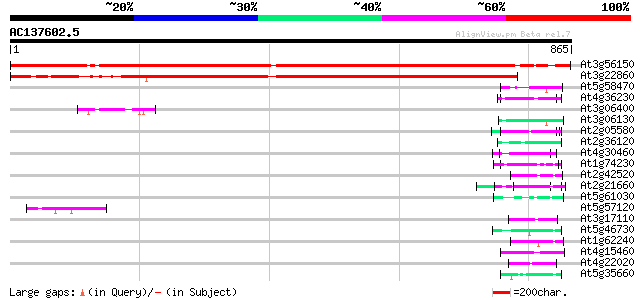
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137602.5 - phase: 0
(865 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g56150 PROBABLE EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SU... 1150 0.0
At3g22860 eukaryotic translation initiation factor 3 (subunit 8) 844 0.0
At5g58470 RNA/ssDNA-binding protein - like 55 2e-07
At4g36230 putative glycine-rich cell wall protein 52 1e-06
At3g06400 putative ATPase (ISW2-like) 51 3e-06
At3g06130 unknown protein 51 3e-06
At2g05580 unknown protein 50 7e-06
At2g36120 unknown protein 49 9e-06
At4g30460 glycine-rich protein 49 2e-05
At1g74230 putative RNA-binding protein 49 2e-05
At2g42520 putative ATP-dependent RNA helicase 48 3e-05
At2g21660 glycine-rich RNA binding protein 48 3e-05
At5g61030 RNA-binding protein - like 47 6e-05
At5g57120 unknown protein 46 8e-05
At3g17110 hypothetical protein 46 8e-05
At5g46730 unknown protein 46 1e-04
At1g62240 unknown protein (At1g62240) 45 2e-04
At4g15460 glycine-rich protein like, predicted GPI-anchored protein 44 3e-04
At4g22020 glycine-rich protein 44 4e-04
At5g35660 unknown protein 44 5e-04
>At3g56150 PROBABLE EUKARYOTIC TRANSLATION INITIATION FACTOR 3
SUBUNIT 8
Length = 900
Score = 1150 bits (2976), Expect = 0.0
Identities = 593/867 (68%), Positives = 692/867 (79%), Gaps = 33/867 (3%)
Query: 1 MASTVDQIKNAIKINDWVSLQETFDKINKQLDKVMRVIESQKIPNLYIKALVMLEDFLAQ 60
M TVDQ+KNA+KINDWVSLQE FDK+NKQL+KVMR+ E+ K P LYIK LVMLEDFL +
Sbjct: 63 MTYTVDQMKNAMKINDWVSLQENFDKVNKQLEKVMRITEAVKPPTLYIKTLVMLEDFLNE 122
Query: 61 ASSNKDAKKKMSPTNAKAFNSMKQKLKKNNKQYEDLIIKFRETPQSEEEKFEDDEDSDQY 120
A +NK+AKKKMS +N+KA NSMKQKLKKNNK YED I K+RE P+ EEEK +D+D D
Sbjct: 123 ALANKEAKKKMSTSNSKALNSMKQKLKKNNKLYEDDINKYREAPEVEEEKQPEDDDDDD- 181
Query: 121 GSDDDEIIEPDQLKKPEPVSDSETSELGNDRPGDDDDAPWDQKLRKKDRLLEKMFMKKPS 180
DDD+ +E D + D T + G+D D+ W++ L KKD+LLEK+ K P
Sbjct: 182 --DDDDEVEDDD----DSSIDGPTVDPGSDVDEPTDNLTWEKMLSKKDKLLEKLMNKDPK 235
Query: 181 EITWDTVNNKFKEILEARGRKGTGRFEQVEQFTFLTKVAKTPAQKLQILFSVVSAQFDVN 240
EITWD VN KFKEI+ ARG+KGT RFE V+Q T LTK+AKTPAQKL+ILFSV+SAQFDVN
Sbjct: 236 EITWDWVNKKFKEIVAARGKKGTARFELVDQLTHLTKIAKTPAQKLEILFSVISAQFDVN 295
Query: 241 PGLSGHMPINVWKKCVQNMLVILDILVQHPNIKVDDSVELDENESKKGDDYDGPIHVWGN 300
PGLSGHMPINVWKKCV NML ILDILV++ NI VDD+VE DENE+ K DYDG I VWGN
Sbjct: 296 PGLSGHMPINVWKKCVLNMLTILDILVKYSNIVVDDTVEPDENETSKPTDYDGKIRVWGN 355
Query: 301 LVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEPLFVVLAQNVQEYLESIGDFKASSKVA 360
LVAFLE++D EFFKSLQCIDPHTREYVERLRDEP+F+ LAQN+Q+Y E +GDFKA++KVA
Sbjct: 356 LVAFLERVDTEFFKSLQCIDPHTREYVERLRDEPMFLALAQNIQDYFERMGDFKAAAKVA 415
Query: 361 LKRVELIYYKPHEVYEATRKLAELTVDGDNGEEVSEESKGFVDTRIIAPFVVTPELVARK 420
L+RVE IYYKP EVY+A RKLAEL + + EE EES F+V PE+V RK
Sbjct: 416 LRRVEAIYYKPQEVYDAMRKLAELVEEEEETEEAKEESGPPTS------FIVVPEVVPRK 469
Query: 421 PTFPENSRTLMDVLVSLIYKYGDERTKARAMLCDIYHHALLDEFAVARDLLLMSHLQENV 480
PTFPE+SR +MD+LVSLIY+ GDERTKARAMLCDI HHAL+D F ARDLLLMSHLQ+N+
Sbjct: 470 PTFPESSRAMMDILVSLIYRNGDERTKARAMLCDINHHALMDNFVTARDLLLMSHLQDNI 529
Query: 481 HHMDISTQILFNRAMSQLGLCAFRVGLVSEAHGCLSELYSGGRVKELLAQGVSQNRYHEK 540
HMDISTQILFNR M+QLGLCAFR G+++E+H CLSELYSG RV+ELLAQGVSQ+RYHEK
Sbjct: 530 QHMDISTQILFNRTMAQLGLCAFRAGMITESHSCLSELYSGQRVRELLAQGVSQSRYHEK 589
Query: 541 TPEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLL 600
TPEQER+ERRRQMPYHMH+NLELLE+VHL AMLLEVPNMAAN HDAKR++ISKNFRRLL
Sbjct: 590 TPEQERMERRRQMPYHMHLNLELLEAVHLICAMLLEVPNMAANSHDAKRRVISKNFRRLL 649
Query: 601 EVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFVKNRDTVLEMLKDK 660
E+SE+Q FT PPE VRDHVMAATR L GDFQKAF+++ SLEVW+ +KNRD++L+M+KD+
Sbjct: 650 EISERQAFTAPPENVRDHVMAATRALTKGDFQKAFEVLNSLEVWRLLKNRDSILDMVKDR 709
Query: 661 IKEEALRTYLFTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEELHASWDQPTGCI 720
IKEEALRTYLFT+SSSY+SLS+DQL FD+S P+ HSIVS+MMINEELHASWDQPT CI
Sbjct: 710 IKEEALRTYLFTYSSSYESLSLDQLAKMFDVSEPQVHSIVSKMMINEELHASWDQPTRCI 769
Query: 721 IFRNVELSRVQALAFQLTEKLSILAESNERASEARLGGGGLDLPPRRRD-GQDYAAAAAG 779
+F V+ SR+Q+LAFQLTEKLSILAESNERA E+R GGGGLDL RRRD QDYA AA+G
Sbjct: 770 VFHEVQHSRLQSLAFQLTEKLSILAESNERAMESRTGGGGLDLSSRRRDNNQDYAGAASG 829
Query: 780 GGGGTSSGGRWQD-LSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSS 838
G GG WQD +Y Q RQG+ R+GYGG S Q G G +RG G G
Sbjct: 830 G------GGYWQDKANYGQGRQGN-RSGYGG-GRSSGQNGQWSGQNRGGGYAG------- 874
Query: 839 RTQGGSALRGPHGDVSTRMVSL-RGVR 864
+ GS RG D S+RMVSL RGVR
Sbjct: 875 --RVGSGNRGMQMDGSSRMVSLNRGVR 899
>At3g22860 eukaryotic translation initiation factor 3 (subunit 8)
Length = 800
Score = 844 bits (2180), Expect = 0.0
Identities = 461/792 (58%), Positives = 567/792 (71%), Gaps = 73/792 (9%)
Query: 1 MASTVDQIKNAIKINDWVSLQETFDKINKQLDKVMRVIESQKIPNLYIKALVMLEDFLAQ 60
MA+T++ +K+A+ IND V LQETF+K+NKQ+ K S K P LYIK LVMLEDFL +
Sbjct: 58 MANTIENMKHAMNINDCVYLQETFEKLNKQISK------SVKTPTLYIKTLVMLEDFLNE 111
Query: 61 ASSNKDAKKKMSPTNAKAFNSMKQKLKKNNKQYEDLIIKFRETPQSEEEKFEDDEDSDQY 120
N K+KMS +N+KA N+M+QKLKKNN QY++ I +FRE+P+ E
Sbjct: 112 --DNMKTKEKMSTSNSKALNAMRQKLKKNNLQYQEDIKRFRESPEIE------------- 156
Query: 121 GSDDDEIIEPDQLKKPEPVSDSETSELGNDRPGDDDDAPWDQKLRKKDRLLEKMFMKKPS 180
DDDE E E V DS D+ W E +F
Sbjct: 157 --DDDEYEE-------EVVEDSA------------DNVSW-----------EMLFSLDHE 184
Query: 181 EITWDTVNNKFKEILEARGRKGTGRFEQV-------EQFTFLTKVAKTPAQKLQILFSVV 233
EITW+ VN KFKEI AR K ++ ++ LTK+AKTPAQK++ILFSV+
Sbjct: 185 EITWNMVNKKFKEIRAARWSKRRSSSLKLKPGETHAQKHMDLTKIAKTPAQKVEILFSVI 244
Query: 234 SAQFDVNPG-LSGHMPINVWKKCVQNMLVILDILVQHPNIKVDDSVELDENESKKGDDYD 292
SA+F+VN G LSG+MPI+VWKKCV NML ILDILV++ NI VDD+VE DE E+ K YD
Sbjct: 245 SAEFNVNSGGLSGYMPIDVWKKCVVNMLTILDILVKYYNIVVDDTVEPDEKETSKPAAYD 304
Query: 293 GPIHVWGNLVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEPLFVVLAQNVQEYLESIGD 352
G I V GNLVAFLEKI+ EFFKSLQCIDPHT +YVERL+DEP+F+ LAQ++Q+YLE GD
Sbjct: 305 GTIRVSGNLVAFLEKIETEFFKSLQCIDPHTNDYVERLKDEPMFLALAQSIQDYLERTGD 364
Query: 353 FKASSKVALKRVELIYYKPHEVYEATRKLAELTVDGDNGEEVSEESKGFVDTRIIAPFVV 412
KA+SKVA VE IYYKP EV++A KLA+ ++ N E S S F+V
Sbjct: 365 SKAASKVAFILVESIYYKPQEVFDAMSKLADEEIEEANEESGSSSSS----------FIV 414
Query: 413 TPELVARKPTFPENSRTLMDVLVSLIYKYGDERTKARAMLCDIYHHALLDEFAVARDLLL 472
E+V RKPTF ++SR +MD LVS IYK GDERTKARAMLCDIY HAL+D F ARDLLL
Sbjct: 415 VAEIVPRKPTFAKSSRAMMDTLVSFIYKNGDERTKARAMLCDIYQHALMDNFVTARDLLL 474
Query: 473 MSHLQENVHHMDISTQILFNRAMSQLGLCAFRVGLVSEAHGCLSELYSGGRVKELLAQGV 532
MSHLQEN+ HMDISTQILFNR M+QLGLCAFRVG+++E+H CLSELYSG RV+ELL QGV
Sbjct: 475 MSHLQENIQHMDISTQILFNRTMAQLGLCAFRVGMITESHSCLSELYSGNRVRELLGQGV 534
Query: 533 SQNRYHEKTPEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKII 592
SQ+R HEKT EQ +ERR Q+PYHM+INLELLE+V+LT AMLLEVPNMAAN HD+K K I
Sbjct: 535 SQSRDHEKTTEQMLMERRTQIPYHMNINLELLEAVYLTCAMLLEVPNMAANSHDSKHKPI 594
Query: 593 SKNFRRLLEVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFVKNRDT 652
SKN +RLLE SEKQ FT PPE VR HV+AATR LI G+FQ+AF ++ SL++W+ KNRD+
Sbjct: 595 SKNIQRLLEKSEKQAFTAPPENVRVHVIAATRALIKGNFQEAFSVLNSLDIWRLFKNRDS 654
Query: 653 VLEMLKDKIKEEALRTYLFTFSSS-YDSLSVDQLTNFFDLSLPRAHSIVSRMMINEELHA 711
+L+M+K I E ALRTYLFT+SSS Y SLS+ +L FD+S +SIVS+MMIN+EL+A
Sbjct: 655 ILDMVKASISEVALRTYLFTYSSSCYKSLSLAELAKMFDISESHVYSIVSKMMINKELNA 714
Query: 712 SWDQPTGCIIFRNVELSRVQALAFQLTEKLSILAESNERASEARLGGGGLDLPPRRRD-G 770
+WDQPT CI+F V+ +R Q+LAFQ+TEKL+ LAESNE A E++ GG GLD+ RRR+
Sbjct: 715 TWDQPTQCIVFHEVQHNRAQSLAFQITEKLATLAESNESAMESKTGGVGLDMTSRRRENS 774
Query: 771 QDYAAAAAGGGG 782
QDYAAAA+ G
Sbjct: 775 QDYAAAASDNRG 786
>At5g58470 RNA/ssDNA-binding protein - like
Length = 422
Score = 54.7 bits (130), Expect = 2e-07
Identities = 41/102 (40%), Positives = 43/102 (41%), Gaps = 29/102 (28%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG +P DG GGGGG G G AGYGGR S
Sbjct: 9 GGGGAPIPSYGGDGY-------GGGGG----------------YGGGDAGYGGRGASGGG 45
Query: 817 A-GGSGGYSRG-----RGMGGGGYQNSSRTQGGSALRGPHGD 852
+ GG GGY G RG GGGGYQ R GS G GD
Sbjct: 46 SYGGRGGYGGGGGRGNRGGGGGGYQGGDRGGRGSGGGGRDGD 87
Score = 38.5 bits (88), Expect = 0.016
Identities = 39/125 (31%), Positives = 46/125 (36%), Gaps = 31/125 (24%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG R G G GG SGG +D + G + R + N+
Sbjct: 55 GGGG-----RGNRGGGGGGYQGGDRGGRGSGGGGRDGDWRCPNPSCGNVNFA-RRVECNK 108
Query: 817 AG--------------GSGGYSRG-----RGMGGGGYQNS------SRTQGGSALRGPHG 851
G G GGYSRG RG G GG +S SR GGS G +G
Sbjct: 109 CGALAPSGTSSGANDRGGGGYSRGGGDSDRGGGRGGRNDSGRSYESSRYDGGSRSGGSYG 168
Query: 852 DVSTR 856
S R
Sbjct: 169 SGSQR 173
Score = 32.0 bits (71), Expect = 1.5
Identities = 22/70 (31%), Positives = 29/70 (41%), Gaps = 12/70 (17%)
Query: 780 GGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSR 839
GGGG S GG D +G GR G S+ + GG GG Y + S+
Sbjct: 125 GGGGYSRGGGDSD-------RGGGRGGRNDSGRSYESSRYDGG-----SRSGGSYGSGSQ 172
Query: 840 TQGGSALRGP 849
+ GS + P
Sbjct: 173 RENGSYGQAP 182
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 52.0 bits (123), Expect = 1e-06
Identities = 37/100 (37%), Positives = 44/100 (44%), Gaps = 9/100 (9%)
Query: 753 EARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRAL 812
E+R GGGG G +GGGGG +GG +D S+ + + G G GG
Sbjct: 85 ESRAGGGG---GGGGGGGGGGGGQGSGGGGGEGNGGNGKDNSHKRNKSSGGGGGGGG--- 138
Query: 813 SFNQAGGSG-GYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GGSG G RGRG GGGG GG G G
Sbjct: 139 --GGGGGSGNGSGRGRGGGGGGGGGGGGGGGGGGGGGGGG 176
Score = 45.4 bits (106), Expect = 1e-04
Identities = 31/87 (35%), Positives = 37/87 (41%), Gaps = 8/87 (9%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG +D ++GGGGG GG + S +G G G GG
Sbjct: 109 GGGGEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGRGRGGGGGGGGG------- 161
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGG 843
GG GG G G GGGG S +GG
Sbjct: 162 -GGGGGGGGGGGGGGGGGGGGSDGKGG 187
Score = 39.3 bits (90), Expect = 0.010
Identities = 26/76 (34%), Positives = 29/76 (37%), Gaps = 1/76 (1%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GG G D +R GGGGG S G + G G G GG
Sbjct: 116 GGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGRGRGGGGGGGGGGGGGGGGGG-GGGG 174
Query: 817 AGGSGGYSRGRGMGGG 832
GG GG S G+G G G
Sbjct: 175 GGGGGGGSDGKGGGWG 190
Score = 33.1 bits (74), Expect = 0.69
Identities = 25/77 (32%), Positives = 27/77 (34%), Gaps = 13/77 (16%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G+ GGGGG GG G G G GG
Sbjct: 136 GGGGGGGGSGNGSGRGRGGGGGGGGGGGGGGGGG----------GGGGGGGGGGG---GS 182
Query: 817 AGGSGGYSRGRGMGGGG 833
G GG+ G G GG G
Sbjct: 183 DGKGGGWGFGFGWGGDG 199
Score = 30.0 bits (66), Expect = 5.8
Identities = 23/65 (35%), Positives = 27/65 (41%), Gaps = 6/65 (9%)
Query: 801 GSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNS---SRTQGGSALRGPHGDVSTRM 857
G G G GG Q G GG G G GG G NS +++ GG G G +
Sbjct: 90 GGGGGGGGGGGGGGGQGSGGGG---GEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGN 146
Query: 858 VSLRG 862
S RG
Sbjct: 147 GSGRG 151
>At3g06400 putative ATPase (ISW2-like)
Length = 1057
Score = 50.8 bits (120), Expect = 3e-06
Identities = 41/142 (28%), Positives = 65/142 (44%), Gaps = 35/142 (24%)
Query: 105 QSEEEKFEDDEDSDQY---------GSDDDEIIEPDQLKKPEPVSDSETSELGNDRPGDD 155
+ EEE+ +D+E+ D+ GSDDDE+ D+ PVSD E + + +D ++
Sbjct: 14 EEEEERVKDNEEEDEEELEAVARSSGSDDDEVAAADE----SPVSDGEAAPVEDDYEDEE 69
Query: 156 DDAPWDQKLRKKDRLLEKMFMKKPSEITWDTVNNKFKEILEAR--------GRKGTGR-- 205
D+ + R+K RL E +KK K +E+LE++ KG GR
Sbjct: 70 DEEKAEISKREKARLKEMQKLKK----------QKIQEMLESQNASIDADMNNKGKGRLK 119
Query: 206 --FEQVEQFTFLTKVAKTPAQK 225
+Q E F K + +QK
Sbjct: 120 YLLQQTELFAHFAKSDGSSSQK 141
>At3g06130 unknown protein
Length = 473
Score = 50.8 bits (120), Expect = 3e-06
Identities = 38/119 (31%), Positives = 45/119 (36%), Gaps = 24/119 (20%)
Query: 754 ARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALS 813
A L GG P + G+ AA GG GG G QG G+ G GG
Sbjct: 246 ANLAGG-----PAKNGGKGAPAAGGGGAGGGKGAGGGAKGGPGNQNQGGGKNGGGGHPQD 300
Query: 814 FNQAGGSGGYSRG------------------RGMGGGGYQNSSRTQGGS-ALRGPHGDV 853
GG GG + G R MGGGG QN S GG + GP G++
Sbjct: 301 GKNGGGGGGPNAGKKGNGGGGPMAGGVSGGFRPMGGGGPQNMSMPMGGQMGMGGPMGNM 359
>At2g05580 unknown protein
Length = 302
Score = 49.7 bits (117), Expect = 7e-06
Identities = 33/94 (35%), Positives = 39/94 (41%), Gaps = 1/94 (1%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG + G GGG G GG Q QG G+ G GGR +
Sbjct: 143 GGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQG-GQKGGGGRGQGGMK 201
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPH 850
GG GG +G GGGG Q + GG +G H
Sbjct: 202 GGGGGGQGGHKGGGGGGGQGGHKGGGGGGGQGGH 235
Score = 44.3 bits (103), Expect = 3e-04
Identities = 29/92 (31%), Positives = 34/92 (36%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG + GQ G GG GG+ QG + G GG
Sbjct: 154 GGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGGMKGGGGGGQGGHKG 213
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRG 848
GG GG +G GGGG Q + GG G
Sbjct: 214 GGGGGGQGGHKGGGGGGGQGGHKGGGGGGQGG 245
Score = 44.3 bits (103), Expect = 3e-04
Identities = 31/87 (35%), Positives = 33/87 (37%), Gaps = 4/87 (4%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G GGGGG GG QG + G GG +
Sbjct: 216 GGGGQGGHKGGGGGGGQGGHKGGGGGGQGGGGH----KGGGGGQGGHKGGGGGGHVGGGG 271
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGG 843
GG GG G G GGGG S R GG
Sbjct: 272 RGGGGGGRGGGGSGGGGGGGSGRGGGG 298
Score = 43.9 bits (102), Expect = 4e-04
Identities = 31/95 (32%), Positives = 37/95 (38%), Gaps = 2/95 (2%)
Query: 758 GGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQD--LSYSQTRQGSGRAGYGGRALSFN 815
GGG + GQ G GG GGR Q QG + G GG +
Sbjct: 164 GGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGGH 223
Query: 816 QAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPH 850
+ GG GG G GGGG Q +GG +G H
Sbjct: 224 KGGGGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGH 258
Score = 43.5 bits (101), Expect = 5e-04
Identities = 31/95 (32%), Positives = 35/95 (36%), Gaps = 2/95 (2%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG + G GGGGG GG QG + G GG
Sbjct: 191 GGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGGHKG--GGGGGGQGGHKGGGGGGQGGGGH 248
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GG GG +G GGGG+ GG RG G
Sbjct: 249 KGGGGGQGGHKGGGGGGHVGGGGRGGGGGGRGGGG 283
Score = 43.1 bits (100), Expect = 7e-04
Identities = 34/110 (30%), Positives = 41/110 (36%), Gaps = 10/110 (9%)
Query: 743 ILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGS 802
+L E S+ GG G ++ G GGGGG G Q G
Sbjct: 64 VLDGEEEEESKNNQGGIGRGQGGQKGGGGGGQGGQKGGGGGGQGG---------QKGGGG 114
Query: 803 GRAGY-GGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
G+ G GG Q GG GG G+ GGGG Q + GG G G
Sbjct: 115 GQGGQKGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKG 164
Score = 40.8 bits (94), Expect = 0.003
Identities = 28/77 (36%), Positives = 31/77 (39%), Gaps = 15/77 (19%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG + G GGGGG GG +G G G GG
Sbjct: 239 GGGGQGGGGHKGGGGGQGGHKGGGGGGHVGGGG----------RGGGGGGRGGGG----- 283
Query: 817 AGGSGGYSRGRGMGGGG 833
+GG GG GRG GGGG
Sbjct: 284 SGGGGGGGSGRGGGGGG 300
Score = 40.8 bits (94), Expect = 0.003
Identities = 32/103 (31%), Positives = 37/103 (35%), Gaps = 8/103 (7%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGG-----GTSSGGRWQDLSYSQTRQGSGRAGYGGRA 811
GGGG ++ G GGGG G GG+ QG + G GG
Sbjct: 100 GGGGGGQGGQKGGGGGQGGQKGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQ 159
Query: 812 LSFNQAGGSGGYSRGRGMG---GGGYQNSSRTQGGSALRGPHG 851
GG GG G G G GGG Q + GG G G
Sbjct: 160 GGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGGMKG 202
Score = 40.8 bits (94), Expect = 0.003
Identities = 32/102 (31%), Positives = 40/102 (38%), Gaps = 5/102 (4%)
Query: 750 RASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGG 809
R + GGGG ++ G GGGGG GG+ ++G G G GG
Sbjct: 82 RGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGG--QGGQKGGGGGQGGQKGGGGGGQGG 139
Query: 810 RALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
+ GG G +G G GG G Q QGG G G
Sbjct: 140 QK---GGGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGGGQG 178
Score = 39.7 bits (91), Expect = 0.007
Identities = 30/100 (30%), Positives = 35/100 (35%), Gaps = 6/100 (6%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAG------YGGR 810
GGGG GQ GGG G GG + G G+ G GG+
Sbjct: 112 GGGGQGGQKGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQ 171
Query: 811 ALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPH 850
Q G GG +G GGGG GG +G H
Sbjct: 172 KGGGGQGGQKGGGGQGGQKGGGGRGQGGMKGGGGGGQGGH 211
Score = 39.3 bits (90), Expect = 0.010
Identities = 32/97 (32%), Positives = 36/97 (36%), Gaps = 5/97 (5%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG + G GGGGG GG QG G GG ++
Sbjct: 203 GGGGGQGGHKGGGGGGGQGGHKGGGGGGGQGGH---KGGGGGGQGGGGHKGGGGGQGGHK 259
Query: 817 AGGSGGY--SRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GG GG+ GRG GGGG GG G G
Sbjct: 260 GGGGGGHVGGGGRGGGGGGRGGGGSGGGGGGGSGRGG 296
Score = 38.5 bits (88), Expect = 0.016
Identities = 32/97 (32%), Positives = 39/97 (39%), Gaps = 7/97 (7%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG ++ G GGGGG G + Q G G GG+ Q
Sbjct: 131 GGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQ----KGGGGQGGQKGGGGQ 186
Query: 817 AGGSGGYSRGRG-MGGGG--YQNSSRTQGGSALRGPH 850
G GG RG+G M GGG Q + GG +G H
Sbjct: 187 GGQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGGH 223
>At2g36120 unknown protein
Length = 255
Score = 49.3 bits (116), Expect = 9e-06
Identities = 33/99 (33%), Positives = 39/99 (39%), Gaps = 15/99 (15%)
Query: 753 EARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRAL 812
E +GGGG G + A GGGGG GG Y G AGYGG
Sbjct: 77 EGHIGGGG---------GGGHGGGAGGGGGGGPGGG------YGGGSGEGGGAGYGGGEA 121
Query: 813 SFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
+ GG GG G G GGG + G+ G +G
Sbjct: 122 GGHGGGGGGGAGGGGGGGGGAHGGGYGGGQGAGAGGGYG 160
Score = 42.0 bits (97), Expect = 0.001
Identities = 35/113 (30%), Positives = 38/113 (32%), Gaps = 19/113 (16%)
Query: 758 GGGLDLPPRRRDGQDYAAAAAGG---------GGGTSSGGRWQDLSYSQTRQGSGRAGYG 808
GGG G Y AGG GGG GG Y + GYG
Sbjct: 101 GGGYGGGSGEGGGAGYGGGEAGGHGGGGGGGAGGGGGGGGGAHGGGYGGGQGAGAGGGYG 160
Query: 809 GRALSFNQAGGSGGYSRGRGMG-------GGGY---QNSSRTQGGSALRGPHG 851
G + GG GG G G G GGGY + GG A G HG
Sbjct: 161 GGGAGGHGGGGGGGNGGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGHG 213
Score = 41.6 bits (96), Expect = 0.002
Identities = 32/86 (37%), Positives = 35/86 (40%), Gaps = 9/86 (10%)
Query: 775 AAAAGGGGGTSSGGRWQDLSYSQTRQGSGRA-------GYGGRALS--FNQAGGSGGYSR 825
A GGGGG +GG S G G GYGG A + GG GG S
Sbjct: 164 AGGHGGGGGGGNGGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGGGGGSG 223
Query: 826 GRGMGGGGYQNSSRTQGGSALRGPHG 851
G G GGGGY +S G G G
Sbjct: 224 GGGGGGGGYAAASGYGHGGGAGGGEG 249
Score = 40.4 bits (93), Expect = 0.004
Identities = 30/96 (31%), Positives = 34/96 (35%), Gaps = 9/96 (9%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
G GG + G A GGGGG GG Y + GYGG +
Sbjct: 115 GYGGGEAGGHGGGGGGGAGGGGGGGGGAHGGG------YGGGQGAGAGGGYGGGGAGGHG 168
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGD 852
GG GG G GGGG GG G G+
Sbjct: 169 GGGGGGNGGG---GGGGSGEGGAHGGGYGAGGGAGE 201
Score = 39.3 bits (90), Expect = 0.010
Identities = 30/90 (33%), Positives = 32/90 (35%), Gaps = 17/90 (18%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGG---GGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALS 813
GGGG G Y A G GGG +GG G G G GG S
Sbjct: 177 GGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGH-----------GGGGGGGGG---S 222
Query: 814 FNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
GG GGY+ G G GG GG
Sbjct: 223 GGGGGGGGGYAAASGYGHGGGAGGGEGSGG 252
Score = 38.1 bits (87), Expect = 0.021
Identities = 26/74 (35%), Positives = 29/74 (39%), Gaps = 8/74 (10%)
Query: 778 AGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNS 837
+G GGG+ GG G G G GG AGG GG G G GGG +
Sbjct: 60 SGYGGGSGEGG-------GAGGHGEGHIGGGGGGGHGGGAGGGGGGGPGGGYGGGSGEGG 112
Query: 838 SRTQGGSALRGPHG 851
GG G HG
Sbjct: 113 GAGYGGGE-AGGHG 125
Score = 37.0 bits (84), Expect = 0.048
Identities = 28/84 (33%), Positives = 31/84 (36%), Gaps = 13/84 (15%)
Query: 751 ASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGR 810
+ E GGG +G A A G GGG GG GSG G GG
Sbjct: 183 SGEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGGGG------------GSGGGGGGGG 230
Query: 811 ALSFNQAGGSGGYSRGRGMGGGGY 834
+ G GG G G G GGY
Sbjct: 231 GYAAASGYGHGG-GAGGGEGSGGY 253
Score = 35.0 bits (79), Expect = 0.18
Identities = 26/78 (33%), Positives = 35/78 (44%), Gaps = 10/78 (12%)
Query: 776 AAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGR--GMGGGG 833
AA G G S+G L G +GYGG + + GG+GG+ G G GGGG
Sbjct: 36 AAYGVNSGLSAG-----LGVGIGGGPGGGSGYGGGS---GEGGGAGGHGEGHIGGGGGGG 87
Query: 834 YQNSSRTQGGSALRGPHG 851
+ + GG G +G
Sbjct: 88 HGGGAGGGGGGGPGGGYG 105
>At4g30460 glycine-rich protein
Length = 162
Score = 48.5 bits (114), Expect = 2e-05
Identities = 35/92 (38%), Positives = 42/92 (45%), Gaps = 12/92 (13%)
Query: 756 LGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFN 815
+GGGG G A + GGGG+SS S S + G G AG + + +
Sbjct: 50 IGGGG--------SGSGAGAGSGSGGGGSSSSSSSSSSSSSSSGGGGGDAGSEAGSYAGS 101
Query: 816 QAG-GSGGYS---RGRGMGGGGYQNSSRTQGG 843
AG GSGG S RGRG GGGG GG
Sbjct: 102 HAGSGSGGRSGSGRGRGSGGGGGHGGGGGGGG 133
Score = 43.9 bits (102), Expect = 4e-04
Identities = 33/92 (35%), Positives = 40/92 (42%), Gaps = 13/92 (14%)
Query: 745 AESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGG-GGTSSGGRWQDLSYSQTRQGSG 803
+ S+ +S + GGGG D + YA + AG G GG S GR R G
Sbjct: 73 SSSSSSSSSSSSGGGGGDAGS---EAGSYAGSHAGSGSGGRSGSGR--------GRGSGG 121
Query: 804 RAGYGGRALSFNQAGGSGGYSRGRGMG-GGGY 834
G+GG GG GG G G G GGGY
Sbjct: 122 GGGHGGGGGGGGGRGGGGGSGNGEGYGEGGGY 153
Score = 40.8 bits (94), Expect = 0.003
Identities = 27/92 (29%), Positives = 34/92 (36%), Gaps = 11/92 (11%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG ++ GG G+ +G SY+ + GSG G G
Sbjct: 66 GGGGSSSSSSSSSSSSSSSGGGGGDAGSEAG------SYAGSHAGSGSGGRSGSGRGRGS 119
Query: 817 A-----GGSGGYSRGRGMGGGGYQNSSRTQGG 843
GG GG GRG GGG +GG
Sbjct: 120 GGGGGHGGGGGGGGGRGGGGGSGNGEGYGEGG 151
>At1g74230 putative RNA-binding protein
Length = 289
Score = 48.5 bits (114), Expect = 2e-05
Identities = 36/98 (36%), Positives = 42/98 (42%), Gaps = 5/98 (5%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GG G P D A GG G +GG + SYS G G GYGG +
Sbjct: 117 GGRGFGGPGGGYGASDGGYGAPAGGYGGGAGGYGGNSSYSGNAGGGG--GYGGNSSYGGN 174
Query: 817 AGGSGG---YSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
AGG GG YS GGGGY ++ GG + G G
Sbjct: 175 AGGYGGNPPYSGNAVGGGGGYGSNFGGGGGYGVAGGVG 212
Score = 42.7 bits (99), Expect = 9e-04
Identities = 38/100 (38%), Positives = 46/100 (46%), Gaps = 9/100 (9%)
Query: 747 SNERASEA-RLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRA 805
S E AS A +L G L RR +YA G GG GG S G+
Sbjct: 84 STEEASNAMQLDGQDLH---GRRIRVNYATERGSGFGGRGFGGPGGGYGASDGGYGAPAG 140
Query: 806 GYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSA 845
GYGG A + GG+ YS G GGGGY +S + GG+A
Sbjct: 141 GYGGGAGGY---GGNSSYS-GNAGGGGGYGGNS-SYGGNA 175
Score = 33.9 bits (76), Expect = 0.40
Identities = 24/75 (32%), Positives = 35/75 (46%), Gaps = 2/75 (2%)
Query: 783 GTSSGGRWQDLSYSQTR-QGSGRAGYGGRALSFNQA-GGSGGYSRGRGMGGGGYQNSSRT 840
G GR ++Y+ R G G G+GG + + GG G + G G G GGY +S
Sbjct: 96 GQDLHGRRIRVNYATERGSGFGGRGFGGPGGGYGASDGGYGAPAGGYGGGAGGYGGNSSY 155
Query: 841 QGGSALRGPHGDVST 855
G + G +G S+
Sbjct: 156 SGNAGGGGGYGGNSS 170
>At2g42520 putative ATP-dependent RNA helicase
Length = 633
Score = 47.8 bits (112), Expect = 3e-05
Identities = 32/82 (39%), Positives = 42/82 (51%), Gaps = 7/82 (8%)
Query: 773 YAAAAA-GGGGGTSSGGRWQDLSYSQTRQ-GSGRAGYGGRALSFNQAGGSGGYSRGRGMG 830
YA+ ++ GGG SGGR+ + + GSGR GYGG + GG GGY G G G
Sbjct: 551 YASRSSFGGGKNRRSGGRFGGRDFRREGSFGSGRGGYGGGGGGY---GGGGGYGGGGGYG 607
Query: 831 GGGYQNSSRTQGGSALRGPHGD 852
GGG GG++ G G+
Sbjct: 608 GGGGYGGG--YGGASSGGYGGE 627
Score = 35.4 bits (80), Expect = 0.14
Identities = 22/59 (37%), Positives = 25/59 (42%), Gaps = 8/59 (13%)
Query: 780 GGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSS 838
G GG SG RW + G G G GG + G GY G GGGG+ N S
Sbjct: 55 GYGGPPSGSRW-----APGGSGVGVGGGGGYRADAGRPGSGSGYG---GRGGGGWNNRS 105
Score = 32.3 bits (72), Expect = 1.2
Identities = 30/83 (36%), Positives = 32/83 (38%), Gaps = 25/83 (30%)
Query: 755 RLGG--GGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRAL 812
R GG GG D RR+G + GGGG GG G G G GG
Sbjct: 564 RSGGRFGGRDF---RREGSFGSGRGGYGGGGGGYGG------------GGGYGGGGG--- 605
Query: 813 SFNQAGGSGGYSRG-RGMGGGGY 834
GG GGY G G GGY
Sbjct: 606 ----YGGGGGYGGGYGGASSGGY 624
>At2g21660 glycine-rich RNA binding protein
Length = 176
Score = 47.8 bits (112), Expect = 3e-05
Identities = 35/87 (40%), Positives = 39/87 (44%), Gaps = 9/87 (10%)
Query: 748 NERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY 807
NE S GGGG R G Y + GGGGG S GG + G G +G
Sbjct: 82 NEAQSRGSGGGGG----HRGGGGGGYRS---GGGGGYSGGGGSYGGGGGRREGGGGYSGG 134
Query: 808 GGRALSFNQAGGSGGYSRGRGMGGGGY 834
GG S + GG G Y GR GGGGY
Sbjct: 135 GGGYSS--RGGGGGSYGGGRREGGGGY 159
Score = 43.9 bits (102), Expect = 4e-04
Identities = 28/80 (35%), Positives = 37/80 (46%), Gaps = 3/80 (3%)
Query: 777 AAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQN 836
A G G GR ++ +Q+R G G+ G ++GG GGYS G G GGG
Sbjct: 65 AIEGMNGQDLDGRSITVNEAQSRGSGGGGGHRGGGGGGYRSGGGGGYSGGGGSYGGG--- 121
Query: 837 SSRTQGGSALRGPHGDVSTR 856
R +GG G G S+R
Sbjct: 122 GGRREGGGGYSGGGGGYSSR 141
Score = 43.5 bits (101), Expect = 5e-04
Identities = 38/134 (28%), Positives = 52/134 (38%), Gaps = 11/134 (8%)
Query: 721 IFRNVELSRVQALAFQLTEKLSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGG 780
I + E R + F + + ++ E + L G + + + G GG
Sbjct: 39 IINDRETGRSRGFGFVTFKDEKAMKDAIEGMNGQDLDGRSITVNEAQSRGSGGGGGHRGG 98
Query: 781 GGG---TSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNS 837
GGG + GG + S G R G GG +GG GGYS RG GGG Y
Sbjct: 99 GGGGYRSGGGGGYSGGGGSYGGGGGRREGGGG------YSGGGGGYS-SRGGGGGSY-GG 150
Query: 838 SRTQGGSALRGPHG 851
R +GG G G
Sbjct: 151 GRREGGGGYGGGEG 164
Score = 38.9 bits (89), Expect = 0.013
Identities = 27/80 (33%), Positives = 32/80 (39%), Gaps = 12/80 (15%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAA---GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALS 813
GGGG R G Y+ GGGG GG + + +G G YGG
Sbjct: 99 GGGGY----RSGGGGGYSGGGGSYGGGGGRREGGGGYSGGGGGYSSRGGGGGSYGG---- 150
Query: 814 FNQAGGSGGYSRGRGMGGGG 833
+ G GGY G G G GG
Sbjct: 151 -GRREGGGGYGGGEGGGYGG 169
Score = 38.1 bits (87), Expect = 0.021
Identities = 31/81 (38%), Positives = 36/81 (44%), Gaps = 30/81 (37%)
Query: 757 GGGGLDLPPRRRDGQDYAA-----AAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRA 811
GGGG RR G Y+ ++ GGGGG+ GGR R+G G GYGG
Sbjct: 120 GGGG-----RREGGGGYSGGGGGYSSRGGGGGSYGGGR---------REGGG--GYGG-- 161
Query: 812 LSFNQAGGSGGYSRGRGMGGG 832
G GG G G GGG
Sbjct: 162 -------GEGGGYGGSGGGGG 175
Score = 33.1 bits (74), Expect = 0.69
Identities = 16/37 (43%), Positives = 18/37 (48%)
Query: 754 ARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRW 790
+R GGGG RR G Y GG GG+ GG W
Sbjct: 140 SRGGGGGSYGGGRREGGGGYGGGEGGGYGGSGGGGGW 176
>At5g61030 RNA-binding protein - like
Length = 309
Score = 46.6 bits (109), Expect = 6e-05
Identities = 36/108 (33%), Positives = 43/108 (39%), Gaps = 31/108 (28%)
Query: 747 SNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAG 806
+N+R S GGGG GGGGG G + G G G
Sbjct: 116 ANDRTSGGGFGGGGY----------------GGGGGGYGGSGGY----------GGGAGG 149
Query: 807 YGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVS 854
YGG S GG+GGY G G GG N++ GGS G GD +
Sbjct: 150 YGG---SGGYGGGAGGYGGNSGGGYGG--NAAGGYGGSGAGGYGGDAT 192
Score = 40.4 bits (93), Expect = 0.004
Identities = 41/108 (37%), Positives = 45/108 (40%), Gaps = 19/108 (17%)
Query: 747 SNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAG 806
S+E AS A G DL R +YA GGG G G G G
Sbjct: 90 SSEAASSAIQALDGRDLHGRVVK-VNYANDRTSGGGFGGGG------------YGGGGGG 136
Query: 807 YGGRALSFNQAGGSGGY--SRGRGMGGGGY-QNSSRTQGGSALRGPHG 851
YGG S GG+GGY S G G G GGY NS GG+A G G
Sbjct: 137 YGG---SGGYGGGAGGYGGSGGYGGGAGGYGGNSGGGYGGNAAGGYGG 181
Score = 38.9 bits (89), Expect = 0.013
Identities = 27/83 (32%), Positives = 37/83 (44%), Gaps = 2/83 (2%)
Query: 774 AAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRG--MGG 831
A++A G GR ++Y+ R G G GG GGSGGY G G G
Sbjct: 94 ASSAIQALDGRDLHGRVVKVNYANDRTSGGGFGGGGYGGGGGGYGGSGGYGGGAGGYGGS 153
Query: 832 GGYQNSSRTQGGSALRGPHGDVS 854
GGY + GG++ G G+ +
Sbjct: 154 GGYGGGAGGYGGNSGGGYGGNAA 176
Score = 29.3 bits (64), Expect = 9.9
Identities = 25/74 (33%), Positives = 31/74 (41%), Gaps = 13/74 (17%)
Query: 778 AGGGGGTSSG--------GRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGS-GGYSRGRG 828
AGGG G+S G G S +G +GYG S N G S GG++
Sbjct: 197 AGGGYGSSGGFGSSGNTYGEGSSASAGAVGDYNGSSGYG----SANTYGSSNGGFAGDSQ 252
Query: 829 MGGGGYQNSSRTQG 842
GG NSS+ G
Sbjct: 253 FGGSPVGNSSQFGG 266
>At5g57120 unknown protein
Length = 330
Score = 46.2 bits (108), Expect = 8e-05
Identities = 34/139 (24%), Positives = 68/139 (48%), Gaps = 19/139 (13%)
Query: 26 KINKQLDKVMRVIESQKIPNLYIKALVMLEDFLAQASSNKDAK------KKMSPTNAKAF 79
K K + + V E +K+ + ++ED + + K+ K +K+ T+A
Sbjct: 93 KKKKNKETKVEVTEEEKVK----ETDAVIEDGVKEKKKKKETKVKVTEEEKVKETDAVIE 148
Query: 80 NSMKQKLKKNNKQY-------EDLIIKFRETPQSEE--EKFEDDEDSDQYGSDDDEIIEP 130
+ +K+K KK +K ++ + K R+ + EE E+ EDD++ + ++ ++E
Sbjct: 149 DGVKEKKKKKSKSKSVEADDDKEKVSKKRKRSEPEETKEETEDDDEESKRRKKEENVVEN 208
Query: 131 DQLKKPEPVSDSETSELGN 149
D+ + PV ++ET E GN
Sbjct: 209 DEGVQETPVKETETKENGN 227
>At3g17110 hypothetical protein
Length = 175
Score = 46.2 bits (108), Expect = 8e-05
Identities = 34/80 (42%), Positives = 37/80 (45%), Gaps = 9/80 (11%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSG---RAGYGGRALSFNQAGGSGGYSRG 826
G + GGGGG GG D S S + GSG +GYGG N GG GG RG
Sbjct: 45 GGNGGGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYGSGSGYGGG----NNGGGGGGGGRG 100
Query: 827 RGMGG--GGYQNSSRTQGGS 844
G GG GG N S GS
Sbjct: 101 GGGGGGNGGSGNGSGYGSGS 120
Score = 40.4 bits (93), Expect = 0.004
Identities = 27/60 (45%), Positives = 29/60 (48%), Gaps = 6/60 (10%)
Query: 779 GGGGGTSSGGRWQDLSY---SQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMG-GGGY 834
GGGGG +GG Y S G+GR G GG GG GG SRG G G G GY
Sbjct: 100 GGGGGGGNGGSGNGSGYGSGSGYGSGAGRGGGGGG--GGGGGGGGGGGSRGNGSGYGSGY 157
Score = 40.4 bits (93), Expect = 0.004
Identities = 30/90 (33%), Positives = 33/90 (36%), Gaps = 7/90 (7%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G Y + + GGG GG R G G G GG
Sbjct: 63 GGGGDGSGSGSGYGSGYGSGSGYGGGNNGGGGG------GGGRGGGGGGGNGGSGNGSGY 116
Query: 817 AGGSG-GYSRGRGMGGGGYQNSSRTQGGSA 845
GSG G GRG GGGG GG +
Sbjct: 117 GSGSGYGSGAGRGGGGGGGGGGGGGGGGGS 146
Score = 38.1 bits (87), Expect = 0.021
Identities = 27/80 (33%), Positives = 29/80 (35%), Gaps = 15/80 (18%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
G GG G Y + A GGGG GG G G G G R N
Sbjct: 106 GNGGSGNGSGYGSGSGYGSGAGRGGGGGGGGG------------GGGGGGGGSRG---NG 150
Query: 817 AGGSGGYSRGRGMGGGGYQN 836
+G GY G G G GG N
Sbjct: 151 SGYGSGYGEGYGSGYGGGDN 170
Score = 36.2 bits (82), Expect = 0.081
Identities = 26/73 (35%), Positives = 33/73 (44%), Gaps = 6/73 (8%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGM 829
G + GG GG GG S + + GSG +GYG A + GG GG G G
Sbjct: 88 GNNGGGGGGGGRGGGGGGGNGG--SGNGSGYGSG-SGYGSGA---GRGGGGGGGGGGGGG 141
Query: 830 GGGGYQNSSRTQG 842
GGGG + + G
Sbjct: 142 GGGGSRGNGSGYG 154
Score = 33.9 bits (76), Expect = 0.40
Identities = 25/76 (32%), Positives = 29/76 (37%), Gaps = 23/76 (30%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGM 829
G Y + + GG GG GG G G GG GG GG G G
Sbjct: 36 GSGYGSGSGGGNGGGGGGG--------------GGGGGGG--------GGGGGDGSGSGS 73
Query: 830 G-GGGYQNSSRTQGGS 844
G G GY + S GG+
Sbjct: 74 GYGSGYGSGSGYGGGN 89
Score = 33.1 bits (74), Expect = 0.69
Identities = 19/42 (45%), Positives = 21/42 (49%), Gaps = 1/42 (2%)
Query: 801 GSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQG 842
GSG +GYG + N GG GG G G GGGG S G
Sbjct: 34 GSG-SGYGSGSGGGNGGGGGGGGGGGGGGGGGGGDGSGSGSG 74
Score = 29.6 bits (65), Expect = 7.6
Identities = 24/68 (35%), Positives = 27/68 (39%), Gaps = 18/68 (26%)
Query: 777 AAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQN 836
A G G G+ SG GSG G G GG GG G G GGGG +
Sbjct: 28 ALGRGSGSGSG------------YGSGSGGGNGGG------GGGGGGGGGGGGGGGGDGS 69
Query: 837 SSRTQGGS 844
S + GS
Sbjct: 70 GSGSGYGS 77
>At5g46730 unknown protein
Length = 268
Score = 45.8 bits (107), Expect = 1e-04
Identities = 43/123 (34%), Positives = 46/123 (36%), Gaps = 29/123 (23%)
Query: 745 AESNERASEARLGGGGLDLPPRRRDGQDYAAAA---AGGGGGTSSGGRWQDLSYSQTRQ- 800
A S AS A GGGG Y AA AGGGGG S GG
Sbjct: 95 ASSGGYASGAGEGGGG-----------GYGGAAGGHAGGGGGGSGGGGGSAYGAGGEHAS 143
Query: 801 ----------GSGRAGYGGRAL--SFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRG 848
G+G +GYGG A GG GG S G GG GY GG+ G
Sbjct: 144 GYGNGAGEGGGAGASGYGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGGE--GGGAGGGG 201
Query: 849 PHG 851
HG
Sbjct: 202 SHG 204
Score = 42.4 bits (98), Expect = 0.001
Identities = 28/74 (37%), Positives = 34/74 (45%), Gaps = 7/74 (9%)
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGG-YQNS 837
GGG G +GG + + GSG G GG A S SGGY+ G G GGGG Y +
Sbjct: 62 GGGSGEGAGGGYGGAEGYASGGGSGHGGGGGGAAS------SGGYASGAGEGGGGGYGGA 115
Query: 838 SRTQGGSALRGPHG 851
+ G G G
Sbjct: 116 AGGHAGGGGGGSGG 129
Score = 41.6 bits (96), Expect = 0.002
Identities = 28/81 (34%), Positives = 31/81 (37%), Gaps = 2/81 (2%)
Query: 773 YAAAAAGGGGG--TSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMG 830
Y A GGGGG GG ++ + G G G G S AGG GG G G
Sbjct: 160 YGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGGEGGGAGGGGSHGGAGGYGGGGGGGSGG 219
Query: 831 GGGYQNSSRTQGGSALRGPHG 851
GG Y GG G G
Sbjct: 220 GGAYGGGGAHGGGYGSGGGEG 240
Score = 40.4 bits (93), Expect = 0.004
Identities = 32/81 (39%), Positives = 35/81 (42%), Gaps = 16/81 (19%)
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRG-----RGMGG-- 831
GGGG SSGG Y+ G GYGG A AGG GG S G G GG
Sbjct: 90 GGGGAASSGG------YASGAGEGGGGGYGGAA--GGHAGGGGGGSGGGGGSAYGAGGEH 141
Query: 832 -GGYQNSSRTQGGSALRGPHG 851
GY N + GG+ G G
Sbjct: 142 ASGYGNGAGEGGGAGASGYGG 162
Score = 40.4 bits (93), Expect = 0.004
Identities = 27/60 (45%), Positives = 30/60 (50%), Gaps = 9/60 (15%)
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRA---GYGGRALSFNQAGG-SGGYSRGRGMGGGGY 834
GGGGG+ GG + GSG GYGG A AGG GG+ G G GGGGY
Sbjct: 212 GGGGGSGGGGAYGGGGAHGGGYGSGGGEGGGYGGGA-----AGGYGGGHGGGGGHGGGGY 266
Score = 38.5 bits (88), Expect = 0.016
Identities = 31/97 (31%), Positives = 35/97 (35%), Gaps = 18/97 (18%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G Y GG GG S G G GYGG +
Sbjct: 173 GGGGGGSAGGAHGGSGYGGGEGGGAGGGGSHG--------------GAGGYGGGGGGGSG 218
Query: 817 AGGS--GGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GG+ GG + G G G GG + GG A G G
Sbjct: 219 GGGAYGGGGAHGGGYGSGGGEGGG--YGGGAAGGYGG 253
Score = 37.4 bits (85), Expect = 0.036
Identities = 30/77 (38%), Positives = 34/77 (43%), Gaps = 6/77 (7%)
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRA---GYGGRALSFNQAGGSG-GYSRGRGMGGGGY 834
GG GG SSGG + S GSG GYGG A + GGSG G G GGY
Sbjct: 43 GGSGGVSSGGYGGE-SGGGYGGGSGEGAGGGYGG-AEGYASGGGSGHGGGGGGAASSGGY 100
Query: 835 QNSSRTQGGSALRGPHG 851
+ + GG G G
Sbjct: 101 ASGAGEGGGGGYGGAAG 117
Score = 34.3 bits (77), Expect = 0.31
Identities = 23/57 (40%), Positives = 24/57 (41%), Gaps = 9/57 (15%)
Query: 800 QGSGRAGYGGRALSFNQAGGSGGYSRG--RGMGGGGYQNSSRTQGGSALRGPHGDVS 854
Q SG G+GG GGSGG S G G GGGY S G G G S
Sbjct: 32 QASGGGGHGG-------GGGSGGVSSGGYGGESGGGYGGGSGEGAGGGYGGAEGYAS 81
Score = 32.7 bits (73), Expect = 0.90
Identities = 28/85 (32%), Positives = 34/85 (39%), Gaps = 18/85 (21%)
Query: 772 DYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGG 831
+Y A+ GGG G G G GYGG + GG GG S G G GG
Sbjct: 29 NYGQASGGGGHGGGGGSG-----------GVSSGGYGGES-----GGGYGGGS-GEGAGG 71
Query: 832 G-GYQNSSRTQGGSALRGPHGDVST 855
G G + GGS G G ++
Sbjct: 72 GYGGAEGYASGGGSGHGGGGGGAAS 96
>At1g62240 unknown protein (At1g62240)
Length = 227
Score = 44.7 bits (104), Expect = 2e-04
Identities = 31/88 (35%), Positives = 39/88 (44%), Gaps = 7/88 (7%)
Query: 772 DYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALS------FNQAGGSGGYSR 825
+Y+A+A GGG G SGG S + GSG G + + GG GG
Sbjct: 139 EYSASAGGGGSGEGSGGGGGGDGSSGSGSGSGSGSGSGTGTASGPDVYMHVEGGGGGGGG 198
Query: 826 GRGMGGGGYQNSSRTQGGSALRGPHGDV 853
G G GGGG S + GS G G+V
Sbjct: 199 GGGGGGGGVDGSG-SGSGSGSGGGSGNV 225
Score = 38.5 bits (88), Expect = 0.016
Identities = 30/82 (36%), Positives = 33/82 (39%), Gaps = 5/82 (6%)
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRA-GYG---GRALSFNQAGGS-GGYSRGRGMGGGG 833
GGGGG GG S GSG GYG GR S + S GG G G GGGG
Sbjct: 98 GGGGGGGGGGGGSGGSNGSFFNGSGSGTGYGSGDGRVSSSGEYSASAGGGGSGEGSGGGG 157
Query: 834 YQNSSRTQGGSALRGPHGDVST 855
+ S G + G T
Sbjct: 158 GGDGSSGSGSGSGSGSGSGTGT 179
Score = 38.5 bits (88), Expect = 0.016
Identities = 23/73 (31%), Positives = 32/73 (43%), Gaps = 3/73 (4%)
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALS---FNQAGGSGGYSRGRGMGGGGYQ 835
GGGGG G ++ + G+G GR S ++ + G GG G G GGGG
Sbjct: 102 GGGGGGGGSGGSNGSFFNGSGSGTGYGSGDGRVSSSGEYSASAGGGGSGEGSGGGGGGDG 161
Query: 836 NSSRTQGGSALRG 848
+S G + G
Sbjct: 162 SSGSGSGSGSGSG 174
Score = 36.6 bits (83), Expect = 0.062
Identities = 26/79 (32%), Positives = 33/79 (40%), Gaps = 7/79 (8%)
Query: 769 DGQDYAAAAAGGGGG---TSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSR 825
DG+ AA GGGGG + GG + + + G G G + +A GG
Sbjct: 39 DGEPICAAGGGGGGGGGASVEGGTEKGIDDNANGYGDG----NGNGNAHGRADCPGGIVV 94
Query: 826 GRGMGGGGYQNSSRTQGGS 844
G G GGGG GGS
Sbjct: 95 GGGGGGGGGGGGGGGSGGS 113
Score = 34.7 bits (78), Expect = 0.24
Identities = 24/77 (31%), Positives = 29/77 (37%), Gaps = 12/77 (15%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG DG + + +G G G+ +G Y G G G GG
Sbjct: 156 GGGG--------DGSSGSGSGSGSGSGSGTGTASGPDVYMHVEGGGGGGGGGGG----GG 203
Query: 817 AGGSGGYSRGRGMGGGG 833
GG G G G G GG
Sbjct: 204 GGGVDGSGSGSGSGSGG 220
Score = 33.9 bits (76), Expect = 0.40
Identities = 22/73 (30%), Positives = 26/73 (35%), Gaps = 12/73 (16%)
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSS 838
GGGGG GG G G G FN +G GY G G + S+
Sbjct: 95 GGGGGGGGGG------------GGGGGSGGSNGSFFNGSGSGTGYGSGDGRVSSSGEYSA 142
Query: 839 RTQGGSALRGPHG 851
GG + G G
Sbjct: 143 SAGGGGSGEGSGG 155
Score = 30.4 bits (67), Expect = 4.5
Identities = 32/98 (32%), Positives = 35/98 (35%), Gaps = 17/98 (17%)
Query: 759 GGLDLPPRRRDGQDYAAAAAGGGGGTSSG--GRWQDLSYSQTRQGSG------------R 804
G D P G GGGGG S G G + + S S T GSG
Sbjct: 84 GRADCPGGIVVGGGGGGGGGGGGGGGSGGSNGSFFNGSGSGTGYGSGDGRVSSSGEYSAS 143
Query: 805 AGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQG 842
AG GG S +GG GG G G G S G
Sbjct: 144 AGGGG---SGEGSGGGGGGDGSSGSGSGSGSGSGSGTG 178
>At4g15460 glycine-rich protein like, predicted GPI-anchored protein
Length = 148
Score = 44.3 bits (103), Expect = 3e-04
Identities = 36/101 (35%), Positives = 43/101 (41%), Gaps = 10/101 (9%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG+ + A+GGGG S GG +G G AG GG
Sbjct: 45 GGGGVHVSVGGAHASVGGGHASGGGGHASVGGGHASGGGGHAVEGGGHAGGGG------- 97
Query: 817 AGGSGGYSRGRGMG-GGGYQNSSRTQ-GGSALRGPHGDVST 855
GG G G G+G GGG + T+ GGS L P D ST
Sbjct: 98 -GGHGEEEGGHGIGRGGGMVHRPATRNGGSTLTIPFVDGST 137
Score = 36.6 bits (83), Expect = 0.062
Identities = 23/74 (31%), Positives = 31/74 (41%), Gaps = 3/74 (4%)
Query: 776 AAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGG--RALSFNQAGGSGGYS-RGRGMGGG 832
A GGGG GG + + G G A GG ++ A G GG++ G G GG
Sbjct: 36 AKGGGGGAHGGGGVHVSVGGAHASVGGGHASGGGGHASVGGGHASGGGGHAVEGGGHAGG 95
Query: 833 GYQNSSRTQGGSAL 846
G +GG +
Sbjct: 96 GGGGHGEEEGGHGI 109
>At4g22020 glycine-rich protein
Length = 396
Score = 43.9 bits (102), Expect = 4e-04
Identities = 28/74 (37%), Positives = 32/74 (42%), Gaps = 1/74 (1%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGM 829
G A GGGGG GG S + G+G AG GG A GG GG G
Sbjct: 171 GGSSGGAGGGGGGGGGEGGGANGGSGHGSGAGAG-AGVGGAAGGVGGGGGGGGGEGGGAN 229
Query: 830 GGGGYQNSSRTQGG 843
GG G+ + S GG
Sbjct: 230 GGSGHGSGSGAGGG 243
Score = 43.9 bits (102), Expect = 4e-04
Identities = 27/73 (36%), Positives = 31/73 (41%), Gaps = 4/73 (5%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGM 829
G + AA GGGGG GG S+ G G G + F +GG GG G G
Sbjct: 241 GGGVSGAAGGGGGGGGGGGS----GGSKVGGGYGHGSGFGGGVGFGNSGGGGGGGGGGGG 296
Query: 830 GGGGYQNSSRTQG 842
GGGG S G
Sbjct: 297 GGGGGNGSGYGSG 309
Score = 41.2 bits (95), Expect = 0.003
Identities = 29/95 (30%), Positives = 32/95 (33%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GG G G GGGGG GG S + GSG G + +
Sbjct: 115 GGSGHGSGSGAGAGVGGTTGGVGGGGGGGGGGGEGGGSSGGSGHGSGSGAGAGAGVGGSS 174
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
G GG G G GGG S G A G G
Sbjct: 175 GGAGGGGGGGGGEGGGANGGSGHGSGAGAGAGVGG 209
Score = 41.2 bits (95), Expect = 0.003
Identities = 31/99 (31%), Positives = 36/99 (36%), Gaps = 7/99 (7%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAA-------GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGG 809
GGGG + G+ Y A A GGGGG G + + GSG G
Sbjct: 69 GGGGGEGGDGYGHGEGYGAGAGMSGYGGVGGGGGRGGGEGGGSSANGGSGHGSGSGAGAG 128
Query: 810 RALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRG 848
+ GG GG G G GGG S G A G
Sbjct: 129 VGGTTGGVGGGGGGGGGGGEGGGSSGGSGHGSGSGAGAG 167
Score = 40.8 bits (94), Expect = 0.003
Identities = 29/94 (30%), Positives = 33/94 (34%)
Query: 758 GGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQA 817
GGG G + + GGGGG GG + S GSG G+
Sbjct: 266 GGGYGHGSGFGGGVGFGNSGGGGGGGGGGGGGGGGGNGSGYGSGSGYGSGMGKGSGSGGG 325
Query: 818 GGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GG GG G G G G GG A G G
Sbjct: 326 GGGGGGGSGGGNGSGSGSGEGYGMGGGAGTGNGG 359
Score = 40.0 bits (92), Expect = 0.006
Identities = 28/83 (33%), Positives = 32/83 (37%), Gaps = 12/83 (14%)
Query: 766 RRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSR 825
+R G GGGGG G Y G+G +GYGG GG GG R
Sbjct: 56 KRYGGGGGGGGGGGGGGGEGGDGYGHGEGYG---AGAGMSGYGG-------VGGGGG--R 103
Query: 826 GRGMGGGGYQNSSRTQGGSALRG 848
G G GGG N G + G
Sbjct: 104 GGGEGGGSSANGGSGHGSGSGAG 126
Score = 39.3 bits (90), Expect = 0.010
Identities = 37/107 (34%), Positives = 40/107 (36%), Gaps = 13/107 (12%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQG--------SGRAGYG 808
GG G G AA GGGGG GG + SG AG G
Sbjct: 193 GGSGHGSGAGAGAGVGGAAGGVGGGGG-GGGGEGGGANGGSGHGSGSGAGGGVSGAAGGG 251
Query: 809 GRALSFNQAGGS---GGYSRGRGMGGG-GYQNSSRTQGGSALRGPHG 851
G +GGS GGY G G GGG G+ NS GG G G
Sbjct: 252 GGGGGGGGSGGSKVGGGYGHGSGFGGGVGFGNSGGGGGGGGGGGGGG 298
Score = 39.3 bits (90), Expect = 0.010
Identities = 31/96 (32%), Positives = 38/96 (39%), Gaps = 7/96 (7%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG R G+ ++A GG G S G + + G G G GG
Sbjct: 99 GGGG------RGGGEGGGSSANGGSGHGSGSGAGAGVGGTTGGVGGGGGGGGGGGEGGGS 152
Query: 817 AGGSG-GYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
+GGSG G G G G G +S GG G G
Sbjct: 153 SGGSGHGSGSGAGAGAGVGGSSGGAGGGGGGGGGEG 188
Score = 38.9 bits (89), Expect = 0.013
Identities = 27/86 (31%), Positives = 31/86 (35%)
Query: 758 GGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQA 817
GGG+ G GGGGG SG S +GSG G GG +
Sbjct: 276 GGGVGFGNSGGGGGGGGGGGGGGGGGNGSGYGSGSGYGSGMGKGSGSGGGGGGGGGGSGG 335
Query: 818 GGSGGYSRGRGMGGGGYQNSSRTQGG 843
G G G G G GG + GG
Sbjct: 336 GNGSGSGSGEGYGMGGGAGTGNGGGG 361
Score = 38.9 bits (89), Expect = 0.013
Identities = 31/97 (31%), Positives = 35/97 (35%), Gaps = 15/97 (15%)
Query: 773 YAAAAAGGGGGTSSGGRWQDLSYSQTR---QGSGRAGYGGRALSFNQAGGSGGYSR---- 825
Y GGGGG GG D Y G+G +GYGG + GG GG S
Sbjct: 58 YGGGGGGGGGGGGGGGEGGD-GYGHGEGYGAGAGMSGYGGVGGGGGRGGGEGGGSSANGG 116
Query: 826 -------GRGMGGGGYQNSSRTQGGSALRGPHGDVST 855
G G G GG GG G G S+
Sbjct: 117 SGHGSGSGAGAGVGGTTGGVGGGGGGGGGGGEGGGSS 153
Score = 37.4 bits (85), Expect = 0.036
Identities = 28/97 (28%), Positives = 37/97 (37%), Gaps = 7/97 (7%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG + G + + GG G +SGG G G G GG +
Sbjct: 255 GGGGGSGGSKVGGGYGHGSGFGGGVGFGNSGGG------GGGGGGGGGGGGGGNGSGYGS 308
Query: 817 AGGSG-GYSRGRGMGGGGYQNSSRTQGGSALRGPHGD 852
G G G +G G GGGG + GG+ G+
Sbjct: 309 GSGYGSGMGKGSGSGGGGGGGGGGSGGGNGSGSGSGE 345
Score = 35.4 bits (80), Expect = 0.14
Identities = 26/86 (30%), Positives = 33/86 (38%), Gaps = 13/86 (15%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYG-GRALSFNQAGGSGGYSRGRG 828
G A GGGGG GG +G +G+G G G +GG G G
Sbjct: 172 GSSGGAGGGGGGGGGEGGG------------ANGGSGHGSGAGAGAGVGGAAGGVGGGGG 219
Query: 829 MGGGGYQNSSRTQGGSALRGPHGDVS 854
GGG ++ G + G G VS
Sbjct: 220 GGGGEGGGANGGSGHGSGSGAGGGVS 245
Score = 35.4 bits (80), Expect = 0.14
Identities = 31/100 (31%), Positives = 36/100 (36%), Gaps = 14/100 (14%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G A GG G S G + + G G G GG N
Sbjct: 180 GGGG---------GGGEGGGANGGSGHGSGAGAGAGVGGAAGGVGGGGGGGGGEGGGANG 230
Query: 817 AGGSG-GYSRGRGM----GGGGYQNSSRTQGGSALRGPHG 851
G G G G G+ GGGG GGS + G +G
Sbjct: 231 GSGHGSGSGAGGGVSGAAGGGGGGGGGGGSGGSKVGGGYG 270
Score = 35.0 bits (79), Expect = 0.18
Identities = 27/80 (33%), Positives = 32/80 (39%), Gaps = 4/80 (5%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GG G G + GGGG GG + S + G G G GG A + N
Sbjct: 300 GGNGSGYGSGSGYGSGMGKGSGSGGGGGGGGGGSGGGNGSGSGSGEG-YGMGGGAGTGNG 358
Query: 817 AGGSGGYSRGRGMG---GGG 833
GG G+ G G G GGG
Sbjct: 359 GGGGVGFGMGIGFGIGIGGG 378
Score = 34.7 bits (78), Expect = 0.24
Identities = 33/107 (30%), Positives = 39/107 (35%), Gaps = 18/107 (16%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G ++GG G S G + G+G G GG
Sbjct: 141 GGGG---------GGGEGGGSSGGSGHGSGSGAGAGAGVGGSSGGAGGGGGGGGGEGGGA 191
Query: 817 AGGSG---GYSRGRGMGG-----GGYQNSSRTQGGSALRGP-HGDVS 854
GGSG G G G+GG GG +GG A G HG S
Sbjct: 192 NGGSGHGSGAGAGAGVGGAAGGVGGGGGGGGGEGGGANGGSGHGSGS 238
Score = 32.0 bits (71), Expect = 1.5
Identities = 27/95 (28%), Positives = 32/95 (33%), Gaps = 11/95 (11%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G + A GG G + GG G G G GG +
Sbjct: 220 GGGGEGGGANGGSGHGSGSGAGGGVSGAAGGGGG----------GGGGGGSGGSKVGGGY 269
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GSG + G G G G GG G +G
Sbjct: 270 GHGSG-FGGGVGFGNSGGGGGGGGGGGGGGGGGNG 303
Score = 30.4 bits (67), Expect = 4.5
Identities = 18/51 (35%), Positives = 21/51 (40%), Gaps = 6/51 (11%)
Query: 801 GSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
G G YGG GG GG G G GG GY + G+ + G G
Sbjct: 52 GKGAKRYGG------GGGGGGGGGGGGGEGGDGYGHGEGYGAGAGMSGYGG 96
Score = 30.0 bits (66), Expect = 5.8
Identities = 29/97 (29%), Positives = 33/97 (33%), Gaps = 14/97 (14%)
Query: 754 ARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALS 813
A LGG D D D ++ + G G G R G G G GG
Sbjct: 20 ALLGGSCGDSVENNNDDDDCSSRSWRGCGSFYGKGA--------KRYGGGGGGGGGGG-- 69
Query: 814 FNQAGGSGG--YSRGRGMGGGGYQNSSRTQGGSALRG 848
GG GG Y G G G G + GG RG
Sbjct: 70 --GGGGEGGDGYGHGEGYGAGAGMSGYGGVGGGGGRG 104
>At5g35660 unknown protein
Length = 343
Score = 43.5 bits (101), Expect = 5e-04
Identities = 35/106 (33%), Positives = 42/106 (39%), Gaps = 19/106 (17%)
Query: 757 GGGGLDLPPRRRDGQD-----------YAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRA 805
GGGG +L +G + GGGGG GG WQ G G
Sbjct: 234 GGGGAELENEEINGASEEIQWQGGGGGHGGGWQGGGGGH--GGGWQG---GGGGHGGGWQ 288
Query: 806 GYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
G GGR + GG GG +G G GGG++ GG RG G
Sbjct: 289 GGGGRGGGWKGGGGHGGGWQGGGGRGGGWKGGG---GGGGWRGGGG 331
Score = 42.0 bits (97), Expect = 0.001
Identities = 32/100 (32%), Positives = 39/100 (39%), Gaps = 13/100 (13%)
Query: 757 GGGGLDLPPRRRDG------QDYAAAAAGG-------GGGTSSGGRWQDLSYSQTRQGSG 803
GGGG +L D Q+Y GG GGG GG QG G
Sbjct: 134 GGGGAELETVEIDSASDEDVQNYRGGHGGGWKEGGGQGGGWKGGGGQGGGWKGGGGQGGG 193
Query: 804 RAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
G GG+ + GG GG +G G GGG++ GG
Sbjct: 194 WKGGGGQGGGWKGGGGQGGGWKGGGGQGGGWKGGGGRGGG 233
Score = 39.3 bits (90), Expect = 0.010
Identities = 27/98 (27%), Positives = 40/98 (40%), Gaps = 16/98 (16%)
Query: 748 NERASEARLGGGGLDLPPRRRDGQDYAAAAAGGG--GGTSSGGRWQDLSYSQTRQGSGRA 805
N+ G G++ + + +Y GGG GG GG W+ G
Sbjct: 49 NDAKDSLNKGDVGVEATVEKEEENNYEIDQHGGGWKGGGGHGGGWK-----------GGG 97
Query: 806 GYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
G+GG + GG GG +G G GGG++ +GG
Sbjct: 98 GHGG---GWKGGGGHGGGWKGGGGHGGGWKGGGGGRGG 132
Score = 38.1 bits (87), Expect = 0.021
Identities = 32/93 (34%), Positives = 36/93 (38%), Gaps = 11/93 (11%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLS------YSQTRQGSGRAGYGGR 810
GGGG + G + GGGGG GG WQ G G G GGR
Sbjct: 256 GGGGGHGGGWQGGGGGHGGGWQGGGGG--HGGGWQGGGGRGGGWKGGGGHGGGWQGGGGR 313
Query: 811 ALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
+ GG GG+ RG GGGG GG
Sbjct: 314 GGGWKGGGGGGGW---RGGGGGGGWRGGGGGGG 343
Score = 37.7 bits (86), Expect = 0.028
Identities = 32/101 (31%), Positives = 39/101 (37%), Gaps = 8/101 (7%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGS----GRAGYGGRAL 812
GGGG + GQ GGGG GG +L + S + G GG
Sbjct: 206 GGGGQGGGWKGGGGQ--GGGWKGGGGRGGGGGGGAELENEEINGASEEIQWQGGGGGHGG 263
Query: 813 SFNQAGG--SGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
+ GG GG+ G G GGG+Q GG G HG
Sbjct: 264 GWQGGGGGHGGGWQGGGGGHGGGWQGGGGRGGGWKGGGGHG 304
Score = 37.4 bits (85), Expect = 0.036
Identities = 23/87 (26%), Positives = 32/87 (36%), Gaps = 8/87 (9%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G + G GGG G + + + GG + +
Sbjct: 115 GGGG--------HGGGWKGGGGGRGGGGGGGAELETVEIDSASDEDVQNYRGGHGGGWKE 166
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGG 843
GG GG +G G GGG++ GG
Sbjct: 167 GGGQGGGWKGGGGQGGGWKGGGGQGGG 193
Score = 34.3 bits (77), Expect = 0.31
Identities = 23/109 (21%), Positives = 43/109 (39%)
Query: 735 FQLTEKLSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLS 794
F L + + + + + G +++P + G G ++ + ++ +
Sbjct: 14 FLLVAAVVVAVAAETSVDDKKQGEKQVEIPTVEGKNDAKDSLNKGDVGVEATVEKEEENN 73
Query: 795 YSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
Y + G G G GG + GG GG +G G GGG++ GG
Sbjct: 74 YEIDQHGGGWKGGGGHGGGWKGGGGHGGGWKGGGGHGGGWKGGGGHGGG 122
Score = 33.9 bits (76), Expect = 0.40
Identities = 30/100 (30%), Positives = 36/100 (36%), Gaps = 8/100 (8%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG + GQ GG GG GG Q + + G GR G GG
Sbjct: 186 GGGGQGGGWKGGGGQGGGWKGGGGQGGGWKGGGGQGGGW---KGGGGRGGGGGGGAELEN 242
Query: 817 AGGSGG-----YSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
+G + G G GGG+Q GG G G
Sbjct: 243 EEINGASEEIQWQGGGGGHGGGWQGGGGGHGGGWQGGGGG 282
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,649,743
Number of Sequences: 26719
Number of extensions: 930428
Number of successful extensions: 10211
Number of sequences better than 10.0: 361
Number of HSP's better than 10.0 without gapping: 120
Number of HSP's successfully gapped in prelim test: 248
Number of HSP's that attempted gapping in prelim test: 5559
Number of HSP's gapped (non-prelim): 1692
length of query: 865
length of database: 11,318,596
effective HSP length: 108
effective length of query: 757
effective length of database: 8,432,944
effective search space: 6383738608
effective search space used: 6383738608
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC137602.5


