

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137602.16 - phase: 0 /partial
(167 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
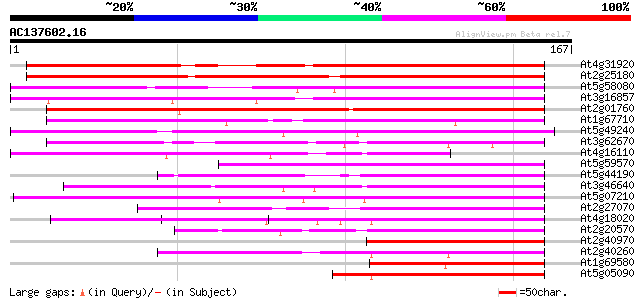
Score E
Sequences producing significant alignments: (bits) Value
At4g31920 predicted protein 145 1e-35
At2g25180 putative two-component response regulator protein 142 8e-35
At5g58080 ARR2 - like protein 126 6e-30
At3g16857 ARR1 protein, putative, 5' partial 120 3e-28
At2g01760 putative two-component response regulator protein 117 2e-27
At1g67710 response regulator 11 (ARR11) 115 1e-26
At5g49240 putative protein 101 2e-22
At3g62670 putative protein 95 2e-20
At4g16110 hypothetical protein 85 2e-17
At5g59570 ARR1 protein - like 72 2e-13
At5g44190 unknown protein 72 2e-13
At3g46640 unknown protein 70 7e-13
At5g07210 putative protein 66 8e-12
At2g27070 putative two-component response regulator protein 65 2e-11
At4g18020 pseudo-response regulator 2 (APRR2) 64 5e-11
At2g20570 GPRI1 protein (gpri1) 62 1e-10
At2g40970 unknown protein 62 2e-10
At2g40260 hypothetical protein 61 3e-10
At1g69580 transfactor, putative 61 3e-10
At5g05090 unknown protein 60 5e-10
>At4g31920 predicted protein
Length = 552
Score = 145 bits (366), Expect = 1e-35
Identities = 76/154 (49%), Positives = 103/154 (66%), Gaps = 17/154 (11%)
Query: 6 NDDLKLVMKAIDVGACDYLLKPVQLKEVKMIWQHVIRKKKTSKRSNDDVPNSDSGNGIDS 65
+ D K VMK + GACDYLLKPV+++E+K IWQHV+RK K K + N +G+G
Sbjct: 98 HSDPKYVMKGVKHGACDYLLKPVRIEELKNIWQHVVRKSKLKKNKS----NVSNGSG--- 150
Query: 66 AVTKSSDNKIEKPSRKRKDGNEDDVEEENEYDHENDGPTMPKKPRFKWSRDLHHKFVVAV 125
+K +RKRK+ E+ EEE E ++ND PT KKPR W+ +LH+KF+ AV
Sbjct: 151 --------NCDKANRKRKEQYEE--EEEEERGNDNDDPTAQKKPRVLWTHELHNKFLAAV 200
Query: 126 NQFGVEKARPKKILNLMNIENLTKEHVASHLQVF 159
+ GVE+A PKKIL+LMN++ LT+E+VASHLQ F
Sbjct: 201 DHLGVERAVPKKILDLMNVDKLTRENVASHLQKF 234
>At2g25180 putative two-component response regulator protein
Length = 573
Score = 142 bits (358), Expect = 8e-35
Identities = 75/154 (48%), Positives = 101/154 (64%), Gaps = 5/154 (3%)
Query: 6 NDDLKLVMKAIDVGACDYLLKPVQLKEVKMIWQHVIRKKKTSKRSNDDVPNSDSGNGIDS 65
+ D K VMK + GACDYLLKPV+++E+K IWQHV+R + R +++ N D +G +
Sbjct: 98 HSDPKYVMKGVTHGACDYLLKPVRIEELKNIWQHVVRSRFDKNRGSNN--NGDKRDGSGN 155
Query: 66 AVTKSSDNKIEKPSRKRKDGNEDDVEEENEYDHENDGPTMPKKPRFKWSRDLHHKFVVAV 125
+SD K +RKRKD +D +E+ + +ND KK R W+ +LH KFV AV
Sbjct: 156 EGVGNSDPNNGKGNRKRKDQYNEDEDEDRD---DNDDSCAQKKQRVVWTVELHKKFVAAV 212
Query: 126 NQFGVEKARPKKILNLMNIENLTKEHVASHLQVF 159
NQ G EKA PKKIL+LMN+E LT+E+VASHLQ F
Sbjct: 213 NQLGYEKAMPKKILDLMNVEKLTRENVASHLQKF 246
>At5g58080 ARR2 - like protein
Length = 632
Score = 126 bits (316), Expect = 6e-30
Identities = 76/168 (45%), Positives = 100/168 (59%), Gaps = 24/168 (14%)
Query: 1 LVFLKNDDLKLVMKAIDVGACDYLLKPVQLKEVKMIWQHVIRKKKTSKRSNDDVPNSDSG 60
L + + D VMK I GACDYL+KPV LKE++ IW HV+ KK K +P S+S
Sbjct: 90 LPVITHSDYDSVMKGIIHGACDYLVKPVGLKELQNIWHHVV--KKNIKSYAKLLPPSES- 146
Query: 61 NGIDSAVTKSSDNKIEKPSRKRKD-----GNEDDVEEENE----YDHENDGPTMPKKPRF 111
+ + SRKRKD G+EDD + E + + + DG KKPR
Sbjct: 147 ------------DSVPSASRKRKDKVNDSGDEDDSDREEDDGEGSEQDGDGSGTRKKPRV 194
Query: 112 KWSRDLHHKFVVAVNQFGVEKARPKKILNLMNIENLTKEHVASHLQVF 159
WS++LH KFV AV Q G++KA PKKIL+LM+IE LT+E+VASHLQ +
Sbjct: 195 VWSQELHQKFVSAVQQLGLDKAVPKKILDLMSIEGLTRENVASHLQKY 242
>At3g16857 ARR1 protein, putative, 5' partial
Length = 569
Score = 120 bits (302), Expect = 3e-28
Identities = 72/182 (39%), Positives = 103/182 (56%), Gaps = 28/182 (15%)
Query: 1 LVFLKNDDLK-LVMKAIDVGACDYLLKPVQLKEVKMIWQHVIRKKKTS----------KR 49
++ + DD K +V+K + GA DYL+KPV+++ +K IWQHV+RK+++ +
Sbjct: 12 VIMMSADDSKSVVLKGVTHGAVDYLIKPVRMEALKNIWQHVVRKRRSEWSVPEHSGSIEE 71
Query: 50 SNDDVPNSDSGNGIDSAVTKSSD------------NKIEKPSRKRKDGNEDDVEEENEYD 97
+ + G G +AV+ D N SRKRKD E E + D
Sbjct: 72 TGERQQQQHRGGGGGAAVSGGEDAVDDNSSSVNEGNNWRSSSRKRKDE-----EGEEQGD 126
Query: 98 HENDGPTMPKKPRFKWSRDLHHKFVVAVNQFGVEKARPKKILNLMNIENLTKEHVASHLQ 157
+++ + KKPR WS +LH +FV AVNQ GVEKA PKKIL LMN+ LT+E+VASHLQ
Sbjct: 127 DKDEDASNLKKPRVVWSVELHQQFVAAVNQLGVEKAVPKKILELMNVPGLTRENVASHLQ 186
Query: 158 VF 159
+
Sbjct: 187 KY 188
>At2g01760 putative two-component response regulator protein
Length = 382
Score = 117 bits (294), Expect = 2e-27
Identities = 66/154 (42%), Positives = 96/154 (61%), Gaps = 7/154 (4%)
Query: 12 VMKAIDVGACDYLLKPVQLKEVKMIWQHVIRKKKTSKR------SNDDVPNSDSGNGIDS 65
VM I+ GACDYL+KP++ +E+K IWQHV+R+K K+ + +D NS S +
Sbjct: 99 VMTGINHGACDYLIKPIRPEELKNIWQHVVRRKCVMKKELRSSQALEDNKNSGSLETVVV 158
Query: 66 AVTKSSDNKIEKPSRKRKDGNEDDVEEENEYDHENDGPTMPKKPRFKWSRDLHHKFVVAV 125
+V++ S+ + K K+K ++NE D D P KK R WS +LH +FV AV
Sbjct: 159 SVSECSEESLMKCRNKKKKKKRSVDRDDNEDDLLLD-PGNSKKSRVVWSIELHQQFVNAV 217
Query: 126 NQFGVEKARPKKILNLMNIENLTKEHVASHLQVF 159
N+ G++KA PK+IL LMN+ L++E+VASHLQ F
Sbjct: 218 NKLGIDKAVPKRILELMNVPGLSRENVASHLQKF 251
>At1g67710 response regulator 11 (ARR11)
Length = 521
Score = 115 bits (287), Expect = 1e-26
Identities = 69/152 (45%), Positives = 91/152 (59%), Gaps = 8/152 (5%)
Query: 12 VMKAIDVGACDYLLKPVQLKEVKMIWQHVIRKKKTSKRSNDDVPNSDSGNGI---DSAVT 68
VMK + GACDYLLKP+++KE+K+IWQHV+RKK R + + I D A
Sbjct: 98 VMKGVQHGACDYLLKPIRMKELKIIWQHVLRKKLQEVRDIEGCGYEGGADWITRYDEAHF 157
Query: 69 KSSDNKIEKPSRKRKDGNEDDVEEENEYDHENDGPTMPKKPRFKWSRDLHHKFVVAVNQF 128
+ +KRKD D E++ D + + KK R WS +LHHKFV AVNQ
Sbjct: 158 LGGGEDVSF-GKKRKD---FDFEKKLLQDESDPSSSSSKKARVVWSFELHHKFVNAVNQI 213
Query: 129 GVE-KARPKKILNLMNIENLTKEHVASHLQVF 159
G + KA PKKIL+LMN+ LT+E+VASHLQ +
Sbjct: 214 GCDHKAGPKKILDLMNVPWLTRENVASHLQKY 245
>At5g49240 putative protein
Length = 292
Score = 101 bits (252), Expect = 2e-22
Identities = 54/164 (32%), Positives = 98/164 (58%), Gaps = 6/164 (3%)
Query: 1 LVFLKNDDLKLVMKAIDVGACDYLLKPVQLKEVKMIWQHVIRKKKTSKRSNDDVPNSDSG 60
++ ++D +K V K + GA DYL+KP++ ++++++++H+++K R V ++
Sbjct: 118 IIISEDDSVKSVKKWMINGAADYLIKPIRPEDLRIVFKHLVKKM----RERRSVVTGEAE 173
Query: 61 NGIDSAVTKSSDNKIEKPSR-KRKDGNEDDVEEENEYDHENDG-PTMPKKPRFKWSRDLH 118
+ D+ I P++ KR E +V EE+ +DH + + KK R W +LH
Sbjct: 174 KAAGEKSSSVGDSTIRNPNKSKRSSCLEAEVNEEDRHDHNDRACASSAKKRRVVWDEELH 233
Query: 119 HKFVVAVNQFGVEKARPKKILNLMNIENLTKEHVASHLQVFFFI 162
F+ AV+ G+E+A PKKIL++M ++ +++E+VASHLQV F I
Sbjct: 234 QNFLNAVDFLGLERAVPKKILDVMKVDYISRENVASHLQVTFLI 277
>At3g62670 putative protein
Length = 426
Score = 95.1 bits (235), Expect = 2e-20
Identities = 61/154 (39%), Positives = 87/154 (55%), Gaps = 18/154 (11%)
Query: 12 VMKAIDVGACDYLLKPVQLKEVKMIWQHVIRKKKTSKRSNDDVPNSDSGNGIDSAVTKSS 71
VM++I GACD+L+KPV + + ++W+HV RK+ + +S D P G V
Sbjct: 126 VMESIKYGACDFLVKPVSKEVIAVLWRHVYRKRMS--KSGLDKP------GESGTVESDP 177
Query: 72 DNKIEKPSRKRKDGNEDDVEEENEYDH-ENDGPTMPKKPRFKWSRDLHHKFVVAVNQFG- 129
D + + NE+ +N DH E PT KKPR +W+ +LHHKF VAV + G
Sbjct: 178 DEYDDLEQDNLYESNEEG--SKNTCDHKEEKSPT--KKPRMQWTPELHHKFEVAVEKMGS 233
Query: 130 VEKARPKKILNLM----NIENLTKEHVASHLQVF 159
+EKA PK IL M N++ LT+ +VASHLQ +
Sbjct: 234 LEKAFPKTILKYMQEELNVQGLTRNNVASHLQKY 267
>At4g16110 hypothetical protein
Length = 644
Score = 85.1 bits (209), Expect = 2e-17
Identities = 50/142 (35%), Positives = 77/142 (54%), Gaps = 17/142 (11%)
Query: 1 LVFLKNDDLKLVMKAIDVGACDYLLKPVQLKEVKMIWQHVIRKKK----TSKRSNDDVPN 56
+V +D +V+K + GA DYL+KPV+++ +K IWQHV+RKK+ S+ S + +
Sbjct: 109 IVMSADDSKSVVLKGVTHGAVDYLIKPVRIEALKNIWQHVVRKKRNEWNVSEHSGGSIED 168
Query: 57 SDSGNGIDSAVTKSSDNKIE-------KPSRKRKDGNEDDVEEENEYDHENDGPTMPKKP 109
+ + +DN + SRKRK+ DD + D + D ++ KKP
Sbjct: 169 TGGDRDRQQQHREDADNNSSSVNEGNGRSSRKRKEEEVDD-----QGDDKEDSSSL-KKP 222
Query: 110 RFKWSRDLHHKFVVAVNQFGVE 131
R WS +LH +FV AVNQ GV+
Sbjct: 223 RVVWSVELHQQFVAAVNQLGVD 244
>At5g59570 ARR1 protein - like
Length = 298
Score = 71.6 bits (174), Expect = 2e-13
Identities = 37/97 (38%), Positives = 53/97 (54%)
Query: 63 IDSAVTKSSDNKIEKPSRKRKDGNEDDVEEENEYDHENDGPTMPKKPRFKWSRDLHHKFV 122
++S+V S K +K ED EE + E+ K+PR W+ LH +FV
Sbjct: 96 VESSVPGSDPKKQKKSDGGEAAAVEDSTAEEGDSGPEDASGKTSKRPRLVWTPQLHKRFV 155
Query: 123 VAVNQFGVEKARPKKILNLMNIENLTKEHVASHLQVF 159
V G++ A PK I+ LMN+E LT+E+VASHLQ +
Sbjct: 156 DVVAHLGIKNAVPKTIMQLMNVEGLTRENVASHLQKY 192
>At5g44190 unknown protein
Length = 386
Score = 71.6 bits (174), Expect = 2e-13
Identities = 43/115 (37%), Positives = 65/115 (56%), Gaps = 14/115 (12%)
Query: 45 KTSKRSNDDVPNSDSGNGIDSAVTKSSDNKIEKPSRKRKDGNEDDVEEENEYDHENDGPT 104
+TS+R + V + G G D + + ++ +K KD D END
Sbjct: 98 ETSER-DVGVCKQEGGGGGDGGFRDKTVRRGKRKGKKSKDCLSD----------END--- 143
Query: 105 MPKKPRFKWSRDLHHKFVVAVNQFGVEKARPKKILNLMNIENLTKEHVASHLQVF 159
+ KKP+ W+ +LH KFV AV Q GV+KA P +IL +MN+++LT+ +VASHLQ +
Sbjct: 144 IKKKPKVDWTPELHRKFVQAVEQLGVDKAVPSRILEIMNVKSLTRHNVASHLQKY 198
>At3g46640 unknown protein
Length = 323
Score = 69.7 bits (169), Expect = 7e-13
Identities = 52/167 (31%), Positives = 77/167 (45%), Gaps = 26/167 (15%)
Query: 17 DVGACDYLLKPVQLKEVKMIWQHVIRKKKTSKRSNDDVPNSDSGNGIDSAVTKSSDNKIE 76
D+ + Y L P L I R + R+++ +S G G T SS+N +E
Sbjct: 31 DLASLSYSLIPPNLAMAFSITPERSRTIQDVNRASETTLSSLRG-GSSGPNTSSSNNNVE 89
Query: 77 KPSR-------------KRKDGNEDD-----------VEEENEYDHENDGPTMPKKPRFK 112
+ R K +G+ DD EE + + G T+ K+PR
Sbjct: 90 EEDRVGSSSPGSDSKKQKTSNGDGDDGGGVDPDSAMAAEEGDSGTEDLSGKTL-KRPRLV 148
Query: 113 WSRDLHHKFVVAVNQFGVEKARPKKILNLMNIENLTKEHVASHLQVF 159
W+ LH +FV V G++ A PK I+ LMN+E LT+E+VASHLQ +
Sbjct: 149 WTPQLHKRFVDVVAHLGIKNAVPKTIMQLMNVEGLTRENVASHLQKY 195
>At5g07210 putative protein
Length = 621
Score = 66.2 bits (160), Expect = 8e-12
Identities = 47/182 (25%), Positives = 78/182 (42%), Gaps = 24/182 (13%)
Query: 2 VFLKNDDLKLVMKAIDVGACDYLLKPVQLKEVKMIWQHVIRKKKTSKRSNDDVPNSDSGN 61
V + + D +++ GA ++ KP+ ++ I+Q + K+ K + N +
Sbjct: 100 VLVMSSDTNKEEESLSCGAMGFIPKPIHPTDLTKIYQFALSNKRNGKSTLSTEQNHKDAD 159
Query: 62 ------------GIDSAVTKSSDNKIEKPSRKRKDGN----EDDVEEENEYDHENDGPT- 104
D TK + + SR N D +N N GP+
Sbjct: 160 VSVPQQITLVPEQADVLKTKRKNCSFKSDSRTVNSTNGSCVSTDGSRKNRKRKPNGGPSD 219
Query: 105 -------MPKKPRFKWSRDLHHKFVVAVNQFGVEKARPKKILNLMNIENLTKEHVASHLQ 157
KK + +W+ LH F+ A+ G++KA PKKIL M++ LT+E+VASHLQ
Sbjct: 220 DGESMSQPAKKKKIQWTDSLHDLFLQAIRHIGLDKAVPKKILAFMSVPYLTRENVASHLQ 279
Query: 158 VF 159
+
Sbjct: 280 KY 281
>At2g27070 putative two-component response regulator protein
Length = 575
Score = 64.7 bits (156), Expect = 2e-11
Identities = 41/121 (33%), Positives = 60/121 (48%), Gaps = 13/121 (10%)
Query: 39 HVIRKKKTSKRSNDDVPNSDSGNGIDSAVTKSSDNKIEKPSRKRKDGNEDDVEEENEYDH 98
+V++ KK + S D +S N + S N+ KP K G DD E ++
Sbjct: 169 YVLKTKKKNCSSKSDTRTVNSTNVSHVSTNGSRKNRKRKP----KGGPSDDGESLSQ--- 221
Query: 99 ENDGPTMPKKPRFKWSRDLHHKFVVAVNQFGVEKARPKKILNLMNIENLTKEHVASHLQV 158
PKK + W+ L F+ A+ G +K PKKIL +MN+ LT+E+VASHLQ
Sbjct: 222 ------PPKKKKIWWTNPLQDLFLQAIQHIGYDKVVPKKILAIMNVPYLTRENVASHLQK 275
Query: 159 F 159
+
Sbjct: 276 Y 276
>At4g18020 pseudo-response regulator 2 (APRR2)
Length = 535
Score = 63.5 bits (153), Expect = 5e-11
Identities = 42/137 (30%), Positives = 66/137 (47%), Gaps = 23/137 (16%)
Query: 46 TSKRSNDDVPNSDSGNGIDSAVTKSSDNKI-----EKPSRKRKDGNEDDV--EEENEYD- 97
T K + +D + +D+ K D+K+ S K ++G D+ E+ + D
Sbjct: 211 TEKDNMEDHQDIGESKSVDTTNRKLDDDKVVVKEERGDSEKEEEGETGDLISEKTDSVDI 270
Query: 98 HENDGPTMP---------------KKPRFKWSRDLHHKFVVAVNQFGVEKARPKKILNLM 142
H+ + T P + + W+ +LH KFV AV Q GV++A P +IL LM
Sbjct: 271 HKKEDETKPINKSSGIKNVSGNKTSRKKVDWTPELHKKFVQAVEQLGVDQAIPSRILELM 330
Query: 143 NIENLTKEHVASHLQVF 159
+ LT+ +VASHLQ F
Sbjct: 331 KVGTLTRHNVASHLQKF 347
Score = 40.0 bits (92), Expect = 6e-04
Identities = 21/65 (32%), Positives = 34/65 (52%)
Query: 13 MKAIDVGACDYLLKPVQLKEVKMIWQHVIRKKKTSKRSNDDVPNSDSGNGIDSAVTKSSD 72
MK I +GA ++L KP+ +++K IWQHV+ K SN + + S + +D
Sbjct: 111 MKCIALGAVEFLQKPLSPEKLKNIWQHVVHKAFNDGGSNVSISLKPVKESVVSMLHLETD 170
Query: 73 NKIEK 77
IE+
Sbjct: 171 MTIEE 175
>At2g20570 GPRI1 protein (gpri1)
Length = 420
Score = 62.0 bits (149), Expect = 1e-10
Identities = 41/116 (35%), Positives = 59/116 (50%), Gaps = 12/116 (10%)
Query: 50 SNDDVPNSDSGNGIDSAVTKSSDNKIEKPS------RKRKDGNEDDVEEENEYDHENDGP 103
S + SG G + V+K D E + RKRK + +N N+G
Sbjct: 95 SQGETTKGSSGKG-EEVVSKRDDVAAETVTYDGDSDRKRKYSSS--ASSKNNRISNNEGK 151
Query: 104 TMPKKPRFKWSRDLHHKFVVAVNQFGVEKARPKKILNLMNIENLTKEHVASHLQVF 159
+K + W+ +LH +FV AV Q GV+KA P +IL LM + LT+ +VASHLQ +
Sbjct: 152 ---RKVKVDWTPELHRRFVEAVEQLGVDKAVPSRILELMGVHCLTRHNVASHLQKY 204
>At2g40970 unknown protein
Length = 248
Score = 61.6 bits (148), Expect = 2e-10
Identities = 27/53 (50%), Positives = 38/53 (70%)
Query: 107 KKPRFKWSRDLHHKFVVAVNQFGVEKARPKKILNLMNIENLTKEHVASHLQVF 159
K+PR W+ LH +FV AV G++ A PK I+ LM++E LT+E+VASHLQ +
Sbjct: 104 KRPRLVWTPQLHKRFVDAVGHLGIKNAVPKTIMQLMSVEGLTRENVASHLQKY 156
>At2g40260 hypothetical protein
Length = 410
Score = 60.8 bits (146), Expect = 3e-10
Identities = 35/120 (29%), Positives = 61/120 (50%), Gaps = 10/120 (8%)
Query: 45 KTSKRSNDDVPNSDSGNGIDSAVTKSSDNKIEKPSRKRKDGNEDDVEEENEYDHENDGPT 104
K + ++ +D D G + +S D S + + G+ ++ + +N G
Sbjct: 20 KATTKNEEDKDEEDDEEGEEDEEERSGDQSPSSNSYEEESGSH-----HHDQNKKNGGSV 74
Query: 105 MP----KKPRFKWSRDLHHKFVVAVNQFG-VEKARPKKILNLMNIENLTKEHVASHLQVF 159
P K PR +W+ +LH F+ AV + G ++A PK +L LMN++ L+ HV SHLQ++
Sbjct: 75 RPYNRSKTPRLRWTPELHICFLQAVERLGGPDRATPKLVLQLMNVKGLSIAHVKSHLQMY 134
>At1g69580 transfactor, putative
Length = 332
Score = 60.8 bits (146), Expect = 3e-10
Identities = 29/53 (54%), Positives = 36/53 (67%), Gaps = 1/53 (1%)
Query: 108 KPRFKWSRDLHHKFVVAVNQF-GVEKARPKKILNLMNIENLTKEHVASHLQVF 159
KPR KW+ DLHHKF+ AVNQ G KA PK ++ +M I LT H+ SHLQ +
Sbjct: 30 KPRLKWTCDLHHKFIEAVNQLGGPNKATPKGLMKVMEIPGLTLYHLKSHLQKY 82
>At5g05090 unknown protein
Length = 266
Score = 60.1 bits (144), Expect = 5e-10
Identities = 29/64 (45%), Positives = 41/64 (63%), Gaps = 1/64 (1%)
Query: 97 DHENDGPTMP-KKPRFKWSRDLHHKFVVAVNQFGVEKARPKKILNLMNIENLTKEHVASH 155
D D P K+PR W+ LH +FV AV G++ A PK I+ LM+++ LT+E+VASH
Sbjct: 69 DSTGDEPARTLKRPRLVWTPQLHKRFVDAVAHLGIKNAVPKTIMQLMSVDGLTRENVASH 128
Query: 156 LQVF 159
LQ +
Sbjct: 129 LQKY 132
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,171,141
Number of Sequences: 26719
Number of extensions: 197891
Number of successful extensions: 1736
Number of sequences better than 10.0: 248
Number of HSP's better than 10.0 without gapping: 149
Number of HSP's successfully gapped in prelim test: 100
Number of HSP's that attempted gapping in prelim test: 1286
Number of HSP's gapped (non-prelim): 416
length of query: 167
length of database: 11,318,596
effective HSP length: 92
effective length of query: 75
effective length of database: 8,860,448
effective search space: 664533600
effective search space used: 664533600
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC137602.16


