

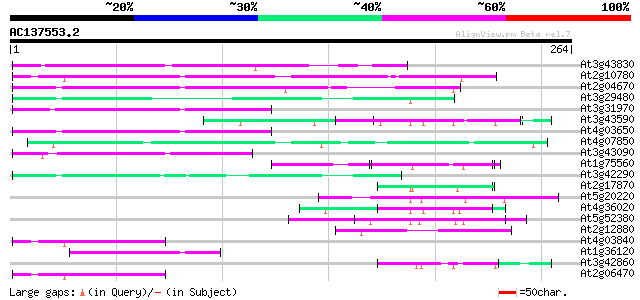
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137553.2 - phase: 0
(264 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g43830 putative protein 78 4e-15
At2g10780 pseudogene 78 6e-15
At2g04670 putative retroelement pol polyprotein 63 2e-10
At3g29480 hypothetical protein 57 8e-09
At3g31970 hypothetical protein 54 9e-08
At3g43590 putative protein 52 2e-07
At4g03650 putative reverse transcriptase 52 3e-07
At4g07850 putative polyprotein 50 9e-07
At3g43090 putative protein 50 1e-06
At1g75560 DNA-binding protein 49 2e-06
At3g42290 putative protein 48 6e-06
At2g17870 putative glycine-rich, zinc-finger DNA-binding protein 46 2e-05
At5g20220 zinc finger protein 45 4e-05
At4g36020 glycine-rich protein 45 4e-05
At5g52380 unknown protein 45 5e-05
At2g12880 pseudogene 45 5e-05
At4g03840 putative transposon protein 42 3e-04
At1g36120 putative reverse transcriptase gb|AAD22339.1 42 3e-04
At3g42860 unknown protein (At3g42860) 42 4e-04
At2g06470 putative retroelement pol polyprotein 42 4e-04
>At3g43830 putative protein
Length = 329
Score = 78.2 bits (191), Expect = 4e-15
Identities = 57/189 (30%), Positives = 86/189 (45%), Gaps = 26/189 (13%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L EA +WW+ + EQ G V W+ F+ EF RY PE+ E++FL L+QG +V
Sbjct: 154 LRGEAQEWWERVKQ-REQVGCVDQWSFFKEEFTRRYLPEETIDDLEMKFLRLQQGTKTVR 212
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELV---SSYM 118
+Y +F L +F + E KF +GLR DI+ + N +LV +S
Sbjct: 213 KYEKEFHSLERF---ERRKRGEHELIHKFISGLRVDIRLCCHVRDFDNMIDLVEKAASLE 269
Query: 119 IYEEDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCG 178
I E+ + +I E+ G G Q + +RR + E+ CY+CG
Sbjct: 270 IGLEEEARYKRIAQEKEAMGSYG-------------QTGHSKRRCQ------EVTCYRCG 310
Query: 179 EKGHKSNVC 187
GH + C
Sbjct: 311 VAGHIARDC 319
>At2g10780 pseudogene
Length = 1611
Score = 77.8 bits (190), Expect = 6e-15
Identities = 65/240 (27%), Positives = 103/240 (42%), Gaps = 26/240 (10%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVV-TWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSV 60
L +A +WW L V ++ G V ++A F EF +YFP + + E +L+L QG+ +V
Sbjct: 220 LEGDAHNWW---LTVEKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTV 276
Query: 61 TEYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIY 120
EY +F L ++ E E ++ +F GLR +I+ + + SELV +
Sbjct: 277 REYDEEFNRLRRYVGRELEE--EQAQLRRFIRGLRIEIRNHCLVRTFNSVSELVERAAMI 334
Query: 121 EEDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEK 180
EE + E R ++ P + +++ ++ + D C CG K
Sbjct: 335 EEGIE-------EERYLNREKAPIRNNQSTKPADKKRKFDKVDNTKSDAKTGECVTCG-K 386
Query: 181 GHKSNVCNGEEKKCFRCGKKWHTIAECKRGD-----------IVCFNCDEEGHLTSQCKK 229
H S C C RCG K H I C + + CF C + GHL +C K
Sbjct: 387 NH-SGTCWKAIGACGRCGSKDHAIQSCPKMEPGQSKVLGEETRTCFYCGKTGHLKRECPK 445
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 62.8 bits (151), Expect = 2e-10
Identities = 57/215 (26%), Positives = 89/215 (40%), Gaps = 31/215 (14%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L +A WW+S+ Q + +WA F EF +YFP++ + E FLEL QG+ SV
Sbjct: 176 LEGDAHLWWRSVTARRRQADM--SWADFVAEFNAKYFPQEALDRMEARFLELTQGEWSVR 233
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYE 121
EY +F L + + + ++ +F GLR D++ + + LV + E
Sbjct: 234 EYDREFNRLLAYAGRGMED--DQAQMRRFLRGLRPDLRVQCRVSQYATKAALVETAAEVE 291
Query: 122 EDTKAHY---KIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCG 178
ED + + +N QQ P P +G++R D
Sbjct: 292 EDLQRQVVGVSPAVQTKNTQQQVTPSKGGKPA-QGQKRKWD------------------- 331
Query: 179 EKGHKSNVCNGEEKKCFRCGKKWHTIAEC-KRGDI 212
H S G F CG HT+A+C +RG +
Sbjct: 332 ---HPSRAGQGGRAGYFSCGSLDHTVADCTERGHL 363
>At3g29480 hypothetical protein
Length = 718
Score = 57.4 bits (137), Expect = 8e-09
Identities = 47/210 (22%), Positives = 76/210 (35%), Gaps = 55/210 (26%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L +A WWKS+ Q + +WA F EF +YFP++ + E FLEL QG+ SV
Sbjct: 89 LEGDAHLWWKSVTTRRRQANM--SWADFVAEFNAKYFPQEALDRMEAHFLELTQGERSVR 146
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYE 121
EY +F ++ ++E S+ +
Sbjct: 147 EYDREF-------------------------------------NRLLVYAECRSTRIPVV 169
Query: 122 EDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKG 181
+ K H ++ + K QG+ + + P R C+ CG
Sbjct: 170 QPKKTHQQVAPSKGGKPAQGQKRSWD--------------HPSRAGQGGRAGCFFCGSLD 215
Query: 182 HKSNVC--NGEEKKCFRCGKKWHTIAECKR 209
HK C E ++ + C K+ H C +
Sbjct: 216 HKVADCTQRAETREYYHCRKRRHLRPNCPK 245
>At3g31970 hypothetical protein
Length = 1329
Score = 53.9 bits (128), Expect = 9e-08
Identities = 37/122 (30%), Positives = 59/122 (48%), Gaps = 4/122 (3%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L +A WW+S+ Q ++WA F EF +YFP++ + E FLEL QG+ SV
Sbjct: 196 LEGDAHLWWRSVTARRRQ--AHMSWADFVAEFNAKYFPQEALDRMEARFLELTQGERSVR 253
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYE 121
EY +F L + + + ++ +F GLR D++ + + LV + E
Sbjct: 254 EYEREFNRLLVYAGRGMQD--DQAQMRRFLRGLRPDLRVRCRVLQYATKAALVETAAEVE 311
Query: 122 ED 123
ED
Sbjct: 312 ED 313
>At3g43590 putative protein
Length = 551
Score = 52.4 bits (124), Expect = 2e-07
Identities = 45/175 (25%), Positives = 67/175 (37%), Gaps = 21/175 (12%)
Query: 92 NGLRADIKRAIGYQKI-RNFSELVSSYMIYEEDTKAHYKIRNERRNKGQQGR------PK 144
NG+++ +K++ +K+ RN E I D + + +G+ PK
Sbjct: 79 NGVKSKVKKSESSKKMKRNKLEADHEIPIVWNDQDEEKVVEEIVKGEGEDDEVERSDEPK 138
Query: 145 PYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVC---NGEEKKCFRCGKKW 201
N ++L R D + CY CGE+GH S C K CF CG
Sbjct: 139 TEETASNLVLKKLLRGARYFDPPDAGWVSCYSCGEQGHTSFNCPTPTKRRKPCFICGSLE 198
Query: 202 HTIAECKRGDIVCFNCDEEGHLTSQC-KKPKKGQASVYDSDYVLTKVMNKCGELG 255
H +C +G C+ C + GH C K K G V +CG+ G
Sbjct: 199 HGAKQCSKGH-DCYICKKTGHRAKDCPDKYKNGSKGA---------VCLRCGDFG 243
Score = 45.8 bits (107), Expect = 2e-05
Identities = 24/101 (23%), Positives = 38/101 (36%), Gaps = 33/101 (32%)
Query: 172 IVCYKCGEKGHKSNVC--------------------NGEEKKCFRCGKKWHTIAEC---- 207
+ CY+CG+ GH C + E +C+RCG++ H EC
Sbjct: 285 VSCYRCGQLGHSGLACGRHYEESNENDSATPERLFNSREASECYRCGEEGHFARECPNSS 344
Query: 208 -------KRGDIVCFNCDEEGHLTSQCKKPKKGQASVYDSD 241
+ +C+ C+ GH +C P Q S D +
Sbjct: 345 SISTSHGRESQTLCYRCNGSGHFAREC--PNSSQVSKRDRE 383
Score = 43.9 bits (102), Expect = 9e-05
Identities = 31/99 (31%), Positives = 42/99 (42%), Gaps = 13/99 (13%)
Query: 154 EQRLNDERRPKRRDDPAEIV-CYKCGEKGHKSNVC---------NGEEKK--CFRCGKKW 201
E ND P+R + E CY+CGE+GH + C +G E + C+RC
Sbjct: 306 ESNENDSATPERLFNSREASECYRCGEEGHFARECPNSSSISTSHGRESQTLCYRCNGSG 365
Query: 202 HTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQASVYDS 240
H EC V D E TS + K + S +DS
Sbjct: 366 HFARECPNSSQVS-KRDRETSTTSHKSRKKNKENSEHDS 403
Score = 32.0 bits (71), Expect = 0.35
Identities = 23/106 (21%), Positives = 31/106 (28%), Gaps = 51/106 (48%)
Query: 173 VCYKCGEKGHKSNVCNGEEKK-------------------------------CFRCGKKW 201
VC +CG+ GH +C E K C+RCG+
Sbjct: 235 VCLRCGDFGHDMILCKYEYSKEDLKDVQCYICKSFGHLCCVEPGNSLSWAVSCYRCGQLG 294
Query: 202 HTIAEC--------------------KRGDIVCFNCDEEGHLTSQC 227
H+ C R C+ C EEGH +C
Sbjct: 295 HSGLACGRHYEESNENDSATPERLFNSREASECYRCGEEGHFAREC 340
>At4g03650 putative reverse transcriptase
Length = 839
Score = 52.0 bits (123), Expect = 3e-07
Identities = 37/122 (30%), Positives = 57/122 (46%), Gaps = 4/122 (3%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L +A WW+S+ Q + +WA F EF +YFP + + E FLEL QG SV
Sbjct: 194 LEGDAHLWWRSVTARRRQADM--SWADFMAEFNAKYFPREALDRMEARFLELTQGVRSVR 251
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYE 121
EY KF L + + + ++ +F GLR +++ + + LV + E
Sbjct: 252 EYDRKFNRLLVYAGRGMED--DQAQMRRFLRGLRPNLRVRCRVSQYATKAALVETAAEVE 309
Query: 122 ED 123
ED
Sbjct: 310 ED 311
>At4g07850 putative polyprotein
Length = 1138
Score = 50.4 bits (119), Expect = 9e-07
Identities = 56/253 (22%), Positives = 89/253 (35%), Gaps = 28/253 (11%)
Query: 9 WWKSLLPVLEQ--DGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVTEYAAK 66
WW ++ + D + TW + R+ P + L QG+ +V EY
Sbjct: 110 WWDQVVTTKRRLGDDSIETWNQLKNIMKRRFVPSHYHRELHQRLRNLVQGNRTVEEY--- 166
Query: 67 FVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYEEDTKA 126
F E+ + + +F GL DI N EL +++E+ K
Sbjct: 167 FKEMETLMLRADVQEECEATMSRFMGGLNRDILDRFEVIHYENLEELFHKAVMFEKQIK- 225
Query: 127 HYKIRNERRNKGQQGRPKP-YSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSN 185
R K KP Y G Q+ E +P + EI KG +
Sbjct: 226 ------RRSAKPSYNSSKPSYQREEKSGFQK---EYKPFVKPKVEEI-----SSKGKEKE 271
Query: 186 VCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQASVYDSDYVLT 245
V + KCF+C H +EC I+ + G + S+ +KP++ +L
Sbjct: 272 VTRTRDLKCFKCHGLGHYASECSNKRIMIIR--DSGEVESEDEKPEESDVEEAPKGELLV 329
Query: 246 -----KVMNKCGE 253
V+NK E
Sbjct: 330 TMRVLSVLNKAEE 342
>At3g43090 putative protein
Length = 357
Score = 50.1 bits (118), Expect = 1e-06
Identities = 36/116 (31%), Positives = 52/116 (44%), Gaps = 9/116 (7%)
Query: 2 LAEEADDWWKSLL---PVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDM 58
L EEA WW +L PV V+ W FR EF ++FP++ E +F EL+Q
Sbjct: 169 LREEASHWWDGVLGNTPVQH----VIFWEDFREEFNRKFFPQEAMDSLEDDFEELRQDTK 224
Query: 59 SVTEYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELV 114
V E + L++F A E S + GLR DI ++ + +LV
Sbjct: 225 KVRENERELSHLSRF--SVRAGRGEQSMIRRLMRGLRPDILTRCASREFFSMIDLV 278
>At1g75560 DNA-binding protein
Length = 257
Score = 49.3 bits (116), Expect = 2e-06
Identities = 24/68 (35%), Positives = 33/68 (48%), Gaps = 7/68 (10%)
Query: 170 AEIVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDI------VCFNCDEEGHL 223
AE C+ C E GH ++ C+ E C CGK H +C D +C NC ++GHL
Sbjct: 91 AESRCWNCREPGHVASNCSNEGI-CHSCGKSGHRARDCSNSDSRAGDLRLCNNCFKQGHL 149
Query: 224 TSQCKKPK 231
+ C K
Sbjct: 150 AADCTNDK 157
Score = 48.5 bits (114), Expect = 4e-06
Identities = 21/64 (32%), Positives = 33/64 (50%), Gaps = 7/64 (10%)
Query: 171 EIVCYKCGEKGHKSNVCN------GEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLT 224
E +C+ CG+ GH++ C+ G+ + C C K+ H A+C D C NC GH+
Sbjct: 111 EGICHSCGKSGHRARDCSNSDSRAGDLRLCNNCFKQGHLAADC-TNDKACKNCRTSGHIA 169
Query: 225 SQCK 228
C+
Sbjct: 170 RDCR 173
Score = 46.6 bits (109), Expect = 1e-05
Identities = 29/104 (27%), Positives = 45/104 (42%), Gaps = 10/104 (9%)
Query: 124 TKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHK 183
++ ++ R+ R + + R + AP + ER P+R + C C GH
Sbjct: 15 SRDRFRSRSPRDRRMRSERVSYHDAPSRR-------EREPRRAFSQGNL-CNNCKRPGHF 66
Query: 184 SNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQC 227
+ C+ C CG H AEC + C+NC E GH+ S C
Sbjct: 67 ARDCSNVSV-CNNCGLPGHIAAECT-AESRCWNCREPGHVASNC 108
Score = 27.3 bits (59), Expect = 8.6
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query: 153 GEQRLNDERRPKRRDD-PAEIVCYKCGEKGHKSNVC 187
G QR R + R+ A I+C+ CG +GH++ C
Sbjct: 209 GMQRGGLSRMSRDREGVSAMIICHNCGGRGHRAYEC 244
>At3g42290 putative protein
Length = 462
Score = 47.8 bits (112), Expect = 6e-06
Identities = 43/183 (23%), Positives = 74/183 (39%), Gaps = 28/183 (15%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L E+A WWKS+ Q + +WA F EF +YF +D + E+ FLEL + SV
Sbjct: 192 LEEDAHLWWKSVTARRRQADM--SWADFVVEFNAKYFLQDELDRMEVRFLELTLVERSVW 249
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYE 121
EY +F + Y + + + ++ +F GLR D++ +S Y +
Sbjct: 250 EYDREFNRVL-VYAGWGMKDGQ-AELRRFMRGLRPDLRVRC----------RMSQYALKA 297
Query: 122 EDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKG 181
+ ++ + K QG+ + ++ R C+ CG
Sbjct: 298 ALVETAAEVAPRKGGKPMQGKKRSWN--------------HLSRAGQGGRAGCFSCGNLA 343
Query: 182 HKS 184
HK+
Sbjct: 344 HKN 346
>At2g17870 putative glycine-rich, zinc-finger DNA-binding protein
Length = 301
Score = 45.8 bits (107), Expect = 2e-05
Identities = 23/72 (31%), Positives = 29/72 (39%), Gaps = 18/72 (25%)
Query: 174 CYKCGEKGHKSNVCN--------GEEKKCFRCGKKWHTIAECKR----------GDIVCF 215
CY CG GH + VC G + C+ CG H +C R G CF
Sbjct: 226 CYTCGGVGHIAKVCTSKIPSGGGGGGRACYECGGTGHLARDCDRRGSGSSGGGGGSNKCF 285
Query: 216 NCDEEGHLTSQC 227
C +EGH +C
Sbjct: 286 ICGKEGHFAREC 297
Score = 43.9 bits (102), Expect = 9e-05
Identities = 22/82 (26%), Positives = 33/82 (39%), Gaps = 27/82 (32%)
Query: 174 CYKCGEKGHKSNVC----------------NGEEKKCFRCGKKWHTIAECKR-------- 209
C+ CGE GH + C +G E +C+ CG H +C++
Sbjct: 96 CFNCGEVGHMAKDCDGGSGGKSFGGGGGRRSGGEGECYMCGDVGHFARDCRQSGGGNSGG 155
Query: 210 ---GDIVCFNCDEEGHLTSQCK 228
G C++C E GHL C+
Sbjct: 156 GGGGGRPCYSCGEVGHLAKDCR 177
Score = 38.9 bits (89), Expect = 0.003
Identities = 20/77 (25%), Positives = 27/77 (34%), Gaps = 23/77 (29%)
Query: 174 CYKCGEKGHKSNVCNGEE---------------KKCFRCGKKWHTIAECKR--------G 210
CY CGE GH + C G C+ CG H +C++ G
Sbjct: 163 CYSCGEVGHLAKDCRGGSGGNRYGGGGGRGSGGDGCYMCGGVGHFARDCRQNGGGNVGGG 222
Query: 211 DIVCFNCDEEGHLTSQC 227
C+ C GH+ C
Sbjct: 223 GSTCYTCGGVGHIAKVC 239
Score = 37.7 bits (86), Expect = 0.006
Identities = 21/78 (26%), Positives = 26/78 (32%), Gaps = 16/78 (20%)
Query: 174 CYKCGEKGHKSNVCN--------GEEKKCFRCGKKWHTIAECKR--------GDIVCFNC 217
CY CG GH + C G C+ CG H C G C+ C
Sbjct: 198 CYMCGGVGHFARDCRQNGGGNVGGGGSTCYTCGGVGHIAKVCTSKIPSGGGGGGRACYEC 257
Query: 218 DEEGHLTSQCKKPKKGQA 235
GHL C + G +
Sbjct: 258 GGTGHLARDCDRRGSGSS 275
Score = 35.8 bits (81), Expect = 0.024
Identities = 19/82 (23%), Positives = 29/82 (35%), Gaps = 26/82 (31%)
Query: 174 CYKCGEKGHKSNVCN-----------GEEKKCFRCGKKWHTIAECK-------------- 208
CY CG+ GH + C G + C+ CG+ H +C+
Sbjct: 132 CYMCGDVGHFARDCRQSGGGNSGGGGGGGRPCYSCGEVGHLAKDCRGGSGGNRYGGGGGR 191
Query: 209 -RGDIVCFNCDEEGHLTSQCKK 229
G C+ C GH C++
Sbjct: 192 GSGGDGCYMCGGVGHFARDCRQ 213
Score = 33.1 bits (74), Expect = 0.16
Identities = 17/74 (22%), Positives = 25/74 (32%), Gaps = 16/74 (21%)
Query: 178 GEKGHKSNVCNGEEKKCFRCGKKWHTIAEC----------------KRGDIVCFNCDEEG 221
G K N G CF CG+ H +C G+ C+ C + G
Sbjct: 80 GSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKSFGGGGGRRSGGEGECYMCGDVG 139
Query: 222 HLTSQCKKPKKGQA 235
H C++ G +
Sbjct: 140 HFARDCRQSGGGNS 153
>At5g20220 zinc finger protein
Length = 270
Score = 45.1 bits (105), Expect = 4e-05
Identities = 34/136 (25%), Positives = 55/136 (40%), Gaps = 31/136 (22%)
Query: 146 YSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVC-----NGEEK-KCFRCGK 199
+S PVN+ ++ L+ + + C CG++GH+ + C N + K +C CG
Sbjct: 123 FSNPVNREQRSLSMKG--------TKFYCKNCGQEGHRRHYCPELGTNADRKFRCRGCGG 174
Query: 200 KWHTIAECKRGDIV-----------CFNCDEEGHLTSQCKKPK------KGQASVYDSDY 242
K H C + + C C E GH + C+KP G+ S D
Sbjct: 175 KGHNRRTCPKSKSIVTKGISTRYHKCGICGERGHNSRTCRKPTGVNPSCSGENSGEDGVG 234
Query: 243 VLTKVMNKCGELGIRV 258
+T C ++G V
Sbjct: 235 KITYACGFCKKMGHNV 250
>At4g36020 glycine-rich protein
Length = 299
Score = 45.1 bits (105), Expect = 4e-05
Identities = 32/123 (26%), Positives = 47/123 (38%), Gaps = 26/123 (21%)
Query: 137 KGQQGRPKPYS--APVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVC------N 188
+G G+ K + AP ++ N+ R R CY CGE GH S C
Sbjct: 63 QGSDGKTKAVNVTAPGGGSLKKENNSRGNGARRGGGGSGCYNCGELGHISKDCGIGGGGG 122
Query: 189 GEEKK------CFRCGKKWHTIAEC------------KRGDIVCFNCDEEGHLTSQCKKP 230
G E++ C+ CG H +C K G+ C+ C + GH+ C +
Sbjct: 123 GGERRSRGGEGCYNCGDTGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDCTQK 182
Query: 231 KKG 233
G
Sbjct: 183 SVG 185
Score = 44.7 bits (104), Expect = 5e-05
Identities = 21/64 (32%), Positives = 31/64 (47%), Gaps = 10/64 (15%)
Query: 174 CYKCGEKGHKSNVCNGEEKK---CFRCGKKWHTIAEC-KRG------DIVCFNCDEEGHL 223
CY CG GH + C + + C++CG H +C +RG D C+ C +EGH
Sbjct: 232 CYSCGGVGHIARDCATKRQPSRGCYQCGGSGHLARDCDQRGSGGGGNDNACYKCGKEGHF 291
Query: 224 TSQC 227
+C
Sbjct: 292 AREC 295
Score = 40.4 bits (93), Expect = 0.001
Identities = 25/97 (25%), Positives = 36/97 (36%), Gaps = 22/97 (22%)
Query: 174 CYKCGEKGHKSNVC------------NGEEKKCFRCGKKWHTIAEC---KRGDIVCFNCD 218
CY CG+ GH + C G C+ CG H +C ++ C+ C
Sbjct: 200 CYTCGDVGHFARDCTQKVAAGNVRSGGGGSGTCYSCGGVGHIARDCATKRQPSRGCYQCG 259
Query: 219 EEGHLTSQCKKPKKGQASVYDSDYVLTKVMNKCGELG 255
GHL C + G ++ Y KCG+ G
Sbjct: 260 GSGHLARDCDQRGSGGGGNDNACY-------KCGKEG 289
Score = 39.7 bits (91), Expect = 0.002
Identities = 21/80 (26%), Positives = 31/80 (38%), Gaps = 26/80 (32%)
Query: 174 CYKCGEKGHKSNVC----NGEEKK--------CFRCGKKWHTIAEC-------------- 207
CY CG+ GH + C NG+++ C+ CG H +C
Sbjct: 134 CYNCGDTGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDCTQKSVGNGDQRGAV 193
Query: 208 KRGDIVCFNCDEEGHLTSQC 227
K G+ C+ C + GH C
Sbjct: 194 KGGNDGCYTCGDVGHFARDC 213
>At5g52380 unknown protein
Length = 268
Score = 44.7 bits (104), Expect = 5e-05
Identities = 21/84 (25%), Positives = 37/84 (44%), Gaps = 13/84 (15%)
Query: 163 PKRRDDPAEIVCYKCGEKGHKSNVC------NGEEKKCFRCGKKWHTIAECK-------R 209
P++ + +C +C +GH C + E+K C+ CG H+++ C
Sbjct: 90 PEKSEWERNKICLQCRRRGHSLKNCPEKNNESSEKKLCYNCGDTGHSLSHCPYPMEDGGT 149
Query: 210 GDIVCFNCDEEGHLTSQCKKPKKG 233
CF C +GH++ C + K G
Sbjct: 150 KFASCFICKGQGHISKNCPENKHG 173
Score = 42.4 bits (98), Expect = 3e-04
Identities = 27/125 (21%), Positives = 55/125 (43%), Gaps = 14/125 (11%)
Query: 132 NERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDD--PAEIVCYKCGEKGHKSNVCNG 189
N+++ K + K + ++ ++ + R P R P E C+ C K H + +C
Sbjct: 33 NKKKKKKSLFKKKKPGSSTDRPQRTGSSTRHPLRVPGMKPGE-GCFICHSKTHIAKLCPE 91
Query: 190 EE-----KKCFRCGKKWHTIAECKRGDI------VCFNCDEEGHLTSQCKKPKKGQASVY 238
+ K C +C ++ H++ C + +C+NC + GH S C P + + +
Sbjct: 92 KSEWERNKICLQCRRRGHSLKNCPEKNNESSEKKLCYNCGDTGHSLSHCPYPMEDGGTKF 151
Query: 239 DSDYV 243
S ++
Sbjct: 152 ASCFI 156
>At2g12880 pseudogene
Length = 119
Score = 44.7 bits (104), Expect = 5e-05
Identities = 25/90 (27%), Positives = 38/90 (41%), Gaps = 21/90 (23%)
Query: 154 EQRLNDERRPK-------RRDDPAEIVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAE 206
++R ND R + RR+D CYKCG+ GH + C H + +
Sbjct: 9 QERDNDRERTRASYNNDRRRNDYDPRACYKCGKLGHFARSC--------------HVVTQ 54
Query: 207 CKRGDIVCFNCDEEGHLTSQCKKPKKGQAS 236
I C+ C EEGH ++ C + Q +
Sbjct: 55 PTTAYITCYFCSEEGHRSNGCPNKRTDQVN 84
Score = 33.1 bits (74), Expect = 0.16
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 7/40 (17%)
Query: 170 AEIVCYKCGEKGHKSNVCNGE-------EKKCFRCGKKWH 202
A I CY C E+GH+SN C + + C+ CG + H
Sbjct: 58 AYITCYFCSEEGHRSNGCPNKRTDQVNPKGHCYWCGNQDH 97
>At4g03840 putative transposon protein
Length = 973
Score = 42.4 bits (98), Expect = 3e-04
Identities = 25/73 (34%), Positives = 41/73 (55%), Gaps = 4/73 (5%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVV-TWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSV 60
L +A +WW L V ++ G V ++A F EF +YFP + + E +L+L QG+ +V
Sbjct: 185 LERDAHNWW---LTVEKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTV 241
Query: 61 TEYAAKFVELAKF 73
EY +F L ++
Sbjct: 242 REYDEEFNRLRRY 254
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 42.4 bits (98), Expect = 3e-04
Identities = 23/71 (32%), Positives = 39/71 (54%), Gaps = 2/71 (2%)
Query: 29 FRREFLNRYFPEDVRGKKEIEFLELKQGDMSVTEYAAKFVELAKFYPHYSAETAEFSKCI 88
F EF +YFP++ + E FLEL +G++SV EY +F L + + + ++
Sbjct: 157 FVAEFNAKYFPQEALDRMEARFLELTKGELSVREYGREFKRLLVYAGRGMED--DQAQMR 214
Query: 89 KFENGLRADIK 99
+F GLR D++
Sbjct: 215 RFLRGLRPDLR 225
>At3g42860 unknown protein (At3g42860)
Length = 372
Score = 41.6 bits (96), Expect = 4e-04
Identities = 21/58 (36%), Positives = 30/58 (51%), Gaps = 6/58 (10%)
Query: 174 CYKCGEKGHKSNVCNGE-EKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKP 230
CYKCG++GH S C G+ + F+ G+ T GD C+ C + GH + C P
Sbjct: 306 CYKCGKQGHWSRDCTGQSSNQQFQSGQAKST---SSTGD--CYKCGKAGHWSRDCTSP 358
Score = 40.8 bits (94), Expect = 8e-04
Identities = 29/115 (25%), Positives = 43/115 (37%), Gaps = 37/115 (32%)
Query: 174 CYKCGEKGHKSNVCNGEEK-----------KCFRCGKKWHTIAEC--------------- 207
CYKCG++GH + C + CF+CGK H +C
Sbjct: 239 CYKCGKEGHWARDCTVQSDTGPVKSTSAAGDCFKCGKPGHWSRDCTAQSGNPKYEPGQMK 298
Query: 208 -KRGDIVCFNCDEEGHLTSQC------KKPKKGQASVYDSDYVLTKVMNKCGELG 255
C+ C ++GH + C ++ + GQA S T KCG+ G
Sbjct: 299 SSSSSGECYKCGKQGHWSRDCTGQSSNQQFQSGQAKSTSS----TGDCYKCGKAG 349
Score = 27.7 bits (60), Expect = 6.6
Identities = 11/27 (40%), Positives = 14/27 (51%)
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKK 200
CYKCG+ GH S C + GK+
Sbjct: 342 CYKCGKAGHWSRDCTSPAQTTNTPGKR 368
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 41.6 bits (96), Expect = 4e-04
Identities = 25/73 (34%), Positives = 41/73 (55%), Gaps = 4/73 (5%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVV-TWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSV 60
L +A +WW L V ++ G V ++A F EF +YFP + + E +L+L QG+ +V
Sbjct: 185 LEGDAHNWW---LTVEKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTV 241
Query: 61 TEYAAKFVELAKF 73
EY +F L ++
Sbjct: 242 REYDEEFNRLRRY 254
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,422,349
Number of Sequences: 26719
Number of extensions: 295731
Number of successful extensions: 1258
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 987
Number of HSP's gapped (non-prelim): 215
length of query: 264
length of database: 11,318,596
effective HSP length: 97
effective length of query: 167
effective length of database: 8,726,853
effective search space: 1457384451
effective search space used: 1457384451
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC137553.2


