

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137553.12 + phase: 0
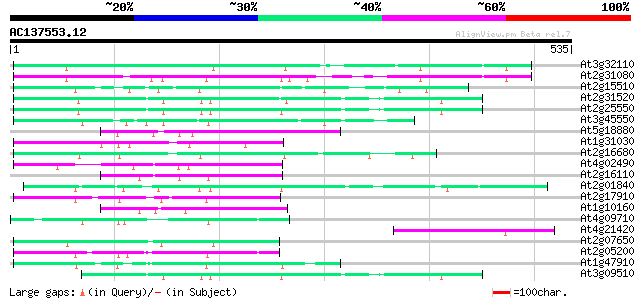
(535 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g32110 non-LTR reverse transcriptase, putative 116 4e-26
At2g31080 putative non-LTR retroelement reverse transcriptase 113 2e-25
At2g15510 putative non-LTR retroelement reverse transcriptase 106 4e-23
At2g31520 putative non-LTR retroelement reverse transcriptase 105 8e-23
At2g25550 putative non-LTR retroelement reverse transcriptase 103 3e-22
At3g45550 putative protein 95 1e-19
At5g18880 SAE1-S9-protein - like 92 7e-19
At1g31030 F17F8.5 92 7e-19
At2g16680 putative non-LTR retroelement reverse transcriptase 90 3e-18
At4g02490 89 6e-18
At2g16110 putative non-LTR retrolelement reverse transcriptase 87 2e-17
At2g01840 putative non-LTR retroelement reverse transcriptase 87 2e-17
At2g17910 putative non-LTR retroelement reverse transcriptase 86 7e-17
At1g10160 putative reverse transcriptase 85 1e-16
At4g09710 RNA-directed DNA polymerase -like protein 84 3e-16
At4g21420 hypothetical protein 82 7e-16
At2g07650 putative non-LTR retrolelement reverse transcriptase 82 7e-16
At2g05200 putative non-LTR retroelement reverse transcriptase 82 7e-16
At1g47910 reverse transcriptase, putative 82 7e-16
At3g09510 putative non-LTR reverse transcriptase 79 8e-15
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 116 bits (290), Expect = 4e-26
Identities = 120/515 (23%), Positives = 207/515 (39%), Gaps = 41/515 (7%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWD-LFQSEEQWAQVLRSR-----VIRNS 57
VAW +VC EGGLGIRS +N A K+ W L WAQV+RS+ V +
Sbjct: 1378 VAWTRVCLPRREGGLGIRSATAMNKALIAKVGWRVLNDGSSLWAQVVRSKYKVGDVHDRN 1437
Query: 58 TCIHHHVYSSIWSGAKTEFQNLI-DNSNWLVGDGDTINCWLDNWCGET-LVDLFDIDSQQ 115
+ +SS W +++I +W++GDG I W D W ET + D +
Sbjct: 1438 WTVAKSNWSSTWRSVGVGLRDVIWREQHWVIGDGRQIRFWTDRWLSETPIADDSIVPLSL 1497
Query: 116 LNMLPKKLRNYMQNFNWCFPDDILSLFPDMRL-LASKVTIPKHCIRDKLIWKHSNNGELT 174
ML + W + + RL L + + D+L W +++G T
Sbjct: 1498 AQMLCTARDLWRDGTGWDMSQIAPFVTDNKRLDLLAVIVDSVTGAHDRLAWGMTSDGRFT 1557
Query: 175 LQDAY-KFKKTNFPKVNWAK---HIWSPDIPPSKALLVWRFMLNKLPTDDNLMNKGCNLP 230
++ A+ + P+ + + +W P + +W + + T+ +
Sbjct: 1558 VKSAFAMLTNDDSPRQDMSSLYGRVWKVQAPERVRVFLWLVVNQAIMTNSERKRRHLCDS 1617
Query: 231 SMCSLCNNMSETTHHLFLECSFALNLWSWFA--GILNCSINIQSKEDLW-NICNGSWNPQ 287
+C +C E+ H+ +C +W + + E L+ N+ G
Sbjct: 1618 DVCQVCRGGIESILHVLRDCPAMSGIWDRIVPRRLQQSFFTMSLLEWLYSNLRQGLMTEG 1677
Query: 288 CKTVIIASLINIINSIWYARNQRRFQNIKIHWRSSIASIISNASLTGNLSRKVASPSIS- 346
+ ++ ++W+ W+ ++I + R + ++
Sbjct: 1678 SDWSTMFAM-----AVWWG------------WKWRCSNIFGENKTCRDRVRFIKDLAVEV 1720
Query: 347 NFVILKKFCVTLHPPRAPKIIEVIWQPPPLNWTKCNTDGSTSGS--LSSCGGIFRNCNSE 404
+ ++ + L R K I W PP W K NTDG++ G+ L+S GG+ RN
Sbjct: 1721 SIAYSREVELRLSGLRVNKPIR--WTPPMEGWYKINTDGASRGNPGLASAGGVLRNSAGA 1778
Query: 405 MLICFGENTGIGNSLHAELSGAMRAIELAISHQWLNLWLEVDSELVIRAFKNQSIVPWHL 464
F N G ++ AEL G + +A + Q +L LEVDSE+V+ F I H
Sbjct: 1779 WCGGFAVNIGRCSAPLAELWGVYYGLYMAWAKQLTHLELEVDSEVVV-GFLKTGIGETHP 1837
Query: 465 RNRWMNCM--LLTRNMNFIATHIFRERNVCADLLA 497
+ + L+++ +H++RE N AD LA
Sbjct: 1838 LSFLVRLCHNFLSKDWTVRISHVYREANSLADGLA 1872
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 113 bits (283), Expect = 2e-25
Identities = 130/535 (24%), Positives = 222/535 (41%), Gaps = 85/535 (15%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQSEEQ-WAQVLRSR-----VIRNS 57
++W+K+C EGG+G+RS +N A K+ W L Q +E WA+V+R + V S
Sbjct: 702 LSWRKICKPKAEGGIGLRSARDMNKALVAKVGWRLLQDKESLWARVVRKKYKVGGVQDTS 761
Query: 58 TCIHHHVYSSIWSGAKTEFQN-LIDNSNWLVGDGDTINCWLDNW-CGETLVDLFDIDSQQ 115
+SS W + ++ W+ GDG TI WLD W E LV+L
Sbjct: 762 WLKPQPRWSSTWRSVAVGLREVVVKGVGWVPGDGCTIRFWLDRWLLQEPLVEL------G 815
Query: 116 LNMLPKKLRNYMQNFNWC----FPDDILSLF-PD--MRLLASKVTIPKHCIRDKLIWKHS 168
+M+P+ R + W + +IL L+ P+ R L S V D++ WK +
Sbjct: 816 TDMIPEGERIKVAADYWLPGSGWNLEILGLYLPETVKRRLLSVVVQVFLGNGDEISWKGT 875
Query: 169 NNGELTLQDAYKFKKTNF---PKV-NWAKHIWSPDIPPSKALLVWRFMLNKLPTDDNLMN 224
+G T++ AY + + P + ++ IW P + +W N + T+ +
Sbjct: 876 QDGAFTVRSAYSLLQGDVGDRPNMGSFFNRIWKLITPERVRVFIWLVSQNVIMTNVERVR 935
Query: 225 KGCNLPSMCSLCNNMSETTHHLFLECSFALNLW----------SWFAGILN--CSINIQS 272
+ + ++CS+CN ET H+ +C +W +F+ L N+
Sbjct: 936 RHLSENAICSVCNGAEETILHVLRDCPAMEPIWRRLLPLRRHHEFFSQSLLEWLFTNMDP 995
Query: 273 KEDLWNICNG-----SWNPQCKTVIIASLINIINSIWYARNQRRFQNIKIHWRSSIASII 327
+ +W G +W +C V +R+ ++ + +A +
Sbjct: 996 VKGIWPTLFGMGIWWAWKWRCCDVF---------------GERKICRDRLKFIKDMAEEV 1040
Query: 328 SNASLTGNLSRKVASPSISNFVILKKFCVTLHPPRAPKIIEVI-WQPPPLNWTKCNTDGS 386
R+V ++ N P ++ +I WQ P W K TDG+
Sbjct: 1041 ----------RRVHVGAVGN------------RPNGVRVERMIRWQVPSDGWVKITTDGA 1078
Query: 387 TSGS--LSSCGGIFRNCNSEMLICFGENTGIGNSLHAELSGAMRAIELAISHQWLNLWLE 444
+ G+ L++ GG RN E L F N G + AEL GA + +A + + L+
Sbjct: 1079 SRGNHGLAAAGGAIRNGQGEWLGGFALNIGSCAAPLAELWGAYYGLLIAWDKGFRRVELD 1138
Query: 445 VDSELVIRAFKNQSIVPWHLRNRWMNCM--LLTRNMNFIATHIFRERNVCADLLA 497
+D +LV+ F + + H + + TR+ +H++RE N AD LA
Sbjct: 1139 LDCKLVV-GFLSTGVSNAHPLSFLVRLCQGFFTRDWLVRVSHVYREANRLADGLA 1192
>At2g15510 putative non-LTR retroelement reverse transcriptase
Length = 1138
Score = 106 bits (264), Expect = 4e-23
Identities = 108/476 (22%), Positives = 190/476 (39%), Gaps = 76/476 (15%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQSEE-QWAQVLRSRVIRNSTCIH- 61
V+W+K+C + GGLG R + N A K W + Q +A+ +SR + +
Sbjct: 615 VSWEKLCLAKESGGLGFRDIESFNQALLAKQSWRILQYPSFLFARFFKSRYFDDEEFLEA 674
Query: 62 ------HHVYSSIWSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWCGETLVDLFDID--- 112
+ SI G + + L VG+G ++N W+D W +FDI
Sbjct: 675 DLGVRPSYACCSILFGRELLAKGLRKE----VGNGKSLNVWMDPW-------IFDIAPRL 723
Query: 113 ------SQQLNMLPKKLRNYMQNFNWCFPDDILS--LFP-DMRLLASKVTIPKHCIRDKL 163
S L++ L N+ C+ D+L +P D+ L+ + + + D
Sbjct: 724 PLQRHFSVNLDLKVNDLINFEDR---CWKRDLLEELFYPTDVELITKRNPVVN--MDDFW 778
Query: 164 IWKHSNNGELTLQDAY----------KFKKTNF-PKVNWAKH-IWSPDIPPSKALLVWRF 211
+W H+ GE +++ Y ++ N P N K +WS P + +WR
Sbjct: 779 VWLHAKTGEYSVKSGYWLAFQSNKLDLIQQANLLPSTNGLKEQVWSTKSSPKIKMFLWRI 838
Query: 212 MLNKLPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALNLWSWFAGILNCSINIQ 271
+ LP D ++ +G ++ C +C + E+T+H+ CS A +W+ +G+ Q
Sbjct: 839 LSAALPVADQIIRRGMSIDPRCQICGDEGESTNHVLFTCSMARQVWA-LSGVPTPEFGFQ 897
Query: 272 SKEDLWNICNGSWNPQCKTVIIASLIN-----IINSIWYARNQRRFQNIKIHWRSSIASI 326
+ NI K +++ L+ ++ +W RN+ F I +SI
Sbjct: 898 NASIFANI--QFLFELKKMILVPDLVKRSWPWVLWRLWKNRNKLFFDGITFCPLNSIV-- 953
Query: 327 ISNASLTGNLSRKVASPSISNFVILKKFCVTLHPPRAPKIIEVI--WQPPPLNWTKCNTD 384
K+ ++ F + V+ ++I W+PPP W KCN
Sbjct: 954 ------------KIQEDTLEWFQAQSQIRVS-ESEEGQRVIPFTHSWEPPPEGWVKCNIG 1000
Query: 385 GSTSGSLSSCGG--IFRNCNSEMLICFGEN-TGIGNSLHAELSGAMRAIELAISHQ 437
+ SG CGG + R+ + +++ G N A L AI+ SH+
Sbjct: 1001 SAWSGKKKVCGGAWVLRDEHGSVILHSRRAFNGCSNKKEASLRCIFWAIDSMRSHR 1056
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 105 bits (261), Expect = 8e-23
Identities = 113/471 (23%), Positives = 186/471 (38%), Gaps = 39/471 (8%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQSEEQ-WAQVLRSRVIRNSTCIHH 62
VAWK++ EGGLG R L N A K W L Q +A+V+++R ++ + +
Sbjct: 995 VAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRLIQYPNSLFARVMKARYFKDVSILDA 1054
Query: 63 HV---YSSIWSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWCGETLVDLFDIDSQQLNML 119
V S W+ L + L+GDG I LDN + + M
Sbjct: 1055 KVRKQQSYGWASLLDGIALLKKGTRHLIGDGQNIRIGLDNIVDSHPPRPLNTEETYKEMT 1114
Query: 120 PKKLRNYMQNFNWCFPDDILSLFPDM--RLLASKVTIPKHCIRDKLIWKHSNNGELTLQD 177
L ++ + + D +S F D ++ + K DK+IW ++ GE T++
Sbjct: 1115 INNLFERKGSY-YFWDDSKISQFVDQSDHGFIHRIYLAKSKKPDKIIWNYNTTGEYTVRS 1173
Query: 178 AYKF----KKTNFPKVN-------WAKHIWSPDIPPSKALLVWRFMLNKLPTDDNLMNKG 226
Y TN P +N IW+ I P +WR + L T + L +G
Sbjct: 1174 GYWLLTHDPSTNIPAINPPHGSIDLKTRIWNLPIMPKLKHFLWRALSQALATTERLTTRG 1233
Query: 227 CNLPSMCSLCNNMSETTHHLFLECSFALNLWSWFAG---ILNCSINIQSKEDLWNICNGS 283
+ +C C+ +E+ +H C FA W W + I N ++ +E++ NI N
Sbjct: 1234 MRIDPICPRCHRENESINHALFTCPFATMAW-WLSDSSLIRNQLMSNDFEENISNILNFV 1292
Query: 284 WNPQCKTVIIASLINIINSIWYARNQRRFQNIKIHWRSSIASIISNASLTGNLSRKVASP 343
+ + +I IW ARN F + S +++S + T + S
Sbjct: 1293 QDTTMSDFHKLLPVWLIWRIWKARNNVVFNKFR---ESPSKTVLSAKAETHDWLNATQSH 1349
Query: 344 SISNFVILKKFCVTLHPPRAPKIIEVIWQPPPLNWTKCNTD-GSTSGSLSSCGGIFRNCN 402
KK T P R ++ W+ PP + KCN D G L + GG +
Sbjct: 1350 --------KK---TPSPTRQIAENKIEWRNPPATYVKCNFDAGFDVQKLEATGGWIIRNH 1398
Query: 403 SEMLICFG--ENTGIGNSLHAELSGAMRAIELAISHQWLNLWLEVDSELVI 451
I +G + N L AE + A++ + +++E D + +I
Sbjct: 1399 YGTPISWGSMKLAHTSNPLEAETKALLAALQQTWIRGYTQVFMEGDCQTLI 1449
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 103 bits (256), Expect = 3e-22
Identities = 113/470 (24%), Positives = 185/470 (39%), Gaps = 37/470 (7%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQSEEQ-WAQVLRSRVIRNSTCIHH 62
VAWK++ EGGLG R L N A K W L Q +A+V+++R ++ + +
Sbjct: 1221 VAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRLIQYPNSLFARVMKARYFKDVSILDA 1280
Query: 63 HV---YSSIWSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWCGETLVDLFDIDSQQLNML 119
V S W+ L + L+GDG I LDN + + M
Sbjct: 1281 KVRKQQSYGWASLLDGIALLKKGTRHLIGDGQNIRIGLDNIVDSHPPRPLNTEETYKEMT 1340
Query: 120 PKKLRNYMQNFNWCFPDDILSLFPDM--RLLASKVTIPKHCIRDKLIWKHSNNGELTLQD 177
L ++ + + D +S F D ++ + K DK+IW ++ GE T++
Sbjct: 1341 INNLFERKGSY-YFWDDSKISQFVDQSDHGFIHRIYLAKSKKPDKIIWNYNTTGEYTVRS 1399
Query: 178 AYKF----KKTNFPKVN-------WAKHIWSPDIPPSKALLVWRFMLNKLPTDDNLMNKG 226
Y TN P +N IW+ I P +WR + L T + L +G
Sbjct: 1400 GYWLLTHDPSTNIPAINPPHGSIDLKTRIWNLPIMPKLKHFLWRALSQALATTERLTTRG 1459
Query: 227 CNLPSMCSLCNNMSETTHHLFLECSFALNLW--SWFAGILNCSINIQSKEDLWNICNGSW 284
+ C C+ +E+ +H C FA W S + I N ++ +E++ NI N
Sbjct: 1460 MRIDPSCPRCHRENESINHALFTCPFATMAWRLSDSSLIRNQLMSNDFEENISNILNFVQ 1519
Query: 285 NPQCKTVIIASLINIINSIWYARNQRRFQNIKIHWRSSIASIISNASLTGNLSRKVASPS 344
+ + +I IW ARN F + S +++S + T + S
Sbjct: 1520 DTTMSDFHKLLPVWLIWRIWKARNNVVFNKFR---ESPSKTVLSAKAETHDWLNATQSH- 1575
Query: 345 ISNFVILKKFCVTLHPPRAPKIIEVIWQPPPLNWTKCNTD-GSTSGSLSSCGGIFRNCNS 403
KK T P R ++ W+ PP + KCN D G L + GG +
Sbjct: 1576 -------KK---TPSPTRQIAENKIEWRNPPATYVKCNFDAGFDVQKLEATGGWIIRNHY 1625
Query: 404 EMLICFG--ENTGIGNSLHAELSGAMRAIELAISHQWLNLWLEVDSELVI 451
I +G + N L AE + A++ + +++E D + +I
Sbjct: 1626 GTPISWGSMKLAHTSNPLEAETKALLAALQQTWIRGYTQVFMEGDCQTLI 1675
>At3g45550 putative protein
Length = 851
Score = 94.7 bits (234), Expect = 1e-19
Identities = 100/411 (24%), Positives = 162/411 (39%), Gaps = 52/411 (12%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQSEEQ-WAQVLRSRVIRNSTCIHH 62
VAWK++ EGGLG R L N A K W + Q +A+V+++R ++++ I
Sbjct: 460 VAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRIIQYPNSLFARVMKARYFKDNSIIDA 519
Query: 63 HVYSSI---WSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWCGETLVDLFD----IDSQQ 115
S WS + L + +++GDG TI +DN +VD + +Q
Sbjct: 520 KTRSQQSYGWSSLLSGIALLRKGTRYVIGDGKTIRLGIDN-----VVDSHPPRPLLTDEQ 574
Query: 116 LNMLPKKLRNYMQN--FNWCFPDDILSLFPDM--RLLASKVTIPKHCIRDKLIWKHSNNG 171
N L L N Q+ + C+ + L F D ++ + D+LIW +++ G
Sbjct: 575 HNGL--SLDNLFQHRGHSRCWDNAKLQTFVDQSDHDYIKRIYLSTRSKTDRLIWSYNSTG 632
Query: 172 ELTLQDAYKFKKTNFPK------------VNWAKHIWSPDIPPSKALLVWRFMLNKLPTD 219
+ T++ Y + T+ P V+ IW+ I P +WR + LPT
Sbjct: 633 DYTVRSGY-WLSTHDPSNTIPTMAKPHGSVDLKTKIWNLPIMPKLKHFLWRILSKALPTT 691
Query: 220 DNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALNLWSWF-AGILNCSINIQSKEDLWN 278
D L +G + C C +E+ +H C FA W + SI + ED N
Sbjct: 692 DRLTTRGMRIDPGCPRCRRENESINHALFTCPFATMAWRLSDTPLYRSSILSNNIED--N 749
Query: 279 ICNGSWNPQCKTVIIASLIN---IINSIWYARNQRRFQNIKIHWRSSIASIISNASLTGN 335
I N Q T+ + + ++ IW ARN F N++ S +++ + T
Sbjct: 750 ISNILLLLQNTTITDSQKLIPFWLLWRIWKARNNVVFNNLR---ESPSITVVRAKAETNE 806
Query: 336 LSRKVASPSISNFVILKKFCVTLHPPRAPKIIEVIWQPPPLNWTKCNTDGS 386
+ P R W P + + KCN D S
Sbjct: 807 WLNATQTQGPRRL-----------PKRTTAAGNTTWVKPQMPYIKCNFDAS 846
>At5g18880 SAE1-S9-protein - like
Length = 295
Score = 92.0 bits (227), Expect = 7e-19
Identities = 69/240 (28%), Positives = 108/240 (44%), Gaps = 15/240 (6%)
Query: 87 VGDGDTINCWLDNWC--GETLVDLFDIDSQQLNMLPK-KLRNYMQNFNWCFP---DDILS 140
+G+G++ + W D W G+ L L +QL + ++ +N +W P D
Sbjct: 16 MGNGESASFWYDAWTDFGQLLTFLGAAGPRQLRIRQDARVVEASRNGDWFLPAARSDNSQ 75
Query: 141 LFPDMRLLASKVTIPKHCIR--DKLIWKHSNNGEL---TLQDAYKFKKTNFPKVNWAKHI 195
LF L A + H R D +W+++ L + +D ++ + + P V WAK +
Sbjct: 76 LF----LAALTMAPVPHESRGQDSFLWRNAAGSYLPSFSSRDTWEQIRVHSPTVPWAKVV 131
Query: 196 WSPDIPPSKALLVWRFMLNKLPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALN 255
W + P +L+ W L +LPT D L G N+PS LC+N ET HLF ECSF+L
Sbjct: 132 WFKEYIPRFSLITWMSFLERLPTRDRLRGWGMNIPSSWVLCSNGDETHAHLFFECSFSLA 191
Query: 256 LWSWFAGILNCSINIQSKEDLWNICNGSWNPQCKTVIIASLINIINSIWYARNQRRFQNI 315
+W +FA S I T++ L + + +W RN R F +I
Sbjct: 192 IWEFFASKFRPSPPFGLPAASSWILQLPLRSHSTTILKLLLQSAVYHVWKERNARIFTSI 251
>At1g31030 F17F8.5
Length = 872
Score = 92.0 bits (227), Expect = 7e-19
Identities = 68/276 (24%), Positives = 119/276 (42%), Gaps = 25/276 (9%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQ-SEEQWAQVLRSRVIRNSTCIHH 62
++W VC EGGLG+R+L N S +K+ W + S W + + +IR +
Sbjct: 507 ISWDIVCKPKAEGGLGLRNLKEANDVSCLKLVWRIISNSNSLWTKWVAEYLIRKKSIWSL 566
Query: 63 HVYSSIWSGAKTEFQNLIDNSNWL----VGDGDTINCWLDNWCG-----ETLVDLFDID- 112
+S+ S + + D + VG+G++ + W D+W +T+ D ID
Sbjct: 567 KQSTSMGSWIWRKILKIRDVAKSFSRVEVGNGESASFWYDHWSAHGRLIDTVGDKGTIDL 626
Query: 113 --SQQLNMLPKKLRNYMQNFNWCFPDDILSLFPDMRLLASKVTIPKHCIRDKLIWKHSNN 170
++ ++ R + ++I + R+ S D ++W+ N+
Sbjct: 627 GIPREASVADAWTRRSRRRHRTSLLNEIEEMMAYQRIHHSDA-------EDTVLWRGKND 679
Query: 171 ---GELTLQDAYKFKKTNFPKVNWAKHIWSPDIPPSKALLVWRFMLNKLPTDDNLM--NK 225
+ +D + K V+W K +W P AL W + N+LPT D ++ N
Sbjct: 680 VFKPHFSTRDTWHLIKATSSTVSWHKGVWFRHATPKYALCTWLAIHNRLPTGDRMLKWNS 739
Query: 226 GCNLPSMCSLCNNMSETTHHLFLECSFALNLWSWFA 261
++ C LC N S+T HLF CS+A +W+ A
Sbjct: 740 SGSVSGNCVLCTNNSKTLEHLFFSCSYASTVWAALA 775
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 90.1 bits (222), Expect = 3e-18
Identities = 100/438 (22%), Positives = 161/438 (35%), Gaps = 69/438 (15%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQ-SEEQWAQVLRSRVIRNSTCIHH 62
+ KK+ GG G + L C N A K W LF S+ +Q+ +SR N+ ++
Sbjct: 791 IGGKKLTLPKSLGGFGFKDLQCFNQALLAKQAWRLFSDSKSIVSQIFKSRYFMNTDFLNA 850
Query: 63 HVY---SSIWSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWCGE-------TLVDLFDID 112
S W + L L+G+G+ N W+D W + L L +I
Sbjct: 851 RQGTRPSYTWRSILYGRELLNGGLKRLIGNGEQTNVWIDKWLFDGHSRRPMNLHSLMNIH 910
Query: 113 SQQLNMLPKKLRNY-MQNFNWCFPDDILSLFPDMRLLASKVTIPKHCIRDKLIWKHSNNG 171
+ +++ RN+ ++ F + + L R L S D W +NNG
Sbjct: 911 MKVSHLIDPLTRNWNLKKLTELFHEKDVQLIMHQRPLISS--------EDSYCWAGTNNG 962
Query: 172 ELTLQDAYK----------FKKTN-FPKVN-WAKHIWSPDIPPSKALLVWRFMLNKLPTD 219
T++ Y+ FK+ + +P VN +WS + P + +W+ + L +
Sbjct: 963 LYTVKSGYERSSRETFKNLFKEADVYPSVNPLFDKVWSLETVPKIKVFMWKALKGALAVE 1022
Query: 220 DNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALNLWSWF-----AGILNCSINIQSKE 274
D L ++G C C ET +HL +C FA +W+ A SI
Sbjct: 1023 DRLRSRGIRTADGCLFCKEEIETINHLLFQCPFARQVWALSLIQAPATGFGTSIFSNINH 1082
Query: 275 DLWNICNGSWNPQCKTVIIASLINIINSIWYARNQRRFQNIKIHWRSSIASIISNASLTG 334
+ N N +TV ++ IW RN+ FQ + +A +L
Sbjct: 1083 VIQNSQNFGIPRHMRTVSPW----LLWEIWKNRNKTLFQGTGLTSSEIVAKAYEECNLWI 1138
Query: 335 NLSRKVA---SPSISNFVILKKFCVTLHPPRAPKIIEVIWQPPPLNWTKCN--TDGSTSG 389
N K + SPS E W PPP KCN S
Sbjct: 1139 NAQEKSSGGVSPS-----------------------EHKWNPPPAGELKCNIGVAWSRQK 1175
Query: 390 SLSSCGGIFRNCNSEMLI 407
L+ + R+ ++L+
Sbjct: 1176 QLAGVSWVLRDSMGQVLL 1193
>At4g02490
Length = 657
Score = 89.0 bits (219), Expect = 6e-18
Identities = 68/276 (24%), Positives = 114/276 (40%), Gaps = 44/276 (15%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQSEEQ---------WAQVLRSRVI 54
++W VC+ + GGLG++ L N +K+ W LF + W ++ + R +
Sbjct: 401 ISWDIVCSSKESGGLGLKRLSSWNKVLALKLIWLLFTASGSLWVSWVRWVWRKLCKLREV 460
Query: 55 RNSTCIHHHVYSSIWSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWCGE-TLVDLFDIDS 113
I VG G T W DNW G L+ L +
Sbjct: 461 ARPFVICE------------------------VGSGITARFWQDNWTGHGPLIHLTGLTG 496
Query: 114 QQL--NMLPKKLRNYMQNFNWCFPDDILSLFPDMRLLASKVTIPKHCI----RDKLIWKH 167
QL + +R+ ++N +W S P + LL S + + + D +WK
Sbjct: 497 PQLVGLSITSVVRDAIRNDDWWIASS-RSRNPVILLLKSLLPPVGNLVDCEHDDSYLWKV 555
Query: 168 SN---NGELTLQDAYKFKKTNFPKVNWAKHIWSPDIPPSKALLVWRFMLNKLPTDDNLMN 224
+ + + + D ++ + V+W K +W + P A + W N+L T D L +
Sbjct: 556 GDRVPSSKFSTADTWRALQPFSVSVSWHKAVWFTNQVPKHAFISWVTAWNRLHTRDRLRS 615
Query: 225 KGCNLPSMCSLCNNMSETTHHLFLECSFALNLWSWF 260
G +P+ C LCN + ET HLF C F+ +W++F
Sbjct: 616 WGLIVPAECVLCNLVDETRDHLFFACRFSSRIWTFF 651
>At2g16110 putative non-LTR retrolelement reverse transcriptase
Length = 323
Score = 87.0 bits (214), Expect = 2e-17
Identities = 56/184 (30%), Positives = 85/184 (45%), Gaps = 11/184 (5%)
Query: 87 VGDGDTINCWLDNWCGE-TLVDLFDIDSQQLNMLPKK--LRNYMQNFNWCFPDDILSLFP 143
+G G+T + W DNW G+ L+DL + + +P +R+ ++ NW S P
Sbjct: 41 IGSGETASFWQDNWTGQGPLIDLTGTNGPRSVGMPLNAVVRDALRGDNWWLSSS-RSRNP 99
Query: 144 DMRLLASKVTIPKHCIR----DKLIWKHSNNGELTLQDAYKFKKTNFPK---VNWAKHIW 196
+ LL S + + I D WK ++ + A K P V W K +W
Sbjct: 100 SIALLKSVLPSSESMIECQHDDVYKWKPDHHAPSNIFSASKTWTALNPDGVLVPWQKSVW 159
Query: 197 SPDIPPSKALLVWRFMLNKLPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALNL 256
D P A + W +L T D L G N+P++C LCN + ET HLF +C F+ +
Sbjct: 160 FKDRIPKHAFICWVAAWKRLHTRDRLTQWGLNIPTVCVLCNVVDETHDHLFFQCQFSNEI 219
Query: 257 WSWF 260
WS+F
Sbjct: 220 WSFF 223
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 87.0 bits (214), Expect = 2e-17
Identities = 118/538 (21%), Positives = 205/538 (37%), Gaps = 67/538 (12%)
Query: 14 DEGGLGIRSLVCLNAASNMKICWDLFQSEEQW-AQVLRSRVIRNSTCIH-----HHVYSS 67
+EG LG + L N A K W + + + A++ + N+T + H Y
Sbjct: 1193 NEGDLGFKDLHQFNRALLAKQAWRILTNPQSLLARLYKGLYYPNTTYLRANKGGHASYG- 1251
Query: 68 IWSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWCGETLVD------LFDIDSQQLNMLPK 121
W+ + L +GDG T W D W TL + D D + ++ +
Sbjct: 1252 -WNSIQEGKLLLQQGLRVRLGDGQTTKIWEDPWL-PTLPPRPARGPILDEDMKVADLWRE 1309
Query: 122 KLRNYMQNFNWCFPDDILS---LFPDMRLLASKVTIPKHCIRDKLIWKHSNNGELTLQDA 178
R + D ++ L P+ + LA + + + RD W ++ N + T++
Sbjct: 1310 NKREW---------DPVIFEGVLNPEDQQLAKSLYLSNYAARDSYKWAYTRNTQYTVRSG 1360
Query: 179 Y----KFKKTNFPKVN-------WAKHIWSPDIPPSKALLVWRFMLNKLPTDDNLMNKGC 227
Y T +N + IW I P +WR + L T L N+
Sbjct: 1361 YWVATHVNLTEEEIINPLEGDVPLKQEIWRLKITPKIKHFIWRCLSGALSTTTQLRNRNI 1420
Query: 228 NLPSMCSLCNNMSETTHHLFLECSFALNLW--SWFAGILNCSINIQSKEDLWNICNGSWN 285
C C N ET +H+ CS+A +W + F+G +E++ I G N
Sbjct: 1421 PADPTCQRCCNADETINHIIFTCSYAQVVWRSANFSGSNRLCFTDNLEENIRLILQGKKN 1480
Query: 286 PQCKTVIIASLINIINSIWYARNQRRFQNI-KIHWRSSIASIISNASLTGNLSRKVASPS 344
+ I+ +W +RN+ FQ + + W+ + + T + V +
Sbjct: 1481 QNLPILNGLMPFWIMWRLWKSRNEYLFQQLDRFPWK---VAQKAEQEATEWVETMVNDTA 1537
Query: 345 ISNFVILKKFCVTLHPPRAPKIIEVIWQPPPLNWTKCNTD-GSTSG-SLSSCGGIFRNCN 402
IS+ T P W PP + KCN D G G +S G I R+CN
Sbjct: 1538 ISH--------NTAQSNDRPLSRSKQWSSPPEGFLKCNFDSGYVQGRDYTSTGWILRDCN 1589
Query: 403 SEMLICFGENTGIG------NSLHAELSGAMRAIELAISHQWLNLWLEVDSELVIRAFKN 456
+L ++G ++L AE G + A+++ + +W E D+ L + N
Sbjct: 1590 GRVL-----HSGCAKLQQSYSALQAEALGFLHALQMVWIRGYCYVWFEGDN-LELTNLIN 1643
Query: 457 QSIVPWHLRNRWMNCMLLTRNMNFIAT-HIFRERNVCADLLAFTGHNLDNFTVWMNVP 513
++ L + + F + ++ RERN+ AD L +++ + +VP
Sbjct: 1644 KTEDHHLLETLLYDIRFWMTKLPFSSIGYVNRERNLAADKLTKYANSMSSLYETFHVP 1701
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 85.5 bits (210), Expect = 7e-17
Identities = 64/284 (22%), Positives = 116/284 (40%), Gaps = 42/284 (14%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQSEEQ-WAQVLRSRVIRNSTCIH- 61
++W+++ D+GG G + L C N A K W + Q + +++V +SR NS +
Sbjct: 817 LSWQRLTLPKDQGGFGFKDLQCFNQALLAKQAWRVLQEKGSLFSRVFQSRYFSNSDFLSA 876
Query: 62 ------HHVYSSIWSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWCGE-------TLVDL 108
+ + SI G + Q L ++G+G W D W +
Sbjct: 877 TRGSRPSYAWRSILFGRELLMQGL----RTVIGNGQKTFVWTDKWLHDGSNRRPLNRRRF 932
Query: 109 FDIDSQQLNMLPKKLRNYMQNFNWCFPDDILSLFP--DMRLLASKVTIPKHCIRDKLIWK 166
++D + ++ RN+ N + LFP D+ ++ + P D W
Sbjct: 933 INVDLKVSQLIDPTSRNWNLNM-------LRDLFPWKDVEIILKQR--PLFFKEDSFCWL 983
Query: 167 HSNNGELTLQDAYKFKKTNF-----------PKVNWA-KHIWSPDIPPSKALLVWRFMLN 214
HS+NG +++ Y+F P VN IW+ P + +W+ +
Sbjct: 984 HSHNGLYSVKTGYEFLSKQVHHRLYQEAKVKPSVNSLFDKIWNLHTAPKIRIFLWKALHG 1043
Query: 215 KLPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALNLWS 258
+P +D L +G C +C+ +ET +H+ EC A +W+
Sbjct: 1044 AIPVEDRLRTRGIRSDDGCLMCDTENETINHILFECPLARQVWA 1087
>At1g10160 putative reverse transcriptase
Length = 1118
Score = 84.7 bits (208), Expect = 1e-16
Identities = 54/190 (28%), Positives = 87/190 (45%), Gaps = 13/190 (6%)
Query: 87 VGDGDTINCWLDNWCGE-TLVDLFDIDSQQLNMLPKK--LRNYMQNFNWCFPDD-----I 138
VG G T + W DNW L+ L L LP +R+ +++ W +
Sbjct: 830 VGSGVTASFWHDNWTDHGPLLHLTGPAGPLLAGLPLNSVVRDALRDDTWRISSSRSRNPV 889
Query: 139 LSLFPDMRLLASKVTIPKHCIRDKLIWK---HSNNGELTLQDAYKFKKTNFPKVNWAKHI 195
++L R+L S ++ D +WK H+ + + D + + + + V W K +
Sbjct: 890 ITLL--QRVLPSAASLIDCPHDDTYLWKIGHHAPSNRFSTADTWSYLQPSSTSVLWHKAV 947
Query: 196 WSPDIPPSKALLVWRFMLNKLPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALN 255
W D P +A + W N+L T D L G ++P C LCN++ E+ HLF C F+
Sbjct: 948 WFKDHVPKQAFICWVVAHNRLHTRDRLRRWGFSIPPTCVLCNDLDESREHLFFRCQFSSE 1007
Query: 256 LWSWFAGILN 265
+WS+F LN
Sbjct: 1008 IWSFFMRALN 1017
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 83.6 bits (205), Expect = 3e-16
Identities = 72/296 (24%), Positives = 107/296 (35%), Gaps = 56/296 (18%)
Query: 1 MVTVAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQSEEQW-AQVLRSRVIRNSTC 59
M V+W K+ +EGGLG R + K+ W + + ++VL + S+
Sbjct: 777 MAWVSWDKLTLPINEGGLGFREI-------EAKLSWRILKEPHSLLSRVLLGKYCNTSSF 829
Query: 60 IHHHVYSSI----WSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWCG------------E 103
+ S W G L W +G GD+IN W + W E
Sbjct: 830 MDCSASPSFASHGWRGILAGRDLLRKGLGWSIGQGDSINVWTEAWLSPSSPQTPIGPPTE 889
Query: 104 TLVDL-------FDIDSQQLNMLPKKLRNYMQNFNWCFPDDILSLFPDMRLLASKVTIPK 156
T DL D+ S + + K L Y K+TI
Sbjct: 890 TNKDLSVHDLICHDVKSWNVEAIRKHLPQYEDQIR-------------------KITINA 930
Query: 157 HCIRDKLIWKHSNNGELTLQDAYKFKKTN-FPK----VNWAKHIWSPDIPPSKALLVWRF 211
++D L+W +GE T + Y K N FP NW K+IW P +W+
Sbjct: 931 LPLQDSLVWLPVKSGEYTTKTGYALAKLNSFPASQLDFNWQKNIWKIHTSPKVKHFLWKA 990
Query: 212 MLNKLPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALNLWSWFAGILNCS 267
M LP + L + C C +E++ HL L C +A +W + N S
Sbjct: 991 MKGALPVGEALSRRNIEAEVTCKRC-GQTESSLHLMLLCPYAKKVWELAPVLFNPS 1045
>At4g21420 hypothetical protein
Length = 229
Score = 82.0 bits (201), Expect = 7e-16
Identities = 48/156 (30%), Positives = 80/156 (50%), Gaps = 3/156 (1%)
Query: 367 IEVIWQPPPLNWTKCNTDGSTSGS-LSSCGGIFRNCNSEMLICFGENTGIGNSLHAELSG 425
++V+W+ P K N DGS +S GG+FRN +E L+ + E+ G S AE +
Sbjct: 59 VKVVWKKPENGRIKLNFDGSRGREGQASIGGVFRNHKAEFLLGYSESIGEATSTMAEFAA 118
Query: 426 AMRAIELAISHQWLNLWLEVDSELVIRAFKNQSIVPWHLRNRWMNC--MLLTRNMNFIAT 483
R +ELA+ + +LWLE D+++++ + + N+ +N +++ N + +
Sbjct: 119 LKRGLELALENGLTDLWLEGDAKIIMDIISRRGRLRCEKTNKHVNYIKVVMPELNNCVLS 178
Query: 484 HIFRERNVCADLLAFTGHNLDNFTVWMNVPTCITEP 519
H++RE N AD LA GH + VW P I P
Sbjct: 179 HVYREGNRVADKLAKLGHQFQDPKVWRVQPPEIVLP 214
>At2g07650 putative non-LTR retrolelement reverse transcriptase
Length = 732
Score = 82.0 bits (201), Expect = 7e-16
Identities = 62/271 (22%), Positives = 108/271 (38%), Gaps = 21/271 (7%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQS-EEQWAQVLRSRV----IRNST 58
++WK+VC EGGLGIR +N A K+ W L Q WA+++R +R+
Sbjct: 364 ISWKRVCKPRSEGGLGIRKAQDMNKALLSKVGWRLIQDYHSLWARIMRCNYRVQDVRDGA 423
Query: 59 CIH-HHVYSSIWSGAKTEFQN-LIDNSNWLVGDGDTINCWLDNWCGETLVDLFDIDSQQL 116
V SS W + +I +W++GDG I W+D W + +
Sbjct: 424 WTKVRSVCSSTWRSVALGMREVVIPGLSWVIGDGREILFWMDKWLTNIPLAELLVQEPPA 483
Query: 117 NMLPKKLRNYMQN-FNWCFPDDILSLFP-----DMRLLASKVTIPKHCIRDKLIWKHSNN 170
+ R+ +N W D+ ++ P + L S V RD++ W S +
Sbjct: 484 GWKGMRARDLRRNGIGW----DMATIAPYISSYNRLQLQSFVLDDITGARDRISWGESQD 539
Query: 171 GELTLQDAYKF-KKTNFPKVNWAK---HIWSPDIPPSKALLVWRFMLNKLPTDDNLMNKG 226
G + Y F + P+ + + +WS P L +W + T+ +
Sbjct: 540 GRFKVSTTYSFLTRDEAPRQDMKRFFDRVWSVTAPERVRLFLWLVAQQAIMTNQERHRRH 599
Query: 227 CNLPSMCSLCNNMSETTHHLFLECSFALNLW 257
+ ++C +C E+ H+ +C +W
Sbjct: 600 LSTTTICQVCKGAKESIIHVLRDCPAMEGIW 630
Score = 34.3 bits (77), Expect = 0.18
Identities = 16/45 (35%), Positives = 27/45 (59%), Gaps = 4/45 (8%)
Query: 368 EVIWQPPPLNWTKCNTDGSTSGS--LSSCGGIFRNCNSEMLICFG 410
+V W+ P W K NTDG++ G+ ++ GG R + E+ +C+G
Sbjct: 661 QVSWRKPNEGWVKLNTDGASHGNPGEATAGGALR--DEEVRLCYG 703
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 82.0 bits (201), Expect = 7e-16
Identities = 67/271 (24%), Positives = 117/271 (42%), Gaps = 26/271 (9%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQSEEQW-AQVLRSRVIRNSTCIH- 61
VAW K+ + GGLG R + N + K+ W L S E +++L + +S+ +
Sbjct: 734 VAWSKLTNPKNAGGLGFRDIERCNDSLLAKLGWRLLNSPESLLSRILLGKYCHSSSFMEC 793
Query: 62 ------HHVYSSIWSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWCG--ETLVDLFDI-- 111
H + SI +G + L + WL+ +G+ ++ W D W + LV +
Sbjct: 794 KLPSQPSHGWRSIIAGREI----LKEGLGWLITNGEKVSIWNDPWLSISKPLVPIGPALR 849
Query: 112 DSQQLNMLPKKLRNYMQNFNWCFPDDILSLFPDMRLLASKVTIPKHCIRDKLIWKHSNNG 171
+ Q L + +N +Q ++W + I + P+ L ++ P DKL W +G
Sbjct: 850 EHQDLRVSALINQNTLQ-WDW---NKIAVILPNYENLIKQLPAPSSRGVDKLAWLPVKSG 905
Query: 172 ELTLQDAYKFKKT-NFP----KVNWAKHIWSPDIPPSKALLVWRFMLNKLPTDDNLMNKG 226
+ T + Y + P + NW ++W P L+W+ + LP L+ +
Sbjct: 906 QYTSRSGYGIASVASIPIPQTQFNWQSNLWKLQTLPKIKHLMWKAAMEALPVGIQLVRRH 965
Query: 227 CNLPSMCSLCNNMSETTHHLFLECSFALNLW 257
+ + C C E+T HLF C FA +W
Sbjct: 966 ISPSAACHRC-GAPESTTHLFFHCEFAAQVW 995
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 82.0 bits (201), Expect = 7e-16
Identities = 86/346 (24%), Positives = 141/346 (39%), Gaps = 56/346 (16%)
Query: 4 VAWKKVCADYDEGGLGIRSLVCLNAASNMKICWDLFQSEEQ-WAQVLRSRVIRNSTCIHH 62
+AW K+C+ +GGLG R++ N+A K W L + + +A+V + R R S +
Sbjct: 623 MAWDKLCSSKSDGGLGFRNVDDFNSALLAKQLWRLITAPDSLFAKVFKGRYFRKSNPLDS 682
Query: 63 -------HVYSSIWSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWC----------GETL 105
+ + S+ S ++ LI VG G +I+ W D W G ++
Sbjct: 683 IKSYSPSYGWRSMISARSLVYKGLIKR----VGSGASISVWNDPWIPAQFPRPAKYGGSI 738
Query: 106 VDLFDIDSQQLNMLPKKLRNYMQNFNWCFPDDILSLF-PDMRLLASKVTIPKHCIRDKLI 164
VD ++ K L + NF W D + LF P+ L S + I + D L
Sbjct: 739 VDP--------SLKVKSLIDSRSNF-WNI-DLLKELFDPEDVPLISALPIGNPNMEDTLG 788
Query: 165 WKHSNNGELTLQDAYKFKKTNF--------PKVNWAK-HIWSPDIPPSKALLVWRFMLNK 215
W + G T++ Y + + P + K +IW PP +W+ +
Sbjct: 789 WHFTKAGNYTVKSGYHTARLDLNEGTTLIGPDLTTLKAYIWKVQCPPKLRHFLWQILSGC 848
Query: 216 LPTDDNLMNKGCNLPSMCSLCNNMSETTHHLFLECSFALNLWS-----WFAGILNCSINI 270
+P +NL +G C C E+ +H +C A +W+ GI +
Sbjct: 849 VPVSENLRKRGILCDKGCVSCGASEESINHTLFQCHPARQIWALSQIPTAPGIFPSNSIF 908
Query: 271 QSKEDL-WNICNGSWNPQCKTVIIASLINIINSIWYARNQRRFQNI 315
+ + L W I +G V A II IW ARN++ F+N+
Sbjct: 909 TNLDHLFWRIPSG--------VDSAPYPWIIWYIWKARNEKVFENV 946
>At3g09510 putative non-LTR reverse transcriptase
Length = 484
Score = 78.6 bits (192), Expect = 8e-15
Identities = 91/401 (22%), Positives = 152/401 (37%), Gaps = 33/401 (8%)
Query: 69 WSGAKTEFQNLIDNSNWLVGDGDTINCWLDNWCGETLVDLFDIDSQQLNMLPKKLRNYMQ 128
W+ L + L+GDG I LDN + + M L
Sbjct: 24 WASLLDGIALLKKGTRHLIGDGQNIRIGLDNIVDSHPPRPLNTEETYKEMTINNLFERKG 83
Query: 129 NFNWCFPDDILSLFPDM--RLLASKVTIPKHCIRDKLIWKHSNNGELTLQDAYKF----K 182
++ + + D +S F D ++ + K DK+IW ++ GE T++ Y
Sbjct: 84 SY-YFWDDSKISQFVDQSDHGFIHRIYLAKSKKPDKIIWNYNTTGEYTVRSGYWLLTHDP 142
Query: 183 KTNFPKVN-------WAKHIWSPDIPPSKALLVWRFMLNKLPTDDNLMNKGCNLPSMCSL 235
TN P +N IW+ I P +WR + L T + L +G + C
Sbjct: 143 STNIPAINPPHGSIDLKTRIWNLPIMPKLKHFLWRALSQALATTERLTTRGMRIDPSCPR 202
Query: 236 CNNMSETTHHLFLECSFALNLW--SWFAGILNCSINIQSKEDLWNICNGSWNPQCKTVII 293
C+ +E+ +H C FA W S + I N ++ +E++ NI N +
Sbjct: 203 CHRENESINHALFTCPFATMAWRLSDSSLIRNQLMSNDFEENISNILNFVQDTTMSDFHK 262
Query: 294 ASLINIINSIWYARNQRRFQNIKIHWRSSIASIISNASLTGNLSRKVASPSISNFVILKK 353
+ +I IW ARN F + S +++S + T + S KK
Sbjct: 263 LLPVWLIWRIWKARNNVVFNKFR---ESPSKTVLSAKAETHDWLNATQSH--------KK 311
Query: 354 FCVTLHPPRAPKIIEVIWQPPPLNWTKCNTD-GSTSGSLSSCGGIFRNCNSEMLICFG-- 410
T P R ++ W+ PP + KCN D G L + GG + I +G
Sbjct: 312 ---TPSPTRQIAENKIEWRNPPATYVKCNFDAGFDVQKLEATGGWIIRNHYGTPISWGSM 368
Query: 411 ENTGIGNSLHAELSGAMRAIELAISHQWLNLWLEVDSELVI 451
+ N L AE + A++ + +++E D + +I
Sbjct: 369 KLAHTSNPLEAETKALLAALQQTWIRGYTQVFMEGDCQTLI 409
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.136 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,643,043
Number of Sequences: 26719
Number of extensions: 611139
Number of successful extensions: 1926
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 95
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 1642
Number of HSP's gapped (non-prelim): 171
length of query: 535
length of database: 11,318,596
effective HSP length: 104
effective length of query: 431
effective length of database: 8,539,820
effective search space: 3680662420
effective search space used: 3680662420
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC137553.12


