

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137546.8 + phase: 0 /pseudo
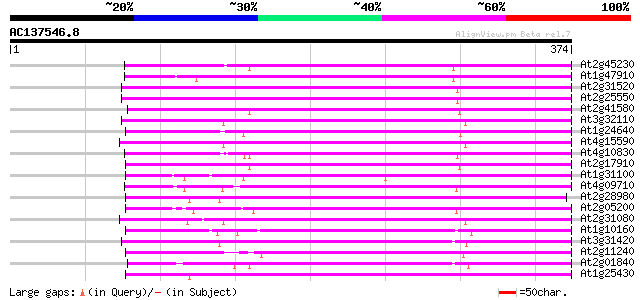
(374 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g45230 putative non-LTR retroelement reverse transcriptase 195 4e-50
At1g47910 reverse transcriptase, putative 183 2e-46
At2g31520 putative non-LTR retroelement reverse transcriptase 182 2e-46
At2g25550 putative non-LTR retroelement reverse transcriptase 182 2e-46
At2g41580 putative non-LTR retroelement reverse transcriptase 182 3e-46
At3g32110 non-LTR reverse transcriptase, putative 181 4e-46
At1g24640 hypothetical protein 181 8e-46
At4g15590 reverse transcriptase like protein 179 2e-45
At4g10830 putative protein 179 2e-45
At2g17910 putative non-LTR retroelement reverse transcriptase 179 2e-45
At1g31100 hypothetical protein 179 2e-45
At4g09710 RNA-directed DNA polymerase -like protein 178 5e-45
At2g28980 putative non-LTR retroelement reverse transcriptase 177 8e-45
At2g05200 putative non-LTR retroelement reverse transcriptase 172 2e-43
At2g31080 putative non-LTR retroelement reverse transcriptase 171 8e-43
At1g10160 putative reverse transcriptase 169 3e-42
At3g31420 hypothetical protein 167 9e-42
At2g11240 pseudogene 166 2e-41
At2g01840 putative non-LTR retroelement reverse transcriptase 162 2e-40
At1g25430 hypothetical protein 160 1e-39
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 195 bits (495), Expect = 4e-50
Identities = 114/305 (37%), Positives = 174/305 (56%), Gaps = 8/305 (2%)
Query: 77 WQQ-SRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRI-EGVEPVRNAIFTHFAQ 134
WQ+ SR W+R GD N+K+FH+ +RR N I ++ E+ R E + +F +
Sbjct: 336 WQEKSRIMWMRNGDRNTKYFHAATKNRRAQNRIQKLIDEEGREWTSDEDLGRVAEAYFKK 395
Query: 135 HFMSHNVVRPSVSNLQFQALSVTE--GEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFG 192
F S +V +V L+ V++ L+ P + +EV+ A + K GPDG+N
Sbjct: 396 LFASEDVGY-TVEELENLTPLVSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNGF 454
Query: 193 FIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSMN 252
++FW + D I + V F R+G + +G+N T I LI K+ +++ DFRPISL +
Sbjct: 455 LYQQFWETMGDQITEMVQAFFRSGSIEEGMNKTNICLIPKILKAEKMTDFRPISLCNVIY 514
Query: 253 KIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEV---VDEAFKLKKDLMLFK 309
K+I KL+ANRL+ ++ S++SETQ+AFVK R I D ILIA+E+ + K ++ + K
Sbjct: 515 KVIGKLMANRLKKILPSLISETQAAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAIK 574
Query: 310 VDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLR 369
D K YD V+W +L + ++ + F W + I ECV + VL+NG+P E RGLR
Sbjct: 575 TDISKAYDRVEWPFLEKAMRGLGFADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGLR 634
Query: 370 QGDPL 374
QGDPL
Sbjct: 635 QGDPL 639
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 183 bits (464), Expect = 2e-46
Identities = 111/304 (36%), Positives = 164/304 (53%), Gaps = 7/304 (2%)
Query: 77 WQQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRIEGVEP--VRNAIFTHFAQ 134
+Q+SR+ W++ GD NSKFFH+L RR N I L ++ I +E ++N ++F
Sbjct: 121 YQKSRSLWMKLGDNNSKFFHALTKQRRARNRITG-LHDENGIWSIEDDDIQNIAVSYFQN 179
Query: 135 HFMSHNVVRPSVSNLQFQALSVTEGEGLIKPFSVD-EVKEAICDYDSFKSSGPDGVNFGF 193
F + N + + Q L L+ + + EV+ A+ K+ GPDG+ F
Sbjct: 180 LFTTANPQVFDEALGEVQVLITDRINDLLTADATECEVRAALFMIHPEKAPGPDGMTALF 239
Query: 194 IKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSMNK 253
++ W ++K D++ V+ F + G K +NTT I LI K + P R+ + RPISL K
Sbjct: 240 FQKSWAIIKSDLLSLVNSFLQEGVFDKRLNTTNICLIPKTERPTRMTELRPISLCNVGYK 299
Query: 254 IIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEV---VDEAFKLKKDLMLFKV 310
+I+K+L RL+ V+ +++SETQSAFV R I D ILIA E+ + K M K
Sbjct: 300 VISKILCQRLKTVLPNLISETQSAFVDGRLISDNILIAQEMFHGLRTNSSCKDKFMAIKT 359
Query: 311 DFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLRQ 370
D K YD V+W ++ +L+ M F W WI C+ T VL+NG P +RGLRQ
Sbjct: 360 DMSKAYDQVEWNFIEALLRKMGFCEKWISWIMWCITTVQYKVLINGQPKGLIIPERGLRQ 419
Query: 371 GDPL 374
GDPL
Sbjct: 420 GDPL 423
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 182 bits (463), Expect = 2e-46
Identities = 105/306 (34%), Positives = 166/306 (53%), Gaps = 6/306 (1%)
Query: 75 ICWQQ-SRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRI-EGVEPVRNAIFTHF 132
I W+Q SRN W++EGD N+ +FH+ +R N + +++ + R+ G + + N F
Sbjct: 490 IYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQDFF 549
Query: 133 AQHFMSHNVVRPSVSNLQFQA-LSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNF 191
F ++ + + F++ ++ T L K FS E+ +AIC K+ GPDG+
Sbjct: 550 TNIFSTNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGPDGLTA 609
Query: 192 GFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSM 251
F K W ++ D++ V +F + IN T I +I K+ NP L+D+RPI+L +
Sbjct: 610 RFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPIALCNVL 669
Query: 252 NKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDE---AFKLKKDLMLF 308
K+I+K L NRL+ + S+VS++Q+AF+ R I D ++IA+EV+ ++ K M
Sbjct: 670 YKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAV 729
Query: 309 KVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGL 368
K D K YD V+W +L ++ F W WI V + SVL+NGSP RG+
Sbjct: 730 KTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGI 789
Query: 369 RQGDPL 374
RQGDPL
Sbjct: 790 RQGDPL 795
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 182 bits (463), Expect = 2e-46
Identities = 105/306 (34%), Positives = 166/306 (53%), Gaps = 6/306 (1%)
Query: 75 ICWQQ-SRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRI-EGVEPVRNAIFTHF 132
I W+Q SRN W++EGD N+ +FH+ +R N + +++ + R+ G + + N F
Sbjct: 716 IYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQDFF 775
Query: 133 AQHFMSHNVVRPSVSNLQFQA-LSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNF 191
F ++ + + F++ ++ T L K FS E+ +AIC K+ GPDG+
Sbjct: 776 TNIFSTNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGPDGLTA 835
Query: 192 GFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSM 251
F K W ++ D++ V +F + IN T I +I K+ NP L+D+RPI+L +
Sbjct: 836 RFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPIALCNVL 895
Query: 252 NKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDE---AFKLKKDLMLF 308
K+I+K L NRL+ + S+VS++Q+AF+ R I D ++IA+EV+ ++ K M
Sbjct: 896 YKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAV 955
Query: 309 KVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGL 368
K D K YD V+W +L ++ F W WI V + SVL+NGSP RG+
Sbjct: 956 KTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGI 1015
Query: 369 RQGDPL 374
RQGDPL
Sbjct: 1016 RQGDPL 1021
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 182 bits (461), Expect = 3e-46
Identities = 106/301 (35%), Positives = 163/301 (53%), Gaps = 5/301 (1%)
Query: 79 QSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRIEGVEPVRNAIFTHFAQHFMS 138
+SR+ W+ GD NSKFF + + + R N++ ++ E+ + V + I F + S
Sbjct: 68 KSRDKWMVGGDKNSKFFQATVKANRVSNSLRFLVDENGNEQTVNREKGKIAVTFFEDLFS 127
Query: 139 HNVVRPSVSNLQFQALSVTE--GEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFGFIKE 196
+ S L+ VTE + L K + E+ +A+ ++ + GPDG F +
Sbjct: 128 SSYPSSMDSVLEGFNKRVTEDMNQDLTKKVNEQEIYKAVFSINAESAPGPDGFTALFFQR 187
Query: 197 FWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSMNKIIA 256
W ++K+ I+ + F + G L + N T + LI K+ P R+ D RPISL M KII+
Sbjct: 188 QWPLVKNQIISDIELFFQTGILPEDWNHTHLCLIPKITKPARMADIRPISLCSVMYKIIS 247
Query: 257 KLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEA---FKLKKDLMLFKVDFE 313
K+L+ RL+ + +VS TQSAFV +R + D I++A+E+V K+ KD M+FK D
Sbjct: 248 KILSARLKKYLPVIVSPTQSAFVAERLVSDNIILAHEIVHNLRTNEKISKDFMVFKTDMS 307
Query: 314 KTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLRQGDP 373
K YD V+W +L +L + F W W+ CV + + SVL+NG P RGLRQGDP
Sbjct: 308 KAYDRVEWPFLKGILLALGFNSTWINWMMACVSSVSYSVLINGQPFGHITPHRGLRQGDP 367
Query: 374 L 374
L
Sbjct: 368 L 368
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 181 bits (460), Expect = 4e-46
Identities = 108/306 (35%), Positives = 160/306 (51%), Gaps = 6/306 (1%)
Query: 75 ICW-QQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVR-IEGVEPVRNAIFTHF 132
+ W Q+SR W GD N+KFFH+ RRR N I + D R + + + ++
Sbjct: 873 VVWMQKSREKWFVHGDRNTKFFHTSTIIRRRRNQIEMLQDNDGRWLSNAQELETHAIDYY 932
Query: 133 AQHFMSHNV--VRPSVSNLQFQALSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVN 190
+ + ++ V + F ALS + L KPFS EV+ AI +K+ GPDG
Sbjct: 933 KRLYSLDDLDAVVEQLPQEGFTALSEADFSSLTKPFSPLEVEGAIRSMGKYKAPGPDGFQ 992
Query: 191 FGFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGS 250
F ++ W V+ + + +FV +F +G + N +VLI+KV P+++ FRPISL
Sbjct: 993 PVFYQQGWEVVGESVTKFVMDFFSSGSFPQETNDVLVVLIAKVLKPEKITQFRPISLCNV 1052
Query: 251 MNKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKLK--KDLMLF 308
+ K I K++ RL+ VI ++ Q++F+ R D I++ EVV + K K ML
Sbjct: 1053 LFKTITKVMVGRLKGVINKLIGPAQTSFIPGRLSTDNIVVVQEVVHSMRRKKGVKGWMLL 1112
Query: 309 KVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGL 368
K+D EK YD + W L + LK P W +WI +CV + +L NG TD F RGL
Sbjct: 1113 KLDLEKAYDRIRWDLLEDTLKAAGLPGTWVQWIMKCVEGPSMRLLWNGEKTDAFKPLRGL 1172
Query: 369 RQGDPL 374
RQGDPL
Sbjct: 1173 RQGDPL 1178
>At1g24640 hypothetical protein
Length = 1270
Score = 181 bits (458), Expect = 8e-46
Identities = 108/305 (35%), Positives = 160/305 (52%), Gaps = 11/305 (3%)
Query: 78 QQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRIEGVEPVRNAIFT-HFAQHF 136
Q+SR WLR G+ NSK+FH+ + R+ I + + ++ E + + +F F
Sbjct: 301 QKSRQKWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNGNMQTSEAAKGEVAAAYFGNLF 360
Query: 137 MSHNVVRPSVSNLQFQAL----SVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFG 192
S N PS F L S E L+ S E+KEA+ + GPDG++
Sbjct: 361 KSSN---PSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSAL 417
Query: 193 FIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSMN 252
F + +W + + + V +F +G + N T + LI K +P + D RPISL +
Sbjct: 418 FFQHYWSTVGNQVTSEVKKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLY 477
Query: 253 KIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAF---KLKKDLMLFK 309
KII+K++A RL+ + +VS+TQSAFV +R I D IL+A+E+V ++ + M K
Sbjct: 478 KIISKIMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVK 537
Query: 310 VDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLR 369
D K YD V+W YL +L + F + W WI CV + T SVL+N P L+RGLR
Sbjct: 538 SDMSKAYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLR 597
Query: 370 QGDPL 374
QGDPL
Sbjct: 598 QGDPL 602
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 179 bits (455), Expect = 2e-45
Identities = 108/306 (35%), Positives = 161/306 (52%), Gaps = 5/306 (1%)
Query: 74 SICWQQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVL-VEDVRIEGVEPVRNAIFTHF 132
++ +Q+SR L GD N+ FFH+ RRR N I + ED + E + ++
Sbjct: 149 TLWFQKSREKLLALGDRNTTFFHTSTVIRRRRNRIEMLKDSEDRWVTEKEALEKLAMDYY 208
Query: 133 AQHFMSHNV--VRPSVSNLQFQALSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVN 190
+ + +V VR ++ F L+ E L +PF+ DEV A+ FK+ GPDG
Sbjct: 209 RKLYSLEDVSVVRGTLPTEGFPRLTREEKNNLNRPFTRDEVVVAVRSMGRFKAPGPDGYQ 268
Query: 191 FGFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGS 250
F ++ W + + + +FV EF +G L K N +VL++KV P+R+ FRP+SL
Sbjct: 269 PVFYQQCWETVGESVSKFVMEFFESGVLPKSTNDVLLVLLAKVAKPERITQFRPVSLCNV 328
Query: 251 MNKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKLK--KDLMLF 308
+ KII K++ RL+ VI ++ Q++F+ R D I++ E V + K K ML
Sbjct: 329 LFKIITKMMVIRLKNVISKLIGPAQASFIPGRLSFDNIVVVQEAVHSMRRKKGRKGWMLL 388
Query: 309 KVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGL 368
K+D EK YD + W +L E L+ W K I ECV S+L NG TD F +RGL
Sbjct: 389 KLDLEKAYDRIRWDFLAETLEAAGLSEGWIKRIMECVAGPEMSLLWNGEKTDSFTPERGL 448
Query: 369 RQGDPL 374
RQGDP+
Sbjct: 449 RQGDPI 454
>At4g10830 putative protein
Length = 1294
Score = 179 bits (455), Expect = 2e-45
Identities = 109/307 (35%), Positives = 168/307 (54%), Gaps = 12/307 (3%)
Query: 77 WQQ-SRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRI-EGVEPVRNAIFTHFAQ 134
WQQ SRN W++EGD N++FFH+ +R N + ++ E+ I G + + F +
Sbjct: 698 WQQKSRNQWMKEGDRNTEFFHACTKTRFSVNRLVTIKDEEGMIYRGDKEIGVHAQEFFTK 757
Query: 135 HFMSHNVVRPSVSNLQFQALS--VTE--GEGLIKPFSVDEVKEAICDYDSFKSSGPDGVN 190
+ S+ RP VS + F VTE + L K S E+ AIC K+ GPDG+
Sbjct: 758 VYESNG--RP-VSIIDFAGFKPIVTEQINDDLTKDLSDLEIYNAICHIGDDKAPGPDGLT 814
Query: 191 FGFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGS 250
F K W ++ D+++ V F R + + IN T I +I K+ NP+ L+D+RPI+L
Sbjct: 815 ARFYKSCWEIVGPDVIKEVKIFFRTSYMKQSINHTNICMIPKITNPETLSDYRPIALCNV 874
Query: 251 MNKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDE---AFKLKKDLML 307
+ KII+K L RL+ + ++VS++Q+AF+ R + D ++IA+E++ ++ + M
Sbjct: 875 LYKIISKCLVERLKGHLDAIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMA 934
Query: 308 FKVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRG 367
K D K YD V+W +L ++ F W KWI V + SVLVNG P +RG
Sbjct: 935 VKTDVSKAYDRVEWNFLETTMRLFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRG 994
Query: 368 LRQGDPL 374
+RQGDPL
Sbjct: 995 IRQGDPL 1001
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 179 bits (455), Expect = 2e-45
Identities = 105/302 (34%), Positives = 162/302 (52%), Gaps = 5/302 (1%)
Query: 78 QQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRIEGVEPVRNAIFTHFAQHFM 137
Q+SR WL GD N+ FFH+ + S R N + +L E+ + + I + F ++
Sbjct: 316 QKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDENDQEFTRNSDKGKIASSFFENLF 375
Query: 138 SHNVVRPSVSNLQFQALSVTE--GEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFGFIK 195
+ + ++L+ VT LI+ + EV A+ + + GPDG F +
Sbjct: 376 TSTYILTHNNHLEGLQAKVTSEMNHNLIQEVTELEVYNAVFSINKESAPGPDGFTALFFQ 435
Query: 196 EFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSMNKII 255
+ W ++K I+ + F G L + N T I LI K+ +PQR++D RPISL + KII
Sbjct: 436 QHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPISLCSVLYKII 495
Query: 256 AKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEA---FKLKKDLMLFKVDF 312
+K+L RL+ + ++VS TQSAFV +R I D IL+A+E++ ++ K+ M FK D
Sbjct: 496 SKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRISKEHMAFKTDM 555
Query: 313 EKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLRQGD 372
K YD V+W +L ++ + F W WI CV + + SVL+NG P RG+RQGD
Sbjct: 556 SKAYDRVEWPFLETMMTALGFNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRGIRQGD 615
Query: 373 PL 374
PL
Sbjct: 616 PL 617
>At1g31100 hypothetical protein
Length = 1090
Score = 179 bits (455), Expect = 2e-45
Identities = 110/309 (35%), Positives = 169/309 (54%), Gaps = 14/309 (4%)
Query: 78 QQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVED--VRIEGVEPVRNAIFTHFAQH 135
Q+SR W+ EGD N+ +FH + SR+ NTI ++++D V+I+ ++ +F+ +
Sbjct: 265 QRSRVTWMGEGDSNTSYFHRMADSRKAVNTI-HIIIDDNGVKIDTQLGIKEHCIEYFS-N 322
Query: 136 FMSHNVVRPSVSNLQFQAL-----SVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVN 190
+ V P + F L S + + L FS ++K A + S K+SGPDG
Sbjct: 323 LLGGEVGPPMLIQEDFDLLLPFRCSHDQKKELAMSFSRQDIKSAFFSFPSNKTSGPDGFP 382
Query: 191 FGFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVG- 249
F KE W V+ ++ VSEF + L K N T +VLI K+ N ++NDFRPIS
Sbjct: 383 VEFFKETWSVIGTEVTDAVSEFFTSSVLLKQWNATTLVLIPKITNASKMNDFRPISCNDF 442
Query: 250 ---SMNKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKLKKDLM 306
++ K+IA+LL NRL+ ++ V+S QSAF+ R + + +L+A E+V + D
Sbjct: 443 GPITLYKVIARLLTNRLQCLLSQVISPFQSAFLPGRFLAENVLLATELVQGYNRQNIDPR 502
Query: 307 -LFKVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLK 365
+ KVD K +DS+ W ++ LK + P + WI +C+ T T SV VNG+ F
Sbjct: 503 GMLKVDLRKAFDSIRWDFIISALKAIGIPDRFVYWITQCISTPTFSVCVNGNTGGFFKST 562
Query: 366 RGLRQGDPL 374
RGLRQG+PL
Sbjct: 563 RGLRQGNPL 571
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 178 bits (451), Expect = 5e-45
Identities = 107/310 (34%), Positives = 167/310 (53%), Gaps = 18/310 (5%)
Query: 77 WQQ-SRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVED---VRIEGVEPVRNAIFTHF 132
W+Q SR WL GD N +FH+ +RR N + ++ED E + + I ++F
Sbjct: 277 WKQWSRVQWLNSGDRNKGYFHATTRTRRMLNNLS--VIEDGSGQEFHEEEQIASTISSYF 334
Query: 133 AQHFMSHN-----VVRPSVSNLQFQALSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPD 187
F + N VV+ ++S + +S E LIK S+ E+KEA+ + K+ GPD
Sbjct: 335 QNIFTTSNNSDLQVVQEALSPI----ISSHCNEELIKISSLLEIKEALFSISADKAPGPD 390
Query: 188 GVNFGFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISL 247
G + F +W +++ D+ + + F + L+ +N T + LI K+ P++++D+RPI+L
Sbjct: 391 GFSASFFHAYWDIIEADVSRDIRSFFVDSCLSPRLNETHVTLIPKISAPRKVSDYRPIAL 450
Query: 248 VGSMNKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVD---EAFKLKKD 304
KI+AK+L RL+ + ++S QSAFV R I D +LI +E++ + K
Sbjct: 451 CNVQYKIVAKILTRRLQPWLSELISLHQSAFVPGRAIADNVLITHEILHFLRVSGAKKYC 510
Query: 305 LMLFKVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPL 364
M K D K YD + W +L EVL + F W +W+ +CV T + S L+NGSP
Sbjct: 511 SMAIKTDMSKAYDRIKWNFLQEVLMRLGFHDKWIRWVMQCVCTVSYSFLINGSPQGSVVP 570
Query: 365 KRGLRQGDPL 374
RGLRQGDPL
Sbjct: 571 SRGLRQGDPL 580
>At2g28980 putative non-LTR retroelement reverse transcriptase
Length = 1529
Score = 177 bits (449), Expect = 8e-45
Identities = 101/301 (33%), Positives = 156/301 (51%), Gaps = 7/301 (2%)
Query: 78 QQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRI----EGVEPVRNAIFTHFA 133
Q+S+ HW+ GD N+ +FH R+ N+I + + E ++ F F
Sbjct: 656 QKSKLHWMNVGDGNNSYFHKAAQVRKMRNSIREIRGPNAETLQTSEEIKGEAERFFNEFL 715
Query: 134 QHFMS--HNVVRPSVSNLQFQALSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNF 191
H + + NL SVT+ L + + +E+++ + + KS GPDG
Sbjct: 716 NRQSGDFHGISVEDLRNLMSYRCSVTDQNILTREVTGEEIQKVLFAMPNNKSPGPDGYTS 775
Query: 192 GFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSM 251
F K W + D + + F G L KG+N T + LI K D + D+RPIS +
Sbjct: 776 EFFKATWSLTGPDFIAAIQSFFVKGFLPKGLNATILALIPKKDEAIEMKDYRPISCCNVL 835
Query: 252 NKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKLK-KDLMLFKV 310
K+I+K+LANRL+L++ S + + QSAFVK+R +++ +L+A E+V + K K+
Sbjct: 836 YKVISKILANRLKLLLPSFILQNQSAFVKERLLMENVLLATELVKDYHKESVTPRCAMKI 895
Query: 311 DFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLRQ 370
D K +DSV W +L L+ ++FP +R WI+ C+ TAT SV VNG F RGLRQ
Sbjct: 896 DISKAFDSVQWQFLLNTLEALNFPETFRHWIKLCISTATFSVQVNGELAGFFGSSRGLRQ 955
Query: 371 G 371
G
Sbjct: 956 G 956
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 172 bits (437), Expect = 2e-43
Identities = 106/307 (34%), Positives = 170/307 (54%), Gaps = 15/307 (4%)
Query: 78 QQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRIEGV-----EPVRNAIFTHF 132
Q+SR WL GD N+ +FH++ +RR N + ++ED I GV + I +F
Sbjct: 233 QRSRVLWLHSGDRNTGYFHAVTRNRRTQNRL--TVMED--INGVAQHEEHQISQIISGYF 288
Query: 133 AQHFMSHNVVRPSVSNLQFQALSVTEGEG--LIKPFSVDEVKEAICDYDSFKSSGPDGVN 190
Q F S + SV + + + V++G+ L + + +EVK+A+ ++ K+ GPDG
Sbjct: 289 QQIFTSESDGDFSVVDEAIEPM-VSQGDNDFLTRIPNDEEVKDAVFSINASKAPGPDGFT 347
Query: 191 FGFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGS 250
GF +W ++ D+ + + F + + +N T I LI K P+++ D+RPI+L
Sbjct: 348 AGFYHSYWHIISTDVGREIRLFFTSKNFPRRMNETHIRLIPKDLGPRKVADYRPIALCNI 407
Query: 251 MNKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVD---EAFKLKKDLML 307
KI+AK++ R++L++ ++SE QSAFV R I D +LI +EV+ + K M
Sbjct: 408 FYKIVAKIMTKRMQLILPKLISENQSAFVPGRVISDNVLITHEVLHFLRTSSAKKHCSMA 467
Query: 308 FKVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRG 367
K D K YD V+W +L +VL+ F +W W+ ECV + + S L+NG+P + RG
Sbjct: 468 VKTDMSKAYDRVEWDFLKKVLQRFGFHSIWIDWVLECVTSVSYSFLINGTPQGKVVPTRG 527
Query: 368 LRQGDPL 374
LRQGDPL
Sbjct: 528 LRQGDPL 534
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 171 bits (432), Expect = 8e-43
Identities = 109/307 (35%), Positives = 161/307 (51%), Gaps = 7/307 (2%)
Query: 74 SICWQQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVR--IEGVEPVRNAIFTH 131
++ +Q+SR ++ GD N+ FFH+ RRR N I S+ +D R + VE A+ T+
Sbjct: 197 TLWFQKSREKYIELGDRNTTFFHTSTIIRRRRNRIESLKGDDDRWVTDKVELEAMAL-TY 255
Query: 132 FAQHFMSHNV--VRPSVSNLQFQALSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGV 189
+ + + +V VR + F ++S E L++ F+ EV A+ FK+ GPDG
Sbjct: 256 YKRLYSLEDVSEVRNMLPTGGFASISEAEKAALLQAFTKAEVVSAVKSMGRFKAPGPDGY 315
Query: 190 NFGFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVG 249
F ++ W + + +FV EF G L N +VLI+KV P+R+ FRP+SL
Sbjct: 316 QPVFYQQCWETVGPSVTRFVLEFFETGVLPASTNDALLVLIAKVAKPERIQQFRPVSLCN 375
Query: 250 SMNKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKLK--KDLML 307
+ KII K++ RL+ VI ++ Q++F+ R +D I++ E V + K K ML
Sbjct: 376 VLFKIITKMMVTRLKNVISKLIGPAQASFIPGRLSIDNIVLVQEAVHSMRRKKGRKGWML 435
Query: 308 FKVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRG 367
K+D EK YD V W +L E L+ W I V + SVL NG TD F RG
Sbjct: 436 LKLDLEKAYDRVRWDFLQETLEAAGLSEGWTSRIMAGVTDPSMSVLWNGERTDSFVPARG 495
Query: 368 LRQGDPL 374
LRQGDPL
Sbjct: 496 LRQGDPL 502
>At1g10160 putative reverse transcriptase
Length = 1118
Score = 169 bits (427), Expect = 3e-42
Identities = 98/307 (31%), Positives = 166/307 (53%), Gaps = 13/307 (4%)
Query: 78 QQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVED-VRIEGVEPVRNAIFTHFAQHF 136
Q+SR WL EGD N++FFH + + + N I + +D R+E V+ ++ + +++ H
Sbjct: 353 QKSRIRWLHEGDANTRFFHRAVIAHQATNLIKFLRGDDGFRVENVDQIKGMLIAYYS-HL 411
Query: 137 M---SHNVVRPSVSNLQ----FQALSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGV 189
+ S NV SV ++ F+ S + P S +E+ + + K+ GPDG
Sbjct: 412 LGIPSENVTPFSVEKIKGLLPFRCDSFLASQLTTIP-SEEEITQVLFSMPRNKAPGPDGF 470
Query: 190 NFGFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVG 249
F E W ++K ++ + EF +G L +G N T I LI KV RL FRP++
Sbjct: 471 PVEFFIEAWAIVKSSVVAAIREFFISGNLPRGFNATAITLIPKVTGADRLTQFRPVACCT 530
Query: 250 SMNKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKLKKDLM--L 307
++ K+I ++++ RL+L I V Q F+K R + + +L+A+E+VD F+ +
Sbjct: 531 TIYKVITRIISRRLKLFIDQAVQANQVGFIKGRLLCENVLLASELVDN-FEADGETTRGC 589
Query: 308 FKVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRG 367
+VD K YD+V+W +L +LK + P+++ WI C+ +A+ S+ NG F K+G
Sbjct: 590 LQVDISKAYDNVNWEFLINILKALDLPLVFIHWIWVCISSASYSIAFNGELIGFFQGKKG 649
Query: 368 LRQGDPL 374
+RQGDP+
Sbjct: 650 IRQGDPM 656
>At3g31420 hypothetical protein
Length = 1491
Score = 167 bits (423), Expect = 9e-42
Identities = 108/309 (34%), Positives = 161/309 (51%), Gaps = 10/309 (3%)
Query: 75 ICWQ-QSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRI-EGVEPVRNAIFTHF 132
I WQ +SR+ WL GD N+++FHS +RR N + SV D I G E + +F
Sbjct: 517 IYWQTKSRSRWLNAGDRNTRYFHSTTKTRRCRNRLLSVQDSDGDICRGDENIAKVAINYF 576
Query: 133 AQHFMS--HNVVRPSVSNLQFQALSVTE-GEGLIKPFSVDEVKEAICDYDSFKSSGPDGV 189
+ S + +R + FQ E E LI+P + E++E++ ++ PDG
Sbjct: 577 DDLYKSTPNTSLRYADVFQGFQQKITDEINEDLIRPVTELEIEESVFSVAPSRTPDPDGF 636
Query: 190 NFGFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVG 249
F ++FW +K ++ V+ F +L + N T + LI KV+ P + FRPI+L
Sbjct: 637 TADFYQQFWPDIKQKVIDEVTRFFERSELDERHNHTNLCLIPKVETPTTIAKFRPIALCN 696
Query: 250 SMNKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKLKK----DL 305
KII+K+L NRL+ +G ++E Q+AFV R I + +IA+EV A K +K
Sbjct: 697 VSYKIISKILVNRLKKHLGGAITENQAAFVPGRLITNNAIIAHEVY-YALKARKRQANSY 755
Query: 306 MLFKVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLK 365
M K D K YD ++W +L E ++ M F W + I CV SVL+NGSP +
Sbjct: 756 MALKTDITKAYDRLEWDFLEETMRQMGFNTKWIERIMICVTMVRFSVLINGSPHGTIKPE 815
Query: 366 RGLRQGDPL 374
RG+R GDPL
Sbjct: 816 RGIRHGDPL 824
>At2g11240 pseudogene
Length = 1044
Score = 166 bits (420), Expect = 2e-41
Identities = 105/306 (34%), Positives = 159/306 (51%), Gaps = 21/306 (6%)
Query: 78 QQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRIEGVEP-VRNAIFTHFAQHF 136
Q+SR WL GD NS +FH++ R N + ED E E + N I +F + F
Sbjct: 172 QRSRQLWLALGDKNSGYFHAITRGRTVINKFSVIEKEDGVPEYEEAGILNVISEYFQKLF 231
Query: 137 MSHNVVRPSVSNLQFQALSVTEGEGLIKPF-----SVDEVKEAICDYDSFKSSGPDGVNF 191
++ R A ++ E IKPF + +E+K A + K+ GPDG +
Sbjct: 232 SANEGAR---------AATIKEA---IKPFISPEQNPEEIKSACFSIHADKAPGPDGFSA 279
Query: 192 GFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSM 251
F + W+ + +I+ + F + L IN T I LI K+ + +R+ D+RPI+L
Sbjct: 280 SFFQSNWMTVGPNIVLEIQSFFSSSTLQPTINKTHITLIPKIQSLKRMVDYRPIALCTVF 339
Query: 252 NKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKL---KKDLMLF 308
KII+KLL+ RL+ ++ ++SE QSAFV KR D +LI +E + L K+ M
Sbjct: 340 YKIISKLLSRRLQPILQEIISENQSAFVPKRASNDNVLITHEALHYLKSLGAEKRCFMAV 399
Query: 309 KVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGL 368
K + K YD ++W ++ V++ M F W WI +C+ T + S L+NGS +RGL
Sbjct: 400 KTNMSKAYDRIEWDFIKLVMQEMGFHQTWISWILQCITTVSYSFLLNGSAQGAVTPERGL 459
Query: 369 RQGDPL 374
RQGDPL
Sbjct: 460 RQGDPL 465
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 162 bits (411), Expect = 2e-40
Identities = 97/306 (31%), Positives = 165/306 (53%), Gaps = 15/306 (4%)
Query: 79 QSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRIEGVEPVRNAIFTHFAQHFMS 138
+SRN W+ GD N+ FF++ R+ N I ++ +G+E R+ A+++ +
Sbjct: 701 KSRNRWMLLGDRNTMFFYASTKLRKSRNRIKAI----TDAQGIENFRDDTIGKVAENYFA 756
Query: 139 HNVVRPSVSN----LQFQALSVTE--GEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFG 192
S+ + A VTE L++ + EV++A+ + ++ G DG
Sbjct: 757 DLFTTTQTSDWEEIISGIAPKVTEQMNHELLQSVTDQEVRDAVFAIGADRAPGFDGFTAA 816
Query: 193 FIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSMN 252
F W ++ +D+ V F + + IN T I LI K+ +P+ ++D+RPISL +
Sbjct: 817 FYHHLWDLIGNDVCLMVRHFFESDVMDNQINQTQICLIPKIIDPKHMSDYRPISLCTASY 876
Query: 253 KIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKLKKD----LMLF 308
KII+K+L RL+ +G V+S++Q+AFV + I D +L+A+E++ + K +++ +
Sbjct: 877 KIISKILIKRLKQCLGDVISDSQAAFVPGQNISDNVLVAHELL-HSLKSRRECQSGYVAV 935
Query: 309 KVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGL 368
K D K YD V+W +L +V+ + F W KWI CV + + VL+NGSP + RG+
Sbjct: 936 KTDISKAYDRVEWNFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVLINGSPYGKIFPSRGI 995
Query: 369 RQGDPL 374
RQGDPL
Sbjct: 996 RQGDPL 1001
>At1g25430 hypothetical protein
Length = 1213
Score = 160 bits (405), Expect = 1e-39
Identities = 96/303 (31%), Positives = 161/303 (52%), Gaps = 6/303 (1%)
Query: 78 QQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRI----EGVEPVRNAIFTHFA 133
Q+SR W EGD N+K+FH + +R N+I ++ + ++ EG+ + + F
Sbjct: 354 QKSRISWFAEGDGNTKYFHRMADARNSSNSISALYDGNGKLVDSQEGILDLCASYFGSLL 413
Query: 134 QHFMSHNVVRPSVSNLQFQ-ALSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFG 192
+ ++ + NL S + L FS ++++ A+ KS GPDG
Sbjct: 414 GDEVDPYLMEQNDMNLLLSYRCSPAQVCELESTFSNEDIRAALFSLPRNKSCGPDGFTAE 473
Query: 193 FIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSMN 252
F + W ++ ++ + EF +G L K N T IVLI K+ NP +DFRPIS + ++
Sbjct: 474 FFIDSWSIVGAEVTDAIKEFFSSGCLLKQWNATTIVLIPKIVNPTCTSDFRPISCLNTLY 533
Query: 253 KIIAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVV-DEAFKLKKDLMLFKVD 311
K+IA+LL +RL+ ++ V+S QSAF+ R + + +L+A ++V + + KVD
Sbjct: 534 KVIARLLTDRLQRLLSGVISSAQSAFLPGRSLAENVLLATDLVHGYNWSNISPRGMLKVD 593
Query: 312 FEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLRQG 371
+K +DSV W ++ L+ ++ P + WI +C+ T T +V +NG F +GLRQG
Sbjct: 594 LKKAFDSVRWEFVIAALRALAIPEKFINWISQCISTPTFTVSINGGNGGFFKSTKGLRQG 653
Query: 372 DPL 374
DPL
Sbjct: 654 DPL 656
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.333 0.145 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,861,372
Number of Sequences: 26719
Number of extensions: 320073
Number of successful extensions: 1328
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 1174
Number of HSP's gapped (non-prelim): 83
length of query: 374
length of database: 11,318,596
effective HSP length: 101
effective length of query: 273
effective length of database: 8,619,977
effective search space: 2353253721
effective search space used: 2353253721
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137546.8


