

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137546.2 - phase: 0
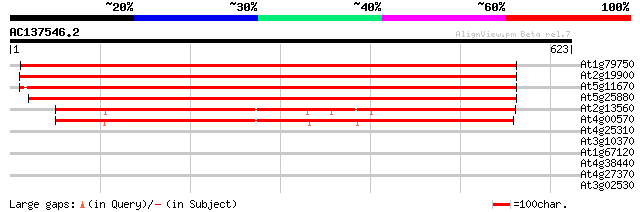
(623 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g79750 malate oxidoreductase like protein 915 0.0
At2g19900 malate oxidoreductase (malic enzyme) 906 0.0
At5g11670 NADP dependent malic enzyme - like protein 899 0.0
At5g25880 malate dehydrogenase - like protein 896 0.0
At2g13560 malate oxidoreductase (malic enzyme) 424 e-119
At4g00570 putative malate oxidoreductase 415 e-116
At4g25310 SRG1-like protein 29 6.9
At3g10370 putative glycerol-3-phosphate dehydrogenase 29 6.9
At1g67120 hypothetical protein 29 6.9
At4g38440 unknown protein 29 9.0
At4g27370 myosin heavy chain - like protein 29 9.0
At3g02530 putative chaperonin 29 9.0
>At1g79750 malate oxidoreductase like protein
Length = 646
Score = 915 bits (2364), Expect = 0.0
Identities = 438/550 (79%), Positives = 507/550 (91%)
Query: 13 SVLDLSPRSAVGGGVEDVYGEDCATEDQLVTPWTFSVASGYSLLRDPQYNKGLAFTEKER 72
S D+ P+S V GGV+DVYGED ATED +TPW+ SVASGY+LLRDP +NKGLAF+ +ER
Sbjct: 68 SAADIVPKSTVSGGVQDVYGEDAATEDMPITPWSLSVASGYTLLRDPHHNKGLAFSHRER 127
Query: 73 DAHYLRGLLPPTVSSQQLQEKKLMHNIRQYEVPLQKYVAMMDLQERNERLFYKLLIDNVE 132
DAHYLRGLLPPTV SQ LQ KK+MH +RQY+VPLQKY+AMMDLQE NERLFYKLLID+VE
Sbjct: 128 DAHYLRGLLPPTVISQDLQVKKIMHTLRQYQVPLQKYMAMMDLQETNERLFYKLLIDHVE 187
Query: 133 ELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPERSIQVIVVTDGER 192
ELLP++YTP VGEACQKYGSI+ RPQGL+ISLKEKGKI EVL+NWPE++IQVIVVTDGER
Sbjct: 188 ELLPVIYTPTVGEACQKYGSIFLRPQGLFISLKEKGKIHEVLRNWPEKNIQVIVVTDGER 247
Query: 193 ILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQKR 252
ILGLGDLGCQGMGIPVGKL+LY+ALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQ+R
Sbjct: 248 ILGLGDLGCQGMGIPVGKLSLYTALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQRR 307
Query: 253 ATGKEYYDLLHEFMTAVKQNYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGTAA 312
ATG+EY +L+HEFMTAVKQNYGEKV++QFEDFANHNAF+LLAKYGTTHLVFNDDIQGTA+
Sbjct: 308 ATGEEYSELMHEFMTAVKQNYGEKVVIQFEDFANHNAFDLLAKYGTTHLVFNDDIQGTAS 367
Query: 313 VVLAGVVASLKLIGGTLPEHTFLFLGAGEAGTGIAELIALEMSKQTKAPIEESRKKIWLV 372
VVLAG++A+L+ +GG+L +H FLFLGAGEAGTGIAELIALE+SK++ P+EE+RK IWLV
Sbjct: 368 VVLAGLIAALRFVGGSLSDHRFLFLGAGEAGTGIAELIALEISKKSHIPLEEARKNIWLV 427
Query: 373 DSKGLIVSSRANSLQHFKKPWAHEHEPVSTLLDAVKIIKPTVLIGSSGVGKTFTKEVVEA 432
DSKGLIVSSR S+QHFKKPWAH+HEP+ L+DAVK IKPTVLIG+SGVG+TFT++VVE
Sbjct: 428 DSKGLIVSSRKESIQHFKKPWAHDHEPIRELVDAVKAIKPTVLIGTSGVGQTFTQDVVET 487
Query: 433 MTEINKIPLILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEYKGKTYYSGQSNN 492
M ++N+ P+IL+LSNPTSQSECTAEEAYTWS+GRAIFASGSPF PVEY+GKT+ GQ+NN
Sbjct: 488 MAKLNEKPIILSLSNPTSQSECTAEEAYTWSQGRAIFASGSPFAPVEYEGKTFVPGQANN 547
Query: 493 AYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISANIA 552
AYIFPGFGLG++MSG IRVHDDMLLAASEALA+++ EE+Y+KG+ YPPF +IRKISA IA
Sbjct: 548 AYIFPGFGLGLIMSGTIRVHDDMLLAASEALAEELMEEHYEKGMIYPPFRNIRKISARIA 607
Query: 553 ANVAAKAYEL 562
A VAAKAYEL
Sbjct: 608 AKVAAKAYEL 617
>At2g19900 malate oxidoreductase (malic enzyme)
Length = 581
Score = 906 bits (2341), Expect = 0.0
Identities = 448/551 (81%), Positives = 502/551 (90%)
Query: 12 ESVLDLSPRSAVGGGVEDVYGEDCATEDQLVTPWTFSVASGYSLLRDPQYNKGLAFTEKE 71
E V + +S+V GGV DVYGED AT + +TPW+ SV+SGYSLLRDP+YNKGLAFTEKE
Sbjct: 2 EKVTNSDLKSSVDGGVVDVYGEDSATIEHNITPWSLSVSSGYSLLRDPRYNKGLAFTEKE 61
Query: 72 RDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYEVPLQKYVAMMDLQERNERLFYKLLIDNV 131
RD HYLRGLLPP V Q+LQEK+L++NIRQY+ PLQKY+A+ +LQERNERLFYKLLIDNV
Sbjct: 62 RDTHYLRGLLPPVVLDQKLQEKRLLNNIRQYQFPLQKYMALTELQERNERLFYKLLIDNV 121
Query: 132 EELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPERSIQVIVVTDGE 191
EELLPIVYTP VGEACQK+GSI++RPQGL+ISLK+KGKIL+VLKNWPER+IQVIVVTDGE
Sbjct: 122 EELLPIVYTPTVGEACQKFGSIFRRPQGLFISLKDKGKILDVLKNWPERNIQVIVVTDGE 181
Query: 192 RILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQK 251
RILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQK
Sbjct: 182 RILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQK 241
Query: 252 RATGKEYYDLLHEFMTAVKQNYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGTA 311
RATG+EY +LL+EFM+AVKQNYGEKVL+QFEDFANHNAFELLAKY THLVFNDDIQGTA
Sbjct: 242 RATGQEYSELLNEFMSAVKQNYGEKVLIQFEDFANHNAFELLAKYSDTHLVFNDDIQGTA 301
Query: 312 AVVLAGVVASLKLIGGTLPEHTFLFLGAGEAGTGIAELIALEMSKQTKAPIEESRKKIWL 371
+VVLAG+V++ KL L EHTFLFLGAGEAGTGIAELIAL MSKQ A +EESRKKIWL
Sbjct: 302 SVVLAGLVSAQKLTNSPLAEHTFLFLGAGEAGTGIAELIALYMSKQMNASVEESRKKIWL 361
Query: 372 VDSKGLIVSSRANSLQHFKKPWAHEHEPVSTLLDAVKIIKPTVLIGSSGVGKTFTKEVVE 431
VDSKGLIV+SR +SLQ FKKPWAHEHEPV LL A+K IKPTVLIGSSGVG++FTKEV+E
Sbjct: 362 VDSKGLIVNSRKDSLQDFKKPWAHEHEPVKDLLGAIKAIKPTVLIGSSGVGRSFTKEVIE 421
Query: 432 AMTEINKIPLILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEYKGKTYYSGQSN 491
AM+ IN+ PLI+ALSNPT+QSECTAEEAYTWS+GRAIFASGSPFDPVEY+GK + S Q+N
Sbjct: 422 AMSSINERPLIMALSNPTTQSECTAEEAYTWSKGRAIFASGSPFDPVEYEGKVFVSTQAN 481
Query: 492 NAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISANI 551
NAYIFPGFGLG+V+SGAIRVHDDMLLAA+EALA QV++ENY+KG+ YP FS IRKISA I
Sbjct: 482 NAYIFPGFGLGLVISGAIRVHDDMLLAAAEALAGQVSKENYEKGMIYPSFSSIRKISAQI 541
Query: 552 AANVAAKAYEL 562
AANVA KAYEL
Sbjct: 542 AANVATKAYEL 552
>At5g11670 NADP dependent malic enzyme - like protein
Length = 588
Score = 899 bits (2324), Expect = 0.0
Identities = 440/552 (79%), Positives = 497/552 (89%), Gaps = 2/552 (0%)
Query: 11 GESVLDLSPRSAVGGGVEDVYGEDCATEDQLVTPWTFSVASGYSLLRDPQYNKGLAFTEK 70
GE V D RS VGGG+ DVYGED AT DQLVTPW SVASGY+L+RDP+YNKGLAFT+K
Sbjct: 10 GEDVAD--NRSGVGGGISDVYGEDSATLDQLVTPWVTSVASGYTLMRDPRYNKGLAFTDK 67
Query: 71 ERDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYEVPLQKYVAMMDLQERNERLFYKLLIDN 130
ERDAHYL GLLPP + SQ +QE+K+MHN+RQY VPLQ+Y+A+MDLQERNERLFYKLLIDN
Sbjct: 68 ERDAHYLTGLLPPVILSQDVQERKVMHNLRQYTVPLQRYMALMDLQERNERLFYKLLIDN 127
Query: 131 VEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPERSIQVIVVTDG 190
VEELLP+VYTP VGEACQKYGSI+++PQGLYISL EKGKILEVLKNWP+R IQVIVVTDG
Sbjct: 128 VEELLPVVYTPTVGEACQKYGSIFRKPQGLYISLNEKGKILEVLKNWPQRGIQVIVVTDG 187
Query: 191 ERILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQ 250
ERILGLGDLGCQGMGIPVGKL+LY+ALGG+RPSACLP+TIDVGTNNEKLLNDEFYIGL+Q
Sbjct: 188 ERILGLGDLGCQGMGIPVGKLSLYTALGGIRPSACLPITIDVGTNNEKLLNDEFYIGLKQ 247
Query: 251 KRATGKEYYDLLHEFMTAVKQNYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGT 310
+RATG+EY + LHEFM AVKQNYGEKVLVQFEDFANHNAF+LL+KY +HLVFNDDIQGT
Sbjct: 248 RRATGQEYAEFLHEFMCAVKQNYGEKVLVQFEDFANHNAFDLLSKYSDSHLVFNDDIQGT 307
Query: 311 AAVVLAGVVASLKLIGGTLPEHTFLFLGAGEAGTGIAELIALEMSKQTKAPIEESRKKIW 370
A+VVLAG++A+ K++G L +HTFLFLGAGEAGTGIAELIAL++SK+T API E+RKKIW
Sbjct: 308 ASVVLAGLIAAQKVLGKKLADHTFLFLGAGEAGTGIAELIALKISKETGAPITETRKKIW 367
Query: 371 LVDSKGLIVSSRANSLQHFKKPWAHEHEPVSTLLDAVKIIKPTVLIGSSGVGKTFTKEVV 430
LVDSKGLIVSSR SLQHFK+PWAHEH+PV L+ AV IKPTVLIG+SGVG+TFTKEVV
Sbjct: 368 LVDSKGLIVSSRKESLQHFKQPWAHEHKPVKDLIGAVNAIKPTVLIGTSGVGQTFTKEVV 427
Query: 431 EAMTEINKIPLILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEYKGKTYYSGQS 490
EAM N+ PLILALSNPTSQ+ECTAE+AYTW++GRAIF SGSPFDPV Y GKTY GQ+
Sbjct: 428 EAMATNNEKPLILALSNPTSQAECTAEQAYTWTKGRAIFGSGSPFDPVVYDGKTYLPGQA 487
Query: 491 NNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISAN 550
NN YIFPG GLG++MSGAIRV DDMLLAASEALA QVTEE+Y GL YPPFS+IR+ISAN
Sbjct: 488 NNCYIFPGLGLGLIMSGAIRVRDDMLLAASEALAAQVTEEHYANGLIYPPFSNIREISAN 547
Query: 551 IAANVAAKAYEL 562
IAA VAAK Y+L
Sbjct: 548 IAACVAAKTYDL 559
>At5g25880 malate dehydrogenase - like protein
Length = 588
Score = 896 bits (2316), Expect = 0.0
Identities = 435/542 (80%), Positives = 494/542 (90%)
Query: 21 SAVGGGVEDVYGEDCATEDQLVTPWTFSVASGYSLLRDPQYNKGLAFTEKERDAHYLRGL 80
S VGGG+ DVYGED AT DQLVTPW SVASGY+L+RDP+YNKGLAFT+KERDAHY+ GL
Sbjct: 18 SGVGGGISDVYGEDSATLDQLVTPWVTSVASGYTLMRDPRYNKGLAFTDKERDAHYITGL 77
Query: 81 LPPTVSSQQLQEKKLMHNIRQYEVPLQKYVAMMDLQERNERLFYKLLIDNVEELLPIVYT 140
LPP V SQ +QE+K+MHN+RQY VPLQ+Y+A+MDLQERNERLFYKLLIDNVEELLP+VYT
Sbjct: 78 LPPVVLSQDVQERKVMHNLRQYTVPLQRYMALMDLQERNERLFYKLLIDNVEELLPVVYT 137
Query: 141 PVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPERSIQVIVVTDGERILGLGDLG 200
P VGEACQKYGSIY+RPQGLYISLKEKGKILEVLKNWP+R IQVIVVTDGERILGLGDLG
Sbjct: 138 PTVGEACQKYGSIYRRPQGLYISLKEKGKILEVLKNWPQRGIQVIVVTDGERILGLGDLG 197
Query: 201 CQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQKRATGKEYYD 260
CQGMGIPVGKL+LY+ALGG+RPSACLP+TIDVGTNNEKLLN+EFYIGL+QKRA G+EY +
Sbjct: 198 CQGMGIPVGKLSLYTALGGIRPSACLPITIDVGTNNEKLLNNEFYIGLKQKRANGEEYAE 257
Query: 261 LLHEFMTAVKQNYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGTAAVVLAGVVA 320
L EFM AVKQNYGEKVLVQFEDFANH+AFELL+KY ++HLVFNDDIQGTA+VVLAG++A
Sbjct: 258 FLQEFMCAVKQNYGEKVLVQFEDFANHHAFELLSKYCSSHLVFNDDIQGTASVVLAGLIA 317
Query: 321 SLKLIGGTLPEHTFLFLGAGEAGTGIAELIALEMSKQTKAPIEESRKKIWLVDSKGLIVS 380
+ K++G +L +HTFLFLGAGEAGTGIAELIAL++SK+T PI+E+RKKIWLVDSKGLIVS
Sbjct: 318 AQKVLGKSLADHTFLFLGAGEAGTGIAELIALKISKETGKPIDETRKKIWLVDSKGLIVS 377
Query: 381 SRANSLQHFKKPWAHEHEPVSTLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMTEINKIP 440
R SLQHFK+PWAH+H+PV LL AV IKPTVLIG+SGVGKTFTKEVVEAM +N+ P
Sbjct: 378 ERKESLQHFKQPWAHDHKPVKELLAAVNAIKPTVLIGTSGVGKTFTKEVVEAMATLNEKP 437
Query: 441 LILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEYKGKTYYSGQSNNAYIFPGFG 500
LILALSNPTSQ+ECTAEEAYTW++GRAIFASGSPFDPV+Y GK + GQ+NN YIFPG G
Sbjct: 438 LILALSNPTSQAECTAEEAYTWTKGRAIFASGSPFDPVQYDGKKFTPGQANNCYIFPGLG 497
Query: 501 LGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKAY 560
LG++MSGAIRV DDMLLAASEALA QVTEEN+ GL YPPF++IRKISANIAA+V AK Y
Sbjct: 498 LGLIMSGAIRVRDDMLLAASEALASQVTEENFANGLIYPPFANIRKISANIAASVGAKTY 557
Query: 561 EL 562
EL
Sbjct: 558 EL 559
>At2g13560 malate oxidoreductase (malic enzyme)
Length = 623
Score = 424 bits (1091), Expect = e-119
Identities = 233/536 (43%), Positives = 333/536 (61%), Gaps = 28/536 (5%)
Query: 52 GYSLLRDPQYNKGLAFTEKERDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYEVP------ 105
G +L DP +NKG AFT ER+ LRGLLPP V + Q + M ++++ E
Sbjct: 46 GLDILHDPWFNKGTAFTMTERNRLDLRGLLPPNVMDSEQQIFRFMTDLKRLEEQARDGPS 105
Query: 106 ----LQKYVAMMDLQERNERLFYKLLIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLY 161
L K+ + L +RNE ++YK+LI+N+EE PIVYTP VG CQ Y +++RP+G+Y
Sbjct: 106 DPNALAKWRILNRLHDRNETMYYKVLINNIEEYAPIVYTPTVGLVCQNYSGLFRRPRGMY 165
Query: 162 ISLKEKGKILEVLKNWPERSIQVIVVTDGERILGLGDLGCQGMGIPVGKLALYSALGGVR 221
S +++G+++ ++ NWP + +IVVTDG RILGLGDLG G+GI VGKL LY A G+
Sbjct: 166 FSAEDRGEMMSMVYNWPAEQVDMIVVTDGSRILGLGDLGVHGIGIAVGKLDLYVAAAGIN 225
Query: 222 PSACLPVTIDVGTNNEKLLNDEFYIGLRQKRATGKEYYDLLHEFMTAVKQNYGEKVLVQF 281
P LPV IDVGTNNEKL ND Y+GL+Q+R +Y D++ EFM AV + V+VQF
Sbjct: 226 PQRVLPVMIDVGTNNEKLRNDPMYLGLQQRRLEDDDYIDVIDEFMEAVYTRW-PHVIVQF 284
Query: 282 EDFANHNAFELLAKYGTTHLVFNDDIQGTAAVVLAGVVASLKLIGGTL---PEHTFLFLG 338
EDF + AF+LL +Y T+ +FNDD+QGTA V +AG++ +++ G + P+ + G
Sbjct: 285 EDFQSKWAFKLLQRYRCTYRMFNDDVQGTAGVAIAGLLGAVRAQGRPMIDFPKMKIVVAG 344
Query: 339 AGEAGTGIAELIALEMSK---QTKAPIEESRKKIWLVDSKGLIVSSRANSLQHFKKPWAH 395
AG AG G+ M++ T+ + ++ + W+VD++GLI R N + +P+A
Sbjct: 345 AGSAGIGVLNAARKTMARMLGNTETAFDSAQSQFWVVDAQGLITEGREN-IDPEAQPFAR 403
Query: 396 EHEPV--------STLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMT-EINKIPLILALS 446
+ + + +TL++ V+ +KP VL+G S VG F+KEV+EAM + P I A+S
Sbjct: 404 KTKEMERQGLKEGATLVEVVREVKPDVLLGLSAVGGLFSKEVLEAMKGSTSTRPAIFAMS 463
Query: 447 NPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEY-KGKTYYSGQSNNAYIFPGFGLGIVM 505
NPT +ECT ++A++ IFASGSPF VE+ G + Q NN Y+FPG GLG ++
Sbjct: 464 NPTKNAECTPQDAFSILGENMIFASGSPFKNVEFGNGHVGHCNQGNNMYLFPGIGLGTLL 523
Query: 506 SGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKAYE 561
SGA V D ML AASE LA ++EE +G+ YPP S IR I+ IAA V +A E
Sbjct: 524 SGAPIVSDGMLQAASECLAAYMSEEEVLEGIIYPPISRIRDITKRIAAAVIKEAIE 579
>At4g00570 putative malate oxidoreductase
Length = 607
Score = 415 bits (1067), Expect = e-116
Identities = 223/526 (42%), Positives = 323/526 (61%), Gaps = 20/526 (3%)
Query: 52 GYSLLRDPQYNKGLAFTEKERDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYE-------- 103
G +L DP +NK F ERD +RGLLPP V + Q + + + R E
Sbjct: 39 GADILHDPWFNKDTGFPLTERDRLGIRGLLPPRVMTCVQQCDRFIESFRSLENNTKGEPE 98
Query: 104 --VPLQKYVAMMDLQERNERLFYKLLIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLY 161
V L K+ + L +RNE L+Y++LIDN+++ PI+YTP VG CQ Y +Y+RP+G+Y
Sbjct: 99 NVVALAKWRMLNRLHDRNETLYYRVLIDNIKDFAPIIYTPTVGLVCQNYSGLYRRPRGMY 158
Query: 162 ISLKEKGKILEVLKNWPERSIQVIVVTDGERILGLGDLGCQGMGIPVGKLALYSALGGVR 221
S K+KG+++ ++ NWP + +IV+TDG RILGLGDLG QG+GIP+GKL +Y A G+
Sbjct: 159 FSAKDKGEMMSMIYNWPAPQVDMIVITDGSRILGLGDLGVQGIGIPIGKLDMYVAAAGIN 218
Query: 222 PSACLPVTIDVGTNNEKLLNDEFYIGLRQKRATGKEYYDLLHEFMTAVKQNYGEKVLVQF 281
P LP+ +DVGTNNEKLL ++ Y+G+RQ R G+EY +++ EFM A + K +VQF
Sbjct: 219 PQRVLPIMLDVGTNNEKLLQNDLYLGVRQPRLEGEEYLEIIDEFMEAAFTRW-PKAVVQF 277
Query: 282 EDFANHNAFELLAKYGTTHLVFNDDIQGTAAVVLAGVVASLKLIGGTLPE---HTFLFLG 338
EDF AF L +Y +FNDD+QGTA V LAG++ +++ G + + + +G
Sbjct: 278 EDFQAKWAFGTLERYRKKFCMFNDDVQGTAGVALAGLLGTVRAQGRPISDFVNQKIVVVG 337
Query: 339 AGEAGTGIAELIALEMSKQTKAPIEESRKKIWLVDSKGLIVSSRAN---SLQHFKKPWAH 395
AG AG G+ ++ +++ E+ K +L+D GL+ + R F K A
Sbjct: 338 AGSAGLGVTKMAVQAVARMAGISESEATKNFYLIDKDGLVTTERTKLDPGAVLFAKNPAE 397
Query: 396 EHEPVSTLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMTEINKI-PLILALSNPTSQSEC 454
E S +++ VK ++P VL+G SGVG F +EV++AM E + P I A+SNPT +EC
Sbjct: 398 IREGAS-IVEVVKKVRPHVLLGLSGVGGIFNEEVLKAMRESDSCKPAIFAMSNPTLNAEC 456
Query: 455 TAEEAYTWSEGRAIFASGSPFDPVEYK-GKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHD 513
TA +A+ + G +FASGSPF+ VE + GK + Q+NN Y+FPG GLG ++SGA V D
Sbjct: 457 TAADAFKHAGGNIVFASGSPFENVELENGKVGHVNQANNMYLFPGIGLGTLLSGARIVTD 516
Query: 514 DMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKA 559
ML AASE LA +T+E +KG+ YP ++IR I+A + A V A
Sbjct: 517 GMLQAASECLASYMTDEEVQKGILYPSINNIRHITAEVGAAVLRAA 562
>At4g25310 SRG1-like protein
Length = 353
Score = 29.3 bits (64), Expect = 6.9
Identities = 28/115 (24%), Positives = 48/115 (41%), Gaps = 17/115 (14%)
Query: 149 KYGSIYKRPQGLYISLKEKGKILEVLKNWPERSIQVIVVTDGERI-LGLGDLGCQGMGIP 207
K+ S+ P L +++ G ILE++ N RSI+ V + E+ L + G G
Sbjct: 253 KWVSVKPLPNALVVNV---GDILEIITNGTYRSIEHRGVVNSEKERLSVASFHNTGFGKE 309
Query: 208 VGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQKRATGKEYYDLL 262
+G + V G + L +E++ GL + GK Y D++
Sbjct: 310 IGPMRSL-------------VERHKGALFKTLTTEEYFHGLFSRELDGKAYLDVM 351
>At3g10370 putative glycerol-3-phosphate dehydrogenase
Length = 629
Score = 29.3 bits (64), Expect = 6.9
Identities = 16/69 (23%), Positives = 32/69 (46%), Gaps = 1/69 (1%)
Query: 485 YYSGQSNNAYIFPGFGLGIVMSG-AIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSD 543
YY GQ N++ + G ++G A+ H +++ ++ K++ + LT F+
Sbjct: 226 YYDGQMNDSRLNVGLACTAALAGAAVLNHAEVVSLITDDATKRIIGARIRNNLTGQEFNS 285
Query: 544 IRKISANIA 552
K+ N A
Sbjct: 286 YAKVVVNAA 294
>At1g67120 hypothetical protein
Length = 5138
Score = 29.3 bits (64), Expect = 6.9
Identities = 14/28 (50%), Positives = 19/28 (67%)
Query: 397 HEPVSTLLDAVKIIKPTVLIGSSGVGKT 424
H V +L A+++ KP +L GS GVGKT
Sbjct: 1576 HRNVLRVLRAMQLSKPILLEGSPGVGKT 1603
>At4g38440 unknown protein
Length = 1465
Score = 28.9 bits (63), Expect = 9.0
Identities = 20/71 (28%), Positives = 32/71 (44%), Gaps = 3/71 (4%)
Query: 11 GESVLDLSPRSAVGGGVEDVYGEDCATEDQLVTPWTFSVASGYSLLRDPQYNKGL---AF 67
GE+ L S G + D ++ TE VTP ++ S+++ P +G A+
Sbjct: 250 GEAKLKKRKHSVQGVSITDETAKNSRTEGHFVTPKVMAIPKEKSVVQKPGIAQGFVWDAW 309
Query: 68 TEKERDAHYLR 78
TE+ A LR
Sbjct: 310 TERVEAARDLR 320
>At4g27370 myosin heavy chain - like protein
Length = 1126
Score = 28.9 bits (63), Expect = 9.0
Identities = 15/47 (31%), Positives = 24/47 (50%)
Query: 64 GLAFTEKERDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYEVPLQKYV 110
G+ +ER + LRG+L + Q ++ HN+R V LQ Y+
Sbjct: 812 GVISVLEERKKYVLRGILGLQKQFRGYQTREYFHNMRNAAVILQSYI 858
>At3g02530 putative chaperonin
Length = 535
Score = 28.9 bits (63), Expect = 9.0
Identities = 38/148 (25%), Positives = 57/148 (37%), Gaps = 20/148 (13%)
Query: 293 LAKYGTTHLVFNDDIQGTAAVVLAGVVASLKLIGGTLPEHTFLFLGAGEAGTGIAELIAL 352
L K G T L+ IQ A+++A + I G T +F+G
Sbjct: 55 LTKDGNT-LLKEMQIQNPTAIMIARTAVAQDDISGDGTTSTVIFIG-------------- 99
Query: 353 EMSKQTKAPIEESRKKIWLVDSKGLIVSSRANSLQHFKKPWAHEHEPVSTLLDAVKIIKP 412
E+ KQ++ I+E LVD + + L FK P EP +L K++
Sbjct: 100 ELMKQSERCIDEGMHPRVLVDGFEIAKRATLQFLDTFKTPVVMGDEPDKEIL---KMVAR 156
Query: 413 TVLIGS--SGVGKTFTKEVVEAMTEINK 438
T L G+ T VV ++ I K
Sbjct: 157 TTLRTKLYEGLADQLTDIVVNSVLCIRK 184
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,682,758
Number of Sequences: 26719
Number of extensions: 596945
Number of successful extensions: 1485
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1463
Number of HSP's gapped (non-prelim): 13
length of query: 623
length of database: 11,318,596
effective HSP length: 105
effective length of query: 518
effective length of database: 8,513,101
effective search space: 4409786318
effective search space used: 4409786318
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC137546.2


