

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137510.5 - phase: 0
(1519 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
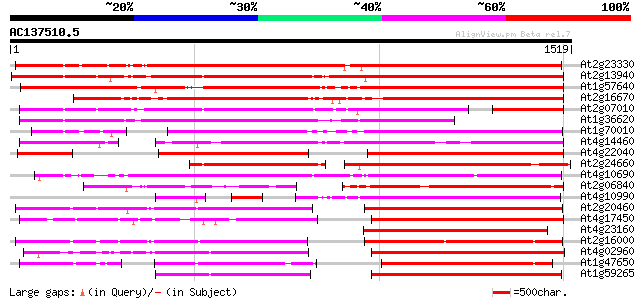
Score E
Sequences producing significant alignments: (bits) Value
At2g23330 putative retroelement pol polyprotein 1395 0.0
At2g13940 putative retroelement pol polyprotein 1379 0.0
At1g57640 1373 0.0
At2g16670 putative retroelement pol polyprotein 1236 0.0
At2g07010 putative retroelement pol polyprotein 1033 0.0
At1g36620 hypothetical protein 902 0.0
At1g70010 hypothetical protein 810 0.0
At4g14460 retrovirus-related like polyprotein 733 0.0
At4g22040 LTR retrotransposon like protein 723 0.0
At2g24660 putative retroelement pol polyprotein 668 0.0
At4g10690 retrotransposon like protein 648 0.0
At2g06840 putative retroelement pol polyprotein 621 e-177
At4g10990 putative retrotransposon polyprotein 540 e-153
At2g20460 putative retroelement pol polyprotein 499 e-141
At4g17450 retrotransposon like protein 499 e-141
At4g23160 putative protein 489 e-138
At2g16000 putative retroelement pol polyprotein 483 e-136
At4g02960 putative polyprotein of LTR transposon 457 e-128
At1g47650 hypothetical protein 447 e-125
At1g59265 polyprotein, putative 446 e-125
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 1395 bits (3611), Expect = 0.0
Identities = 731/1517 (48%), Positives = 988/1517 (64%), Gaps = 72/1517 (4%)
Query: 15 NSSNSEKKVDLVFFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIP 74
++S+S + + ++DNPG +I+ + L+ NY EWS ++ +AK+K GF++G IP
Sbjct: 10 STSSSNTSSTTAYLINASDNPGALISSVVLKENNYAEWSEELQNFLRAKQKLGFIDGSIP 69
Query: 75 KPTTPEKLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARIC 134
KP +L W A+ SM++ W+ +I+P++RST+ + +A LW +L++RF V NG R
Sbjct: 70 KPAADPELSLWIAINSMIVGWIRTSIDPTIRSTVGFVSEASQLWENLRRRFSVGNGVRKT 129
Query: 135 QLKASLGECKQGKGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAED 194
LK + C Q G+ V AY+GRL ++W+EL Y CKCE DI K+ D D
Sbjct: 130 LLKDEIAACTQD-GQPVLAYYGRLIKLWEELQNYKSGRECKCEAA-SDIEKERED----D 183
Query: 195 YLHHFLMGLDGAYATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGEYSIQQNRDNVMAFK 254
+H FL+GLD +++IRS++ +PLP++ Q Y RV+++E+ + + + F
Sbjct: 184 RVHKFLLGLDSRFSSIRSSITDIEPLPDLYQVYSRVVREEQNLNASRTKDVVKTEAIGFS 243
Query: 255 VAPDTRGKSKLVDNSDKFCTHCNREGHDERTCFQIHGFPEWWGDR-PRGGRGSGRGGATT 313
V T + + D S FCTHCNR+GH+ CF +HG+P+WW ++ P+ + S RG +
Sbjct: 244 VQSSTTPRFR--DKSTLFCTHCNRKGHEVTQCFLVHGYPDWWLEQNPQENQPSTRGRGSN 301
Query: 314 GRDTTGRSGGRGRGTYNALARANKATTSSRSIGGNSGQSHAPPHSSEAAGISGITPAQWQ 373
GR G S GRG R++ TT R N+ Q+ AP S + G I AQ
Sbjct: 302 GR---GSSSGRGGN------RSSAPTTRGRG-RANNAQAAAPTVSGD--GNDQI--AQLI 347
Query: 374 QILDALNISKTKDRLHGKNDIS-WIIDTGASHHVTGNFSCLINGKRITNTPVGLPNGKDA 432
+L A S + +RL G ++ +IDTGASHH+TG+ S L++ IT +PV P+GK +
Sbjct: 348 SLLQAQRPSSSSERLSGNTCLTDGVIDTGASHHMTGDCSILVDVFDITPSPVTKPDGKAS 407
Query: 433 TAIQEGSVILDGGLRLNNVLFVPQLTCNLISVTQLIDDSNCIVQFTNALCVIQDRTTRTL 492
A + G+++L +L++VLFVP C LISV++L+ ++ I FT+ C +QDR RTL
Sbjct: 408 QATKCGTLLLHDSYKLHDVLFVPDFDCTLISVSKLLKQTSSIAIFTDTFCFLQDRFLRTL 467
Query: 493 IGAGERIDGLYFFRGV--PKVHALMVEGDSAMDLWHKRLGHPSEKVLKFIPHVSQHSRSK 550
IGAGE +G+Y+F GV P+VH + + DLWH+RLGHPS VL +P ++ S+
Sbjct: 468 IGAGEEREGVYYFTGVLAPRVHKASSDFAISGDLWHRRLGHPSTSVLLSLPECNRSSQGF 527
Query: 551 NN-RPCDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGSYRTRSSCGAQYYLTIVNDYS 609
+ CD C R+KQ R+ FP+S N F L+H D+WG YRT S+ GA Y+LT+V+DYS
Sbjct: 528 DKIDSCDTCFRSKQTREVFPISNNKTMECFSLIHGDVWGPYRTPSTTGAVYFLTLVDDYS 587
Query: 610 RAVWVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTEFNCLRDYFFNNGIVFET 669
R+VW YL+ +KTE+ + NF A +RQF K++K R+DNGTEF CL YF +GI+ +T
Sbjct: 588 RSVWTYLMSSKTEVSQLIKNFCAMSERQFGKQVKAFRTDNGTEFMCLTPYFQTHGILHQT 647
Query: 670 SCVGTPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLTACYLINRTPSSVLNYKT 729
SCV TPQQNGRVERKH+HI+NVARA FQG+LP++FWGE +LTA +LINRTPS+VL KT
Sbjct: 648 SCVDTPQQNGRVERKHRHILNVARACLFQGNLPVKFWGESILTATHLINRTPSAVLKGKT 707
Query: 730 PYEKLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIFVGYPYGKKGWKLYDLES 789
PYE LFG+ P +D ++ FGCLCYAH + R+ DKF SRSRKC+F+GYP+GKK W++YDLE+
Sbjct: 708 PYELLFGERPSYDMLRSFGCLCYAHIRPRNKDKFTSRSRKCVFIGYPHGKKAWRVYDLET 767
Query: 790 KEYIVSRDVKFYEHEFPFDVQLDTTHSTPFIDSEYVTED----IGFETYDASFEGGGASM 845
+ SRDV+F+E +P+ + P V +D I + + + +S
Sbjct: 768 GKIFASRDVRFHEDIYPYATATQSNVPLPPPTPPMVNDDWFLPISTQVDSTNVDSSSSSS 827
Query: 846 ALQDNEQTQLQGGLHGDCNGADVAANEGLSVSGIEGEVEPAHEVDGADVVVMQEDDVE-- 903
Q Q + + + E + I P+ +D + + D E
Sbjct: 828 PAQSGSIDQPPRSIDQSPSTSTNPVPE--EIGSIVPSSSPSRSIDRSTSDLSASDTTELL 885
Query: 904 ----------PGEPEVVAEREAIVASEMGRGMRNKVPNIKLKDFVTHTIRKVKSSK---- 949
PG PE+ +G+G R K ++ LKDFVT+T K K++
Sbjct: 886 STGESSTPSSPGLPEL-----------LGKGCREKKKSVLLKDFVTNTTSKKKTASHNIH 934
Query: 950 ------------SSSAQEDASGTPYPITYFVSCERFSIRHRNFVAAVTAGKEPNNFKEAV 997
S SA + T YP++ F++ +S H F+AA+ EP +FK+A+
Sbjct: 935 SPSQVLPSGLPTSLSADSVSGKTLYPLSDFLTNSGYSANHIAFMAAILDSNEPKHFKDAI 994
Query: 998 KDSGWRDAMRNEIQALEDNETWVMEKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVF 1057
W +AM EI ALE N TW + LP GKKA+ SKWVYK+K++SDG++ER KARLVV
Sbjct: 995 LIKEWCEAMSKEIDALEANHTWDITDLPHGKKAISSKWVYKLKYNSDGTLERHKARLVVM 1054
Query: 1058 GHHQIEGIDYDETFAPVAKMVTVRTFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVYMKVP 1117
G+HQ EG+D+ ETFAPVAK+ TVRT LAVAA K WEVHQMDVHNAFLHGDLEEEVYM++P
Sbjct: 1055 GNHQKEGVDFKETFAPVAKLTTVRTILAVAAAKDWEVHQMDVHNAFLHGDLEEEVYMRLP 1114
Query: 1118 PGFKNTDPNLVCRLKKSLYGLKQAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQI 1177
PGFK +DP+ VCRL+KSLYGLKQAPRCWF+KL TAL+ GF QSY DYSLF+L G+ I
Sbjct: 1115 PGFKCSDPSKVCRLRKSLYGLKQAPRCWFSKLSTALRNIGFTQSYEDYSLFSLKNGDTII 1174
Query: 1178 NVLVYVDDLIIAGNDIAALKIFKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKY 1237
+VLVYVDDLI+AGN++ A+ FK+ L CFHMKDLG LKYFLGLEV+R +G L QRKY
Sbjct: 1175 HVLVYVDDLIVAGNNLDAIDRFKSQLHKCFHMKDLGKLKYFLGLEVSRGPDGFCLSQRKY 1234
Query: 1238 ALEIIDETGLLGAKPADFPMEQHHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYS 1297
AL+I+ ETGLLG KP+ P+ +HKLA ++G +PE YRRL+GR IYL++TRPDL+Y+
Sbjct: 1235 ALDIVKETGLLGCKPSAVPIALNHKLASITGPVFTNPEQYRRLVGRFIYLTITRPDLSYA 1294
Query: 1298 VHILSQFMQKPCEEHWEAALRVVRYLKKHPGQGILLRSDSELKLEGWCDSDWASCPLTRR 1357
VHILSQFMQ P HWEAALR+VRYLK P QGI LRSDS L + +CDSD+ +CPLTRR
Sbjct: 1295 VHILSQFMQAPLVAHWEAALRLVRYLKGSPAQGIFLRSDSSLIINAYCDSDYNACPLTRR 1354
Query: 1358 SLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMAMTTCELKWLKQLLGDLGVSHSQG 1417
SL+ +VV L SP+SWKTKKQ TVS SSAEAEYR+MA T ELKWLK LL DLGV HS
Sbjct: 1355 SLSAYVVYLGDSPISWKTKKQDTVSYSSAEAEYRAMAYTLKELKWLKALLKDLGVHHSSP 1414
Query: 1418 MQLYCDSKSALHIAQNPVFHERTKHIEADCHFVRDAVVAGIICPLYVPTSVQLADIFTKA 1477
M+L+CDS++A+HIA NPVFHERTKHIE+DCH VRDAV+ +I ++ T Q+AD+ TK+
Sbjct: 1415 MKLHCDSEAAIHIAANPVFHERTKHIESDCHKVRDAVLDKLITTEHIYTEDQVADLLTKS 1474
Query: 1478 LGKAQFEFLLRKLGIRD 1494
L + FE LL LG+ D
Sbjct: 1475 LPRPTFERLLSTLGVTD 1491
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 1379 bits (3568), Expect = 0.0
Identities = 735/1532 (47%), Positives = 972/1532 (62%), Gaps = 79/1532 (5%)
Query: 5 SGDGGKKVDDNSSNSEKKVDLVFFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKR 64
SG K S + + + L S+DNPG VI+ ++L G NY++W+ + + QAKR
Sbjct: 10 SGSTTKMGASGGSRVDSLMVSPYTLASSDNPGAVISSVELNGDNYNQWATEMLNALQAKR 69
Query: 65 KYGFVEGKIPKPTTPE-KLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQ 123
K GF+ G IP+P + E+W AV SM++ W+ +IEP +++T+++ DA LW LKQ
Sbjct: 70 KTGFINGTIPRPPPNDPNYENWTAVNSMIVGWIRTSIEPKVKATVTFISDAHLLWKDLKQ 129
Query: 124 RFCVVNGARICQLKASLGECKQGKGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDI 183
RF V N RI Q++A L C+Q G+ V Y+GRLS +W+E Y C C C C
Sbjct: 130 RFSVGNKVRIHQIRAQLSSCRQD-GQAVIEYYGRLSNLWEEYNIYKPVTVCTCGLCRCGA 188
Query: 184 NKQVTDFSAEDYLHHFLMGLDGA-YATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGEYS 242
+ T E+ +H F++GLD + + + + L++ DPLP++ + Y RVI++E+ R
Sbjct: 189 TSEPTKEREEEKIHQFVLGLDESRFGGLCATLINMDPLPSLGEIYSRVIREEQ-RLASVH 247
Query: 243 IQQNRDNVMAFKVAPDTRGKSKLVDNSDK------------------FCTHCNREGHDER 284
+++ ++ + F + VD S C++C R GH+++
Sbjct: 248 VREQKEEAVGFLARREQLDHHSRVDASSSRSEHTGGSRSNSIIKGRVTCSNCGRTGHEKK 307
Query: 285 TCFQIHGFPEWWGDRP--RGGRGSGRGGATTGRDTTGRSGGRGRGTYNALARANKATTSS 342
C+QI GFP+WW +R RG G GRGG G +GGRG+G A AT+S+
Sbjct: 308 ECWQIVGFPDWWSERNGGRGSNGRGRGGR-------GSNGGRGQGQ----VMAAHATSSN 356
Query: 343 RSIGGNSGQSHAPPHSS--EAAGISGITPAQWQQILDALNISKTKDRLHGKNDISWII-D 399
S+ + H S + SG T + DRL GK + II D
Sbjct: 357 SSVFPEFTEEHMRVLSQLVKEKSNSGSTS------------NNNSDRLSGKTKLGDIILD 404
Query: 400 TGASHHVTGNFSCLINGKRITNTPVGLPNGKDATAIQEGSVILDGGLRLNNVLFVPQLTC 459
+GASHH+TG S L N + PVG +G A A+ G + L + L NVLFVP L C
Sbjct: 405 SGASHHMTGTLSSLTNVVPVPPCPVGFADGSKAFALSVGVLTLSNTVSLTNVLFVPSLNC 464
Query: 460 NLISVTQLIDDSNCIVQFTNALCVIQDRTTRTLIGAGERIDGLYFFRGVP--KVHALMVE 517
LISV++L+ + C+ FT+ LC +QDR+++TLIG+GE G+Y+ V K+H V+
Sbjct: 465 TLISVSKLLKQTQCLATFTDTLCFLQDRSSKTLIGSGEERGGVYYLTDVTPAKIHTANVD 524
Query: 518 GDSAMDLWHKRLGHPSEKVLKFIPHVSQHSRSKNNRPCDVCPRAKQHRDSFPLSENNAAS 577
D A LWH+RLGHPS VL +P S+ S + + CDVC RAKQ R+ FP S N
Sbjct: 525 SDQA--LWHQRLGHPSFSVLSSLPLFSKTSSTVTSHSCDVCFRAKQTREVFPESINKTEE 582
Query: 578 LFELVHCDLWGSYRTRSSCGAQYYLTIVNDYSRAVWVYLLCNKTEIETMFLNFVAFVDRQ 637
F L+HCD+WG YR +SCGA Y+LTIV+DYSRAVW YLL K+E+ + NF+ + ++Q
Sbjct: 583 CFSLIHCDVWGPYRVPASCGAVYFLTIVDDYSRAVWTYLLLEKSEVRQVLTNFLKYAEKQ 642
Query: 638 FDKKIKKVRSDNGTEFNCLRDYFFNNGIVFETSCVGTPQQNGRVERKHQHIMNVARALRF 697
F K +K VRSDNGTEF CL YF NGI+ +TSCVGTPQQNGRVERKH+HI+NVARAL F
Sbjct: 643 FGKTVKMVRSDNGTEFMCLSSYFRENGIIHQTSCVGTPQQNGRVERKHRHILNVARALLF 702
Query: 698 QGHLPMQFWGECVLTACYLINRTPSSVLNYKTPYEKLFGKVPKFDNMKIFGCLCYAHNQR 757
Q LP++FWGE +LTA YLINRTPSS+L+ +TPYE L G P + +++FG CY H
Sbjct: 703 QASLPIKFWGESILTAAYLINRTPSSILSGRTPYEVLHGSKPVYSQLRVFGSACYVHRVT 762
Query: 758 RDGDKFASRSRKCIFVGYPYGKKGWKLYDLESKEYIVSRDVKFYEHEFPF-DVQLDTTHS 816
RD DKF RSR CIFVGYP+GKKGWK+YD+E E++VSRDV F E FP+ V T S
Sbjct: 763 RDKDKFGQRSRSCIFVGYPFGKKGWKVYDIERNEFLVSRDVIFREEVFPYAGVNSSTLAS 822
Query: 817 TPFIDSEYVTEDIGFETYDASFEGGGASMALQDNEQTQLQGGLHGDCNGADVAANEGLSV 876
T V+ED + G S+ + E+ C +V + +S
Sbjct: 823 TSL---PTVSEDDDWAIPPLEVRGSIDSV---ETERVV--------CTTDEVVLDTSVSD 868
Query: 877 SGIEG-EVEPAHEVDGADVVVMQEDDVE-PGEPEVVAEREAIVAS--EMGRGMRNKVPNI 932
S I E P + + V P P VV I S + + R P
Sbjct: 869 SEIPNQEFVPDDTPPSSPLSVSPSGSPNTPTTPIVVPVASPIPVSPPKQRKSKRATHPPP 928
Query: 933 KLKDFVTHTIRKVKSSKSSSAQEDASGTP------YPITYFVSCERFSIRHRNFVAAVTA 986
KL D+V + SS + + + + +P+T +VS FS HR ++AA+T
Sbjct: 929 KLNDYVLYNAMYTPSSIHALPADPSQSSTVPGKSLFPLTDYVSDAAFSSSHRAYLAAITD 988
Query: 987 GKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVMEKLPPGKKALGSKWVYKIKHHSDGS 1046
EP +FKEAV+ W DAM E+ ALE N+TW + LPPGK A+GS+WV+K K++SDG+
Sbjct: 989 NVEPKHFKEAVQIKVWNDAMFTEVDALEINKTWDIVDLPPGKVAIGSQWVFKTKYNSDGT 1048
Query: 1047 IERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVAAIKKWEVHQMDVHNAFLHG 1106
+ER KARLVV G+ Q+EG DY ETFAPV +M TVRT L A +WEV+QMDVHNAFLHG
Sbjct: 1049 VERYKARLVVQGNKQVEGEDYKETFAPVVRMTTVRTLLRNVAANQWEVYQMDVHNAFLHG 1108
Query: 1107 DLEEEVYMKVPPGFKNTDPNLVCRLKKSLYGLKQAPRCWFAKLVTALKRYGFVQSYSDYS 1166
DLEEEVYMK+PPGF+++ P+ VCRL+KSLYGLKQAPRCWF KL +L R+GFVQSY DYS
Sbjct: 1109 DLEEEVYMKLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLSDSLLRFGFVQSYEDYS 1168
Query: 1167 LFTLHRGEIQINVLVYVDDLIIAGNDIAALKIFKAYLGVCFHMKDLGVLKYFLGLEVARN 1226
LF+ R I++ VL+YVDDL+I GND L+ FK YL CF MKDLG LKYFLG+EV+R
Sbjct: 1169 LFSYTRNNIELRVLIYVDDLLICGNDGYMLQKFKDYLSRCFSMKDLGKLKYFLGIEVSRG 1228
Query: 1227 HEGIYLCQRKYALEIIDETGLLGAKPADFPMEQHHKLALVSGKPLEDPEPYRRLIGRLIY 1286
EGI+L QRKYAL++I ++G LG++PA P+EQ+H LA G L DP+PYRRL+GRL+Y
Sbjct: 1229 PEGIFLSQRKYALDVIADSGNLGSRPAHTPLEQNHHLASDDGPLLSDPKPYRRLVGRLLY 1288
Query: 1287 LSVTRPDLAYSVHILSQFMQKPCEEHWEAALRVVRYLKKHPGQGILLRSDSELKLEGWCD 1346
L TRP+L+YSVH+L+QFMQ P E H++AALRVVRYLK PGQGILL +D +L LE +CD
Sbjct: 1289 LLHTRPELSYSVHVLAQFMQNPREAHFDAALRVVRYLKGSPGQGILLNADPDLTLEVYCD 1348
Query: 1347 SDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMAMTTCELKWLKQL 1406
SDW SCPLTRRS++ +VVLL SP+SWKTKKQ TVS SSAEAEYR+M+ E+KWL++L
Sbjct: 1349 SDWQSCPLTRRSISAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMSYALKEIKWLRKL 1408
Query: 1407 LGDLGVSHSQGMQLYCDSKSALHIAQNPVFHERTKHIEADCHFVRDAVVAGIICPLYVPT 1466
L +LG+ S +LYCDSK+A+HIA NPVFHERTKHIE+DCH VRDAV GII +V T
Sbjct: 1409 LKELGIEQSTPARLYCDSKAAIHIAANPVFHERTKHIESDCHSVRDAVRDGIITTQHVRT 1468
Query: 1467 SVQLADIFTKALGKAQFEFLLRKLGIRDLHAP 1498
+ QLAD+FTKALG+ QF +L+ KLG+++LH P
Sbjct: 1469 TEQLADVFTKALGRNQFLYLMSKLGVQNLHTP 1500
>At1g57640
Length = 1444
Score = 1373 bits (3554), Expect = 0.0
Identities = 716/1493 (47%), Positives = 977/1493 (64%), Gaps = 95/1493 (6%)
Query: 29 LGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPT--TPEKLEDWK 86
L + DN G VI+ L+ NY+EW+ +T+ ++++K+GF++G IP+P +P+ LEDW
Sbjct: 23 LTAADNSGAVISHPILKTNNYEEWACGFKTALRSRKKFGFLDGTIPQPLDGSPD-LEDWL 81
Query: 87 AVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQG 146
+ ++L++W+ TI+ L + +S+ D A LW +++RF V NG + ++KA L CKQ
Sbjct: 82 TINALLVSWMKMTIDSELLTNISHRDVARDLWEQIRKRFSVSNGPKNQKMKADLATCKQ- 140
Query: 147 KGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAEDYLHHFLMGLDGA 206
+G V Y+G+L++IWD + +Y CKC CIC++ + +D +H +L GL+
Sbjct: 141 EGMTVEGYYGKLNKIWDNINSYRPLRICKCGRCICNLGTDQEKYREDDMVHQYLYGLNET 200
Query: 207 -YATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGEYSIQQNRD-NVMAFKVAPDTRGKSK 264
+ TIRS+L S+ PLP +++ Y V Q+E + S ++ D A ++ P + S+
Sbjct: 201 KFHTIRSSLTSRVPLPGLEEVYNIVRQEEDMVNNRSSNEERTDVTAFAVQMRPRSEVISE 260
Query: 265 LVDNSDKF-----CTHCNREGHDERTCFQIHGFPEWWGDRPRGGRGS-GRGGATTGRDTT 318
NS+K CTHCNR GH CF + G+PEWWGDRPRG S G GR
Sbjct: 261 KFANSEKLQNKKLCTHCNRGGHSPENCFVLIGYPEWWGDRPRGKSNSNGSTSRGRGRFGP 320
Query: 319 GRSGGRGRGTYNALARANKATTSSRSIGGNSGQSHAPPHSSEAAGISGITPAQWQQILDA 378
G +GG+ R TY + +S + S+ +SG+T QW+ ++
Sbjct: 321 GFNGGQPRPTYVNVVMTGPFPSSEHV--------NRVITDSDRDAVSGLTDEQWRGVVKL 372
Query: 379 LNISKTKDRLHGKND--------ISWIIDTGASHHVTGNFSCLINGKRITNTPVGLPNGK 430
LN ++ ++ + SWI+DTGASHH+TGN L + + ++ + L +G
Sbjct: 373 LNAGRSDNKSNAHETQSGTCSLFTSWILDTGASHHMTGNLELLSDMRSMSPVLIILADGN 432
Query: 431 DATAIQEGSVILDGGLRLNNVLFVPQLTCNLISVTQLIDDSNCIVQFTNALCVIQDRTTR 490
A+ EG+V L L L +V +V +L +LISV Q++D+++C+ NA V
Sbjct: 433 KRVAVSEGTVRLGSHLILKSVFYVKELESDLISVGQMMDENHCV----NAAAV------H 482
Query: 491 TLIGAGERIDGLYFFRGVPKVHALMVEGDSAMDLWHKRLGHPSEKVLKFIPH-VSQHSRS 549
T + A DLWH+RLGH S+K++ +P + +
Sbjct: 483 TSVKA-------------------------PFDLWHRRLGHASDKIVNLLPRELLSSGKE 517
Query: 550 KNNRPCDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGSYRTRSSCGAQYYLTIVNDYS 609
CD C RAKQ RD+FPLS+N + F+L+HCD+WG YR S GA+Y+LTIV+DYS
Sbjct: 518 ILENVCDTCMRAKQTRDTFPLSDNRSMDSFQLIHCDVWGPYRAPSYSGARYFLTIVDDYS 577
Query: 610 RAVWVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTEFNCLRDYFFNNGIVFET 669
R VWVYL+ +K+E + +F+A V+RQFD +IK VRSDNGTEF C+R+YF + GI ET
Sbjct: 578 RGVWVYLMTDKSETQKHLKDFIALVERQFDTEIKIVRSDNGTEFLCMREYFLHKGIAHET 637
Query: 670 SCVGTPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLTACYLINRTPSSVLNYKT 729
SCVGTP QNGRVERKH+HI+N+ARALRFQ +LP+QFWGEC+L+A YLINRTPS +L K+
Sbjct: 638 SCVGTPHQNGRVERKHRHILNIARALRFQSYLPIQFWGECILSAAYLINRTPSMLLQGKS 697
Query: 730 PYEKLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIFVGYPYGKKGWKLYDLES 789
PYE L+ PK+ ++++FG LCYAHNQ GDKFA+RSR+C+FVGYP+G+KGW+L+DLE
Sbjct: 698 PYEMLYKTAPKYSHLRVFGSLCYAHNQNHKGDKFAARSRRCVFVGYPHGQKGWRLFDLEE 757
Query: 790 KEYIVSRDVKFYEHEFPFDV----QLDTTHSTPFIDSEYVTEDIGFETYDASFEGGGASM 845
+++ VSRDV F E EFP+ + D + ++ E IG T G
Sbjct: 758 QKFFVSRDVIFQETEFPYSKMSCNEEDERVLVDCVGPPFIEEAIGPRTIIGRNIG---EA 814
Query: 846 ALQDNEQTQLQGGLHGDCNGADVAANEGLSVSGIEGEVEPAHEVDGADVVVMQEDDVEPG 905
+ N T G + + N + +E +S+S ++ + V+
Sbjct: 815 TVGPNVAT---GPIIPEINQESSSPSEFVSLSSLDP--------------FLASSTVQTA 857
Query: 906 EPEVVAEREAIVASEMGRGMRNKVPNIKLKDFVTHTIRKVKSSKSSSAQEDASGTPYPIT 965
+ + + A + ++ R R +KLK+FVT+T+ S S + E +S + YPI
Sbjct: 858 DLPLSSTTPAPI--QLRRSSRQTQKPMKLKNFVTNTV-----SVESISPEASSSSLYPIE 910
Query: 966 YFVSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVMEKLP 1025
+V C RF+ H+ F+AAVTAG EP + EA+ D WR+AM EI++L N+T+ + LP
Sbjct: 911 KYVDCHRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRVNQTFSIVNLP 970
Query: 1026 PGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLA 1085
PGK+ALG+KWVYKIK+ SDG+IER KARLVV G+ Q EG+DYDETFAPVAKM TVR FL
Sbjct: 971 PGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVAKMSTVRLFLG 1030
Query: 1086 VAAIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGFKNTDPNLVCRLKKSLYGLKQAPRCW 1145
VAA + W VHQMDVHNAFLHGDL+EEVYMK+P GF+ DP+ VCRL KSLYGLKQAPRCW
Sbjct: 1031 VAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSLYGLKQAPRCW 1090
Query: 1146 FAKLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIAALKIFKAYLGV 1205
F+KL +ALK+YGF QS SDYSLF+ + I ++VLVYVDDLII+G+ A+ FK+YL
Sbjct: 1091 FSKLSSALKQYGFTQSLSDYSLFSYNNDGIFVHVLVYVDDLIISGSCPDAVAQFKSYLES 1150
Query: 1206 CFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAKPADFPMEQHHKLAL 1265
CFHMKDLG+LKYFLG+EV+RN +G YL QRKY L+II E GLLGA+P+ FP+EQ+HKL+L
Sbjct: 1151 CFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGARPSAFPLEQNHKLSL 1210
Query: 1266 VSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEAALRVVRYLKK 1325
+ L D YRRL+GRLIYL VTRP+L+YSVH L+QFMQ P ++HW AA+RVVRYLK
Sbjct: 1211 STSPLLSDSSRYRRLVGRLIYLVVTRPELSYSVHTLAQFMQNPRQDHWNAAIRVVRYLKS 1270
Query: 1326 HPGQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSS 1385
+PGQGILL S S L++ GWCDSD+A+CPLTRRSLTG+ V L +P+SWKTKKQPTVSRSS
Sbjct: 1271 NPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQLGDTPISWKTKKQPTVSRSS 1330
Query: 1386 AEAEYRSMAMTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVFHERTKHIEA 1445
AEAEYR+MA T EL WLK++L DLGVSH Q M+++ DSKSA+ ++ NPV HERTKH+E
Sbjct: 1331 AEAEYRAMAFLTQELMWLKRVLYDLGVSHVQAMRIFSDSKSAIALSVNPVQHERTKHVEV 1390
Query: 1446 DCHFVRDAVVAGIICPLYVPTSVQLADIFTKALGKAQFEFLLRKLGIRDLHAP 1498
DCHF+RDA++ GII +VP+ QLADI TKALG+ + + LRKLGI D+HAP
Sbjct: 1391 DCHFIRDAILDGIIATSFVPSHKQLADILTKALGEKEVRYFLRKLGILDVHAP 1443
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 1236 bits (3198), Expect = 0.0
Identities = 661/1345 (49%), Positives = 855/1345 (63%), Gaps = 107/1345 (7%)
Query: 174 CKCEGCICDINKQVTDFSAEDYLHHFLMGLDGA-YATIRSNLLSQDPLPNIDQAYQRVIQ 232
CKC GC+CD+ ED +H FL GLD A + T+RS+L+S+ P+ +++ Y V Q
Sbjct: 75 CKCGGCVCDLGALQEKDREEDKVHEFLSGLDDALFRTVRSSLVSRIPVQPLEEVYNIVRQ 134
Query: 233 DERLRQGEYSIQQNRDNVMAFKVAPDTRGKSKLVDNSDK--FCTHCNREGHDERTCFQIH 290
+E L + ++ ++ V AF + D DK C HCNR GH +C+ +
Sbjct: 135 EEDLLRNGANVLDDQREVNAFAAQMRPKLYQGRGDEKDKSMVCKHCNRSGHASESCYAVI 194
Query: 291 GFPEWWGDRPRGGRGSGRGGATTGRDTTGRSGGRGRGTYNALARANKATTSSRSIGGNSG 350
G+PEWWGDRPR R T GR T SGGRGRG A A AN+ T +
Sbjct: 195 GYPEWWGDRPRS-----RSLQTRGRGGTNSSGGRGRG---AAAYANRVTVPNHD---TYE 243
Query: 351 QSHAPPHSSEAAGISGITPAQWQQILDALNISKTKDRLHGKNDISWIIDTGASHHVTGNF 410
Q++ + G++G+T +QW+ I LN K A+ +T
Sbjct: 244 QANYALTDEDRDGVNGLTDSQWRTIKSILNSGKD----------------AATEKLTDMD 287
Query: 411 SCLINGKRITNTPVGLPNGKDATAIQEGSVILDGGLRLNNVLFVPQLTCNLISVTQLIDD 470
LI L +G++ +++EG+V L L + +V +V + +LIS+ QL+D+
Sbjct: 288 PILIV----------LADGRERISVKEGTVRLGSNLVMISVFYVEEFQSDLISIGQLMDE 337
Query: 471 SNCIVQFTNALCVIQDRTTRTLIGAGERIDGLYFFRGVPKVHALMVEGDSAMDLWHKRLG 530
+ C++Q ++ V+QDRT+R ++GAG R+ G + FR ++ V+ + +LWH R+G
Sbjct: 338 NRCVLQMSDRFLVVQDRTSRMVMGAGRRVGGTFHFRSTEIAASVTVKEEKNYELWHSRMG 397
Query: 531 HPSEKVLKFIPHVSQHSRSKN-NRPCDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGS 589
HP+ +V+ IP S S + N+ CDVC RAKQ R+SFPLS N +FEL++CDLWG
Sbjct: 398 HPAARVVSLIPESSVSVSSTHLNKACDVCHRAKQTRNSFPLSINKTLRIFELIYCDLWGP 457
Query: 590 YRTRSSCGAQYYLTIVNDYSRAVWVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDN 649
YRT S GA+Y+LTI++DYSR VW+YLL +K+E NF A DRQF+ KIK VRSDN
Sbjct: 458 YRTPSHTGARYFLTIIDDYSRGVWLYLLNDKSEAPCHLKNFFAMTDRQFNVKIKTVRSDN 517
Query: 650 GTEFNCLRDYFFNNGIVFETSCVGTPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGEC 709
GTEF CL +F G++ E SCV TP++N RVERKH+H++NVARALRFQ +LP+QFWGEC
Sbjct: 518 GTEFLCLTKFFQEQGVIHERSCVATPERNDRVERKHRHLLNVARALRFQANLPIQFWGEC 577
Query: 710 VLTACYLINRTPSSVLNYKTPYEKLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRK 769
VLTA YLINRTPSSVLN TPYE+L K P+FD++++FG LCYAHN+ R GDKFA RSR+
Sbjct: 578 VLTAAYLINRTPSSVLNDSTPYERLHKKQPRFDHLRVFGSLCYAHNRNRGGDKFAERSRR 637
Query: 770 CIFVGYPYGKKGWKLYDLESKEYIVSRDVKFYEHEFPFDVQLDTTHSTPFIDSEYVTEDI 829
C+FVGYP+G+KGW+L+DLE E+ VSRDV F E EFPF + +H I+ E E +
Sbjct: 638 CVFVGYPHGQKGWRLFDLEQNEFFVSRDVVFSELEFPFRI----SHEQNVIEEE--EEAL 691
Query: 830 GFETYDASFEGGGASMALQDNEQTQLQGGLHGDCNGADVAAN------EGLSVSGIEGEV 883
D E ++ Q G C + ++ + E + S ++ EV
Sbjct: 692 WAPIVDGLIE--------EEVHLGQNAGPTPPICVSSPISPSATSSRSEHSTSSPLDTEV 743
Query: 884 EPAHEVDGA---------DVVVMQEDDVEPGEPEVVAEREAIVASEMGRGMRNKVPNIKL 934
P ++ + +P + VA V + RNK P + L
Sbjct: 744 VPTPATSTTSASSPSSPTNLQFLPLSRAKPTTAQAVAP--PAVPPPRRQSTRNKAPPVTL 801
Query: 935 KDFVTHT-IRKVKSSKSSSAQEDASGTPYPITYFVSCERFSIRHRNFVAAVTAGKEPNNF 993
KDFV +T + + SK +S Y + RFS H +VA
Sbjct: 802 KDFVVNTTVCQESPSKLNSIL-------YQLQKRDDTRRFSASHTTYVA----------- 843
Query: 994 KEAVKDSGWRDAMRNEIQALEDNETWVMEKLPPGKKALGSKWVYKIKHHSDGSIERLKAR 1053
I A E+N TW +E LPPGK+A+GS+WVYK+KH+SDGS+ER KAR
Sbjct: 844 ----------------IDAQEENHTWTIEDLPPGKRAIGSQWVYKVKHNSDGSVERYKAR 887
Query: 1054 LVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVY 1113
LV G+ Q EG DY ETFAPVAKM TVR FL VA + WE+HQMDVHNAFLHGDL EEVY
Sbjct: 888 LVALGNKQKEGEDYGETFAPVAKMATVRLFLDVAVKRNWEIHQMDVHNAFLHGDLREEVY 947
Query: 1114 MKVPPGFKNTDPNLVCRLKKSLYGLKQAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRG 1173
MK+PPGF+ + PN VCRL+K+LYGLKQAPRCWF KL TALKRYGF QS +DYSLFTL +G
Sbjct: 948 MKLPPGFEASHPNKVCRLRKALYGLKQAPRCWFEKLTTALKRYGFQQSLADYSLFTLVKG 1007
Query: 1174 EIQINVLVYVDDLIIAGNDIAALKIFKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLC 1233
++I +L+YVDDLII GN A + FK YL CFHMKDLG LKYFLG+EVAR+ GIY+C
Sbjct: 1008 SVRIKILIYVDDLIITGNSQRATQQFKEYLASCFHMKDLGPLKYFLGIEVARSTTGIYIC 1067
Query: 1234 QRKYALEIIDETGLLGAKPADFPMEQHHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPD 1293
QRKYAL+II ETGLLG KPA+FP+EQ+HKL L + L DP+ YRRL+GRLIYL+VTR D
Sbjct: 1068 QRKYALDIISETGLLGVKPANFPLEQNHKLGLSTSPLLTDPQRYRRLVGRLIYLAVTRLD 1127
Query: 1294 LAYSVHILSQFMQKPCEEHWEAALRVVRYLKKHPGQGILLRSDSELKLEGWCDSDWASCP 1353
LA+SVHIL++FMQ+P E+HW AALRVVRYLK PGQG+ LR + ++ GWCDSDWA P
Sbjct: 1128 LAFSVHILARFMQEPREDHWAAALRVVRYLKADPGQGVFLRRSGDFQITGWCDSDWAGDP 1187
Query: 1354 LTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMAMTTCELKWLKQLLGDLGVS 1413
++RRS+TG+ V SP+SWKTKKQ TVS+SSAEAEYR+M+ EL WLKQLL LGVS
Sbjct: 1188 MSRRSVTGYFVQFGDSPISWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQLLFSLGVS 1247
Query: 1414 HSQGMQLYCDSKSALHIAQNPVFHERTKHIEADCHFVRDAVVAGIICPLYVPTSVQLADI 1473
H Q M + CDSKSA++IA NPVFHERTKHIE D HFVRD V G+I P +V T+ QLADI
Sbjct: 1248 HVQPMIMCCDSKSAIYIATNPVFHERTKHIEIDYHFVRDEFVKGVITPRHVGTTSQLADI 1307
Query: 1474 FTKALGKAQFEFLLRKLGIRDLHAP 1498
FTK LG+ F KLGIR+L+AP
Sbjct: 1308 FTKPLGRDCFSAFRIKLGIRNLYAP 1332
Score = 31.2 bits (69), Expect = 4.8
Identities = 16/45 (35%), Positives = 27/45 (59%)
Query: 15 NSSNSEKKVDLVFFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTS 59
N + ++ L + L S +NP VI+ L G NYDEW+ +++T+
Sbjct: 19 NPTVEVRRTILPYDLTSANNPCAVISHPLLTGSNYDEWACSMKTA 63
>At2g07010 putative retroelement pol polyprotein
Length = 1413
Score = 1033 bits (2672), Expect = 0.0
Identities = 560/1245 (44%), Positives = 758/1245 (59%), Gaps = 66/1245 (5%)
Query: 27 FFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPTTPEK-LEDW 85
+ L S+DNPG +I+ + L G NY+EWS + + QAKRK GF+ G I KP E+W
Sbjct: 27 YTLASSDNPGAMISSVMLTGDNYNEWSTKMLNALQAKRKTGFINGSISKPPLDNPDYENW 86
Query: 86 KAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQ 145
+AV SM++ W+ +IEP ++ST+++ DA LW+ LKQRF V N + Q+K L C+Q
Sbjct: 87 QAVNSMIVGWIRASIEPKVKSTVTFICDAHQLWSELKQRFSVGNKVHVHQIKTQLAACRQ 146
Query: 146 GKGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAEDYLHHFLMGLDG 205
G+ V Y+GRL ++W+E Y CKC C C + + E+ +H F++GLD
Sbjct: 147 D-GQPVIDYYGRLCKLWEEFQIYKPITVCKCGLCTCGATLEPSKEREEEKIHQFVLGLDD 205
Query: 206 A-YATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGEYSIQQNRDNVMAF-----KVAPDT 259
+ + + + L++ DP P++ + Y RV+++E+ R I++ + + + F +V D
Sbjct: 206 SRFGGLSATLIAMDPFPSLGEIYSRVVREEQ-RLASVQIREQQQSAIGFLTRQSEVTADG 264
Query: 260 RGKSKLVDNSDK--FCTHCNREGHDERTCFQIHGFPEWWGDRPRGG-RGS---GRGGATT 313
R S ++ + D+ C+HC R GH+++ C+QI GFP+WW +R GG RGS GRGG ++
Sbjct: 265 RTDSSIIKSRDRSVLCSHCGRSGHEKKDCWQIVGFPDWWTERTNGGGRGSSSRGRGGRSS 324
Query: 314 GRDTTGRSGGRGRGTYNALARANKATTSSRSIGGNSGQSHAPPHSSEAAGISGITPAQWQ 373
G + +GR GRG+ T A ATTS+ S TP Q +
Sbjct: 325 GSNNSGR--GRGQVT------AAHATTSNLS------------------PFPEFTPDQLR 358
Query: 374 QILDALNISK--TKDRLHGKNDISWII-DTGASHHVTGNFSCLINGKRITNTPVGLPNGK 430
I + T D+L GK + +I DTGASHH+TG S L N I + VG +G+
Sbjct: 359 VITQMIQNKNNGTSDKLSGKMKLGDVILDTGASHHMTGQLSLLTNIVTIPSCSVGFADGR 418
Query: 431 DATAIQEGSVILDGGLRLNNVLFVPQLTCNLISVTQLIDDSNCIVQFTNALCVIQDRTTR 490
AI G+ L + L+NVL+VP L C+LISV++L+ C+ FT+ +CV+QDR +R
Sbjct: 419 KTFAISMGTFKLSETVSLSNVLYVPALNCSLISVSKLVKQIKCLALFTDTICVLQDRFSR 478
Query: 491 TLIGAGERIDGLYFFR--GVPKVHALMVEGDSAMDLWHKRLGHPSEKVLKFIPHVSQHSR 548
TLIG GE DG+Y+ VH + + D A LWH+RLGHPS VL +P S S
Sbjct: 479 TLIGTGEERDGVYYLTDAATTTVHKVDITTDHA--LWHQRLGHPSFSVLSSLPLFSGSSC 536
Query: 549 SKNNRPCDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGSYRTRSSCGAQYYLTIVNDY 608
S ++R CDVC RAKQ R+ FP S N + F L+HCD+WG YR SSCGA Y+LTIV+D+
Sbjct: 537 SVSSRSCDVCFRAKQTREVFPDSSNKSTDCFSLIHCDVWGPYRVPSSCGAVYFLTIVDDF 596
Query: 609 SRAVWVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTEFNCLRDYFFNNGIVFE 668
SR+VW YLL K+E+ ++ NF+A+ ++QF K +K +RSDNGTEF CL YF GIV +
Sbjct: 597 SRSVWTYLLLAKSEVRSVLTNFLAYTEKQFGKSVKIIRSDNGTEFMCLSSYFKEQGIVHQ 656
Query: 669 TSCVGTPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLTACYLINRTPSSVLNYK 728
TSCVGTPQQNGRVERKH+HI+NV+RAL FQ LP++FWGE V+TA YLINRTPSS+ N
Sbjct: 657 TSCVGTPQQNGRVERKHRHILNVSRALLFQASLPIKFWGEAVMTAAYLINRTPSSIHNGL 716
Query: 729 TPYEKLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIFVGYPYGKKGWKLYDLE 788
+PYE L G P +D +++FG CYAH RD DKF RSR CIFVGYP+G+KGWK+YDL
Sbjct: 717 SPYELLHGCKPDYDQLRVFGSACYAHRVTRDKDKFGERSRLCIFVGYPFGQKGWKVYDLS 776
Query: 789 SKEYIVSRDVKFYEHEFPFDVQLDTTHSTPFIDSEYVTEDIGFETYDASFEGGGASMALQ 848
+ E+IVSRDV F E+ FP+ T TP + +T D + + + G L
Sbjct: 777 TNEFIVSRDVVFRENVFPYATNEGDTIYTPPVTCP-ITYDEDWLPFTTLEDRGSDENYLS 835
Query: 849 DN----EQTQLQGGLHGDCNGADVAANEGLSVSGIEGEVEPAHEVDGADVVVMQEDDVEP 904
D H ++ LS S V P + +V P
Sbjct: 836 DPPVCVTNVSESDTEHDTPQSLPTPVDDPLSPS---TSVTPTQTPTNSSSSTSPSTNVSP 892
Query: 905 GEPEVVAEREAIVASEMGRGMRNKVPNIKLKDFVTH-------TIRKVKSSKSSSAQEDA 957
+ + I + +G R +LKD++ + T + S S S+
Sbjct: 893 PQQDTT---PIIENTPPRQGKRQVQQPARLKDYILYNASCTPNTPHVLSPSTSQSSSSIQ 949
Query: 958 SGTPYPITYFVSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNE 1017
YP+T ++S E FS H+ F+AA+TA EP +FKE VK W DAM E+ ALE N+
Sbjct: 950 GNLQYPLTDYISDECFSAGHKVFLAAITANDEPKHFKEDVKVKVWNDAMYKEVDALEVNK 1009
Query: 1018 TWVMEKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKM 1077
TW + LP GK A+GS+WVYK K ++DG++ER KARLVV G++QIEG DY ETFAPV KM
Sbjct: 1010 TWDIVDLPTGKVAIGSQWVYKTKFNADGTVERYKARLVVQGNNQIEGEDYTETFAPVVKM 1069
Query: 1078 VTVRTFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGFKNTDPNLVCRLKKSLYG 1137
TVRT L + A +WEV+QMDVHNAFLHGDLEEEVYMK+PPGF+++ P+ VCRL+KSLYG
Sbjct: 1070 TTVRTLLRLVAANQWEVYQMDVHNAFLHGDLEEEVYMKLPPGFRHSHPDKVCRLRKSLYG 1129
Query: 1138 LKQAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIAALK 1197
LKQAPRCWF KL ALKR+GF+Q Y DYS F+ I++ VLVYVDDLII GND ++
Sbjct: 1130 LKQAPRCWFKKLSDALKRFGFIQGYEDYSFFSYSCKGIELRVLVYVDDLIICGNDEYMVQ 1189
Query: 1198 IFKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEII 1242
FK YLG CF MKDLG LKYFLG+EV+R + + A+ I+
Sbjct: 1190 KFKEYLGRCFSMKDLGKLKYFLGIEVSRGPDAPREAHLEAAMRIV 1234
Score = 246 bits (627), Expect = 9e-65
Identities = 123/191 (64%), Positives = 152/191 (79%)
Query: 1308 PCEEHWEAALRVVRYLKKHPGQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGWVVLLD 1367
P E H EAA+R+VRYLK PGQGILL ++ +L LE +CDSD+ SCPLTRRSL+ +VVLL
Sbjct: 1222 PREAHLEAAMRIVRYLKGSPGQGILLSANKDLTLEVYCDSDFQSCPLTRRSLSAYVVLLG 1281
Query: 1368 LSPVSWKTKKQPTVSRSSAEAEYRSMAMTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSA 1427
SP+SWKTKKQ TVS SSAEAEYR+M++ E+KWL +LL +LG++ + +L+CDSK+A
Sbjct: 1282 GSPISWKTKKQDTVSHSSAEAEYRAMSVALKEIKWLNKLLKELGITLAAPTRLFCDSKAA 1341
Query: 1428 LHIAQNPVFHERTKHIEADCHFVRDAVVAGIICPLYVPTSVQLADIFTKALGKAQFEFLL 1487
+ IA NPVFHERTKHIE DCH VRDAV GII +V TS QLADIFTKALG+ QF +L+
Sbjct: 1342 ISIAANPVFHERTKHIERDCHSVRDAVRDGIITTHHVRTSEQLADIFTKALGRNQFIYLM 1401
Query: 1488 RKLGIRDLHAP 1498
KLGI++LH P
Sbjct: 1402 SKLGIQNLHTP 1412
>At1g36620 hypothetical protein
Length = 1152
Score = 902 bits (2330), Expect = 0.0
Identities = 504/1197 (42%), Positives = 716/1197 (59%), Gaps = 91/1197 (7%)
Query: 27 FFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPTTPEK-LEDW 85
++L +D+P +V+TP+ L G NY+ W++ R + QAK+K GF++G + KP++ W
Sbjct: 25 YYLHPSDHPHHVLTPMLLNGENYERWAKLTRNNLQAKQKLGFIDGTLTKPSSDSPDYPRW 84
Query: 86 KAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQ 145
SML+ WL +++P ++ ++S D+A +W L+ R+ V N +R+ QLK + C+Q
Sbjct: 85 LQTNSMLVGWLYASLDPQVQKSISVVDNARVMWESLRTRYSVGNASRVHQLKYDIVACRQ 144
Query: 146 GKGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAEDYLHHFLMGLDG 205
G+ + YFG+L +WD+L Y TC C C + + + +H FLMGLD
Sbjct: 145 D-GQTAANYFGKLKVMWDDLDDYEPLLTCCCNRPSCTHRVRQSQRRDHERIHQFLMGLDA 203
Query: 206 A-YATIRSNLL---SQDPLPNIDQAYQRVIQDERLRQGEYSIQQNRDNV-MAFKVAPDTR 260
A + T R+N+L S+D ++D Y +I +ER S ++ D V A + +
Sbjct: 204 AKFGTSRTNILGRLSRDDNISLDSIYSEIIAEERHLTITRSKEERVDAVGFAVQTGVNAI 263
Query: 261 GKSKLVDNSDKFCTHCNREGHDERTCFQIHGFPEWWGDRPRGGRGSGRGGATTGRDTTGR 320
V+N CTHC R H TCF++HG PEW+ ++ G SGRG GR +T R
Sbjct: 264 ASVTRVNNMGP-CTHCGRSNHSADTCFKLHGVPEWYTEK-YGDTSSGRG---RGRSSTPR 318
Query: 321 SGGRGRGTYNALARANKATTSSRSIGGNSGQSHAPPHSSEAAGISGITPAQWQQILDAL- 379
GRGRG N+ +AN A TS H +SE + I G++ W I + L
Sbjct: 319 --GRGRGHGNSY-KANNAQTS-----------HPSSSASEFSDIPGVSKEAWSAIRNLLK 364
Query: 380 -NISKTKDRLHGK-NDISWIIDTGASHHVTGNFSCLINGKRITNTPVGLPNGKDATAIQE 437
+ + + ++L GK N + ++ID+GASHH+TG L I ++ V LPN K A ++
Sbjct: 365 QDTATSSEKLSGKTNCVDFLIDSGASHHMTGFLDLLTEIYEIPHSVVVLPNAKHTIATKK 424
Query: 438 GSVILDGGLRLNNVLFVPQLTCNLISVTQLIDDSNCIVQFTNALCVIQDRTTRTLIGAGE 497
G++IL ++L +VLFVP L+C LISV +L+ + +C FT+ +CVIQDRT++ LIG G
Sbjct: 425 GTLILGANMKLTHVLFVPDLSCTLISVARLLRELHCFAIFTDKVCVIQDRTSKMLIGVGT 484
Query: 498 RIDGLYFFRGVPKV--HALMVEGDSAMDLWHKRLGHPSEKVLKFI-PHVSQHSRSKNNRP 554
+G+Y + V A +V+ + LWH RLGHPS KVL + P + ++
Sbjct: 485 ESNGVYHLQRAEVVATSANVVKWKTNKALWHMRLGHPSSKVLSSVLPSLEDFDSCSSDLK 544
Query: 555 --CDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGSYRTRSSCGAQYYLTIVNDYSRAV 612
CDVC RAKQ R SF S N A F +H D+WG Y+ SSCGA Y+LTIV+D+SRAV
Sbjct: 545 TICDVCVRAKQTRASFSESFNKAEECFSFIHYDVWGPYKHASSCGAHYFLTIVDDHSRAV 604
Query: 613 WVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTEFNCLRDYFFNNGIVFETSCV 672
W++L+ K+E+ ++ F+A RQF+K++K VRS+NGTEF L+ YF GIV + SCV
Sbjct: 605 WIHLMLAKSEVASLLQQFIAMASRQFNKQVKTVRSNNGTEFMSLKSYFAERGIVHQISCV 664
Query: 673 GTPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLTACYLINRTPSSVLNYKTPYE 732
T QQNGRVERKH+HI+NVAR+L FQ LP+ FW E VLTA YLINRTP+ +L+ KTPY+
Sbjct: 665 YTHQQNGRVERKHRHILNVARSLLFQAELPISFWEESVLTAAYLINRTPTPILDGKTPYK 724
Query: 733 KLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIFVGYPYGKKGWKLYDLESKEY 792
L+ + P + ++++FG LC+A DKF R RKCIFVGYP+G+KGW++YD+ES+ +
Sbjct: 725 ILYSQPPSYASLRVFGSLCFARKHTGRLDKFQERGRKCIFVGYPHGQKGWRIYDIESQIF 784
Query: 793 IVSRDVKFYEHEFPF-DVQLDTTHSTP--FIDSEYVTEDIGF-ETYDASFEGGGASMALQ 848
VSRDV F E FPF D + T S+P I S + D F + Y
Sbjct: 785 FVSRDVVFQEDIFPFADKKNKDTFSSPAAVIPSPILPYDDEFLDIYQI------------ 832
Query: 849 DNEQTQLQGGLHGDCNGADVAANEGLSVSGIEGEVEPAHEVDGADVVVMQEDDVEPGEPE 908
DV A L PA ++ +D P P
Sbjct: 833 -----------------GDVPATNPL----------PA---------IIDVNDSPPSSPI 856
Query: 909 VVAEREAIVASEMGRGMRNKVPNIKLKDFVTHTIRKVKSSKSSSAQEDASGT-PYPITYF 967
+ A A + RG+R + N++LKD+ T++ + +S+ D GT YP+ +
Sbjct: 857 ITATPAAASPPPLRRGLRQRQENVRLKDYQTYSAQ----CESTQTLSDNIGTCIYPMANY 912
Query: 968 VSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVMEKLPPG 1027
VS E FS +++F+AA++ P + +A+++ WR+A+ E+ ALED TW + KLP G
Sbjct: 913 VSGEIFSPSNQHFLAAISMVDPPQTYNQAIREKEWRNAVFFEVDALEDQGTWDITKLPQG 972
Query: 1028 KKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVA 1087
KA+GSKWV++IK++S+G++ER KARLV G+HQ EGID+ +TFAPV KM TVR L VA
Sbjct: 973 VKAIGSKWVFRIKYNSNGTVERYKARLVALGNHQKEGIDFTKTFAPVVKMQTVRLLLDVA 1032
Query: 1088 AIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGFKNTDPNLVCRLKKSLYGLKQAPRCWFA 1147
A K WE+HQMDVHNAFLHGDL+E++YMK PPGFK TDP+LVC+LKKS+YGLKQAPRCWF
Sbjct: 1033 AAKDWELHQMDVHNAFLHGDLKEDIYMKPPPGFKTTDPSLVCKLKKSIYGLKQAPRCWFE 1092
Query: 1148 KLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIAALKIFKAYLG 1204
KL T+L ++GF QS DYSLFT RG ++V+VYVDD++I G + K LG
Sbjct: 1093 KLSTSLLKFGFTQSKKDYSLFTSIRGSKVLHVIVYVDDVVICGKAVRENNTSKLALG 1149
>At1g70010 hypothetical protein
Length = 1315
Score = 810 bits (2093), Expect = 0.0
Identities = 447/1081 (41%), Positives = 637/1081 (58%), Gaps = 85/1081 (7%)
Query: 427 PNGKDATAIQEGSVILDGGLRLNNVLFVPQLTCNLISVTQLIDDSNCIVQFTNALCVIQD 486
P+ GSV L L LN+VLF+PQ NL+SV+ L C + F CV+QD
Sbjct: 304 PSSSSTVCPISGSVHLGRHLILNDVLFIPQFKFNLLSVSSLTKSMGCRIWFDETSCVLQD 363
Query: 487 RTTRTLIGAGERIDGLYFFRGVPKVH-----ALMVEGDSAMDLWHKRLGHPSEKVLKFIP 541
T ++G G+++ LY H ++ V ++ DLWHKRLGHPS + L+ +
Sbjct: 364 ATRELMVGMGKQVANLYIVDLDSLSHPGTDSSITVASVTSHDLWHKRLGHPSVQKLQPMS 423
Query: 542 HVSQHSRSKNNRP--CDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGSYRTRSSCGAQ 599
+ + KNN C VC +KQ F N ++ F+L+H D WG + ++ G +
Sbjct: 424 SLLSFPKQKNNTDFHCRVCHISKQKHLPFVSHNNKSSRPFDLIHIDTWGPFSVQTHDGYR 483
Query: 600 YYLTIVNDYSRAVWVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTEFNCLRDY 659
Y+LTIV+DYSRA WVYLL NK+++ T+ FV V+ QF+ IK VRSDN E N +
Sbjct: 484 YFLTIVDDYSRATWVYLLRNKSDVLTVIPTFVTMVENQFETTIKGVRSDNAPELN-FTQF 542
Query: 660 FFNNGIVFETSCVGTPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLTACYLINR 719
+ + GIV SC TPQQN VERKHQHI+NVAR+L FQ H+P+ +WG+C+LTA YLINR
Sbjct: 543 YHSKGIVPYHSCPETPQQNSVVERKHQHILNVARSLFFQSHIPISYWGDCILTAVYLINR 602
Query: 720 TPSSVLNYKTPYEKLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIFVGYPYGK 779
P+ +L K P+E L VP +D++K+FGCLCYA +D KF+ R++ C F+GYP G
Sbjct: 603 LPAPILEDKCPFEVLTKTVPTYDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIGYPSGF 662
Query: 780 KGWKLYDLESKEYIVSRDVKFYEHEFPFDVQLDTTHSTPFIDSEYVTEDIGFETYDASFE 839
KG+KL DLE+ IVSR V F+E F PF+ S+ E+ F
Sbjct: 663 KGYKLLDLETHSIIVSRHVVFHEELF------------PFLGSDLSQEEQNFFP------ 704
Query: 840 GGGASMALQDNEQTQLQGGLHGDCNGADVAANEGLSVSGIEGEVEPAHEVDGADVVVMQE 899
L Q Q H V P+ +++
Sbjct: 705 ------DLNPTPPMQRQSSDH----------------------VNPSDSSSSVEILPSAN 736
Query: 900 DDVEPGEPEVVAEREAIVASEMGRGMRNKVPNIKLKDFVTHTIRKVKSSKSSSAQEDASG 959
EP V + K P L+D+ H++ S
Sbjct: 737 PTNNVPEPSVQTSHR-----------KAKKPAY-LQDYYCHSV--------------VSS 770
Query: 960 TPYPITYFVSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETW 1019
TP+ I F+S +R + + F+A + KEP+N+ EA K WRDAM E LE TW
Sbjct: 771 TPHEIRKFLSYDRINDPYLTFLACLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTW 830
Query: 1020 VMEKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVT 1079
+ LP K+ +G +W++KIK++SDGS+ER KARLV G+ Q EGIDY+ETF+PVAK+ +
Sbjct: 831 EVCSLPADKRCIGCRWIFKIKYNSDGSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNS 890
Query: 1080 VRTFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGFKNTD-----PNLVCRLKKS 1134
V+ L VAA K + Q+D+ NAFL+GDL+EE+YM++P G+ + PN VCRLKKS
Sbjct: 891 VKLLLGVAARFKLSLTQLDISNAFLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKS 950
Query: 1135 LYGLKQAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIA 1194
LYGLKQA R W+ K + L GF+QSY D++ F I + VLVY+DD+IIA N+ A
Sbjct: 951 LYGLKQASRQWYLKFSSTLLGLGFIQSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDA 1010
Query: 1195 ALKIFKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAKPAD 1254
A+ I K+ + F ++DLG LKYFLGLE+ R+ +GI++ QRKYAL+++DETG LG KP+
Sbjct: 1011 AVDILKSQMKSFFKLRDLGELKYFLGLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSS 1070
Query: 1255 FPMEQHHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWE 1314
PM+ A SG + PYRRLIGRL+YL++TRPD+ ++V+ L+QF P + H +
Sbjct: 1071 IPMDPSMVFAHDSGGDFVEVGPYRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQ 1130
Query: 1315 AALRVVRYLKKHPGQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWK 1374
A ++++Y+K GQG+ + SEL+L+ + ++D+ SC +RRS +G+ + L S + WK
Sbjct: 1131 AVYKILQYIKGTIGQGLFYSATSELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWK 1190
Query: 1375 TKKQPTVSRSSAEAEYRSMAMTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNP 1434
++KQ VS+SSAEAEYRS+++ T EL WL L +L V S+ L+CD+++A+HIA N
Sbjct: 1191 SRKQDVVSKSSAEAEYRSLSVATDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNH 1250
Query: 1435 VFHERTKHIEADCHFVRDAVVAGIICPLYVPTSVQLADIFTKALGKAQFEFLLRKLGIRD 1494
VFHERTKHIE+DCH VR+ ++ G+ ++ T +Q+AD FTK L + F L+ K+G+ +
Sbjct: 1251 VFHERTKHIESDCHSVRERLLKGLFELYHINTELQIADPFTKPLYPSHFHRLISKMGLLN 1310
Query: 1495 L 1495
+
Sbjct: 1311 I 1311
Score = 122 bits (305), Expect = 2e-27
Identities = 77/267 (28%), Positives = 130/267 (47%), Gaps = 22/267 (8%)
Query: 58 TSFQAKRKYGFVEGKIPKPTTPEKL-EDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAES 116
TS +AK K GFV+G IPKP + + W+ SM+ +WLLN++ + +++ Y+ A +
Sbjct: 3 TSIEAKNKLGFVDGSIPKPDDDDPYCKIWRRCNSMVKSWLLNSVSKEIYTSILYFPTAAA 62
Query: 117 LWTHLKQRFCVVNGARICQLKASLGECKQGKGEEVSAYFGRLSRIWDELVTYVKKPTCKC 176
+W L RF + R+ +L+ + +QG ++S+Y R +W+EL + P
Sbjct: 63 IWKDLYTRFHKSSLPRLYKLRQQIHSLRQG-NLDLSSYHTRTQTLWEELTSLQAVPRTVE 121
Query: 177 EGCICDINKQVTDFSAEDYLHHFLMGLDGAYATIRSNLLSQDPLPNIDQAYQRVIQDERL 236
+ I +V D FLMGL+ Y T+RS +L + LP++ + + + QDE
Sbjct: 122 DLLIERETNRVID---------FLMGLNDCYDTVRSQILMKKTLPSLSEVFNMIDQDETQ 172
Query: 237 RQGEYSIQQNRDNVMAFKVAPDTRGKSKLVDNSDKF-------CTHCNREGHDERTCFQI 289
R S M V P + S+ N D + C++C+R GH E TC++
Sbjct: 173 RSARISTTPG----MTSSVFPVSNQSSQSALNGDTYQKKERPVCSYCSRPGHVEDTCYKK 228
Query: 290 HGFPEWWGDRPRGGRGSGRGGATTGRD 316
HG+P + + + + S A G +
Sbjct: 229 HGYPTSFKSKQKFVKPSISANAAIGSE 255
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 733 bits (1892), Expect = 0.0
Identities = 430/1118 (38%), Positives = 604/1118 (53%), Gaps = 129/1118 (11%)
Query: 395 SWIIDTGASHHVTGNFSCLINGKRITNTPVGLPNGKDATAIQEGSVILDGGLRLNNVLFV 454
+WIID+GAS HV + + K ++ G+V + L L+NVL V
Sbjct: 481 AWIIDSGASSHVCSDLAMFRELKSVS-----------------GTVHITQKLILHNVLHV 523
Query: 455 PQLTCNLISVTQLIDDSNCIVQFTNALCVIQDRTTRTLIGAGERIDGLYFFR------GV 508
P NL+SV+ L+ +C F C+IQ+ + +IG G LY
Sbjct: 524 PDFKFNLMSVSSLVKTISCSAHFYVDCCLIQELSQGLMIGRGRLYHNLYILETENTSPST 583
Query: 509 PKVHALMVEGDSAMD--LWHKRLGHPSEKVLKFIPHVSQHSRSKNNRPCDVCPRAKQHRD 566
A + G D LWH+RLGHPS VL+ + ++
Sbjct: 584 STPAACLFTGSVLNDGHLWHQRLGHPSSVVLQKLKRLAY--------------------- 622
Query: 567 SFPLSENNAASL-FELVHCDLWGSYRTRSSCGAQYYLTIVNDYSRAVWVYLLCNKTEIET 625
+S NN AS F+LVH D+WG + S G +Y+LT+V+D +R WVY+L NK ++ +
Sbjct: 623 ---ISHNNLASNPFDLVHLDIWGPFSIESIEGFRYFLTVVDDCTRTTWVYMLRNKKDVSS 679
Query: 626 MFLNFVAFVDRQFDKKIKKVRSDNGTEFNCLRDYFFNNGIVFETSCVGTPQQNGRVERKH 685
+F F+ V QF+ KIK +RSDN E + +G++ SC TPQQN VERKH
Sbjct: 680 VFPEFIKLVSTQFNAKIKAIRSDNAPELG-FTEIVKEHGMLHHFSCAYTPQQNSVVERKH 738
Query: 686 QHIMNVARALRFQGHLPMQFWGECVLTACYLINRTPSSVLNYKTPYEKLFGKVPKFDNMK 745
QHI+NVARAL FQ ++PMQ+W +CV TA +LINR PS +LN K+PYE + K P + +K
Sbjct: 739 QHILNVARALLFQSNIPMQYWSDCVTTAVFLINRLPSPLLNNKSPYELILNKQPDYSLLK 798
Query: 746 IFGCLCYAHNQRRDGDKFASRSRKCIFVGYPYGKKGWKLYDLESKEYIVSRDVKFYEHEF 805
FGCLC+ + KF R+R C+F+GYP G KG+K+ DLES VSR+V F EH F
Sbjct: 799 NFGCLCFVSTNAHERTKFTPRARACVFLGYPSGYKGYKVLDLESHSVTVSRNVVFKEHVF 858
Query: 806 PFDVQLDTTHSTPFIDSEYVTEDIGFETYDASFEGGGASMALQDNEQTQLQGGLHGDCNG 865
PF + + + + +M L D + D
Sbjct: 859 PFKTSELLNKAVDMFPNSILPLPAPLHFVE--------TMPLIDEDSLI---PTTTDSRT 907
Query: 866 ADVAANEGLSVSGIEGEVEPAHEVDGADVVVMQEDDVEPGEPEVVAEREAIVASEMGRGM 925
AD A+ S S + + P+ + D+ P ++R S +
Sbjct: 908 ADNHASS--SSSALPSIIPPSSNTETQDID-------SNAVPITRSKRTTRAPSYLSEYH 958
Query: 926 RNKVPNIKLKDFVTHTIRKVKSSKSSSAQEDASGTPYPITYFVSCERFSIRHRNFVAAVT 985
+ VP+I +I + +A TPYPI+ VS ++++ ++++ A
Sbjct: 959 CSLVPSISTLPPTDSSIPIHPLPEIFTASSPKKTTPYPISTVVSYDKYTPLCQSYIFAYN 1018
Query: 986 AGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVMEKLPPGKKALGSKWVYKIKHHSDG 1045
EP F +A+K W E+QA+E N+TW +E LPP K +G KWV+ IK++ DG
Sbjct: 1019 TETEPKTFSQAMKSEKWIRVAVEELQAMELNKTWSVESLPPDKNVVGCKWVFTIKYNPDG 1078
Query: 1046 SIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVAAIKKWEVHQMDVHNAFLH 1105
++ER KARLV G Q EGID+ +TF+PVAK+ + + L +AAI W + QMDV +AFLH
Sbjct: 1079 TVERYKARLVAQGFTQQEGIDFLDTFSPVAKLTSAKMMLGLAAITGWTLTQMDVSDAFLH 1138
Query: 1106 GDLEEEVYMKVPPGFKNT-----DPNLVCRLKKSLYGLKQAPRCWFAKLVTALKRYGFVQ 1160
GDL+EE++M +P G+ PN VCRL KS+YGLKQA R W+ + V AL
Sbjct: 1139 GDLDEEIFMSLPQGYTPPAGTILPPNPVCRLLKSIYGLKQASRQWYKRFVAAL------- 1191
Query: 1161 SYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIAALKIFKAYLGVCFHMKDLGVLKYFLG 1220
VY+DD++IA N+ A ++ KA L F +KDLG ++FLG
Sbjct: 1192 --------------------VYIDDIMIASNNDAEVENLKALLRSEFKIKDLGPARFFLG 1231
Query: 1221 LEVARNHEGIYLCQRKYALEIIDETGLLGAKPADFPMEQHHKLALVSGKPLEDPEPYRRL 1280
L LG KP+ PM+ L G PL +P YR+L
Sbjct: 1232 L--------------------------LGCKPSSIPMDPTLHLVRDMGTPLPNPTAYRKL 1265
Query: 1281 IGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEAALRVVRYLKKHPGQGILLRSDSELK 1340
IGRL+YL++TRPD+ Y+VH LSQF+ P + H +AA +V+RY+K +PGQG++ +D E+
Sbjct: 1266 IGRLLYLTITRPDITYAVHQLSQFISAPSDIHLQAAHKVLRYIKANPGQGLMYSADYEIC 1325
Query: 1341 LEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMAMTTCEL 1400
L G+ D+DWA+C TRRS++G+ + L S +SWK+KKQ SRSS E+EYRSMA TCE+
Sbjct: 1326 LNGFSDADWAACKDTRRSISGFCIYLGTSLISWKSKKQAVASRSSTESEYRSMAQATCEI 1385
Query: 1401 KWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVFHERTKHIEADCHFVRDAVVAGIIC 1460
WL+QLL DL + + +L+CD+KSALH + NPVFHERTKHIE DCH VRD + AG +
Sbjct: 1386 IWLQQLLKDLHIPLTCPAKLFCDNKSALHSSLNPVFHERTKHIEIDCHTVRDQIKAGNLK 1445
Query: 1461 PLYVPTSVQLADIFTKALGKAQFEFLLRKLGIRDLHAP 1498
L+VPT Q ADI TKAL F LLR++ + L P
Sbjct: 1446 ALHVPTENQHADILTKALHPGPFHHLLRQMSLSSLFLP 1483
Score = 128 bits (322), Expect = 2e-29
Identities = 75/281 (26%), Positives = 131/281 (45%), Gaps = 22/281 (7%)
Query: 27 FFLGSNDNPGNVITPIQLR-GLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPTTPEKLED- 84
++L S D+ G ++ +L ++ W R+I + + K GF+ G I KP PE D
Sbjct: 34 YYLHSADHAGLILVSDRLTTASDFHSWRRSILMALNVRNKLGFINGTITKP--PEDHRDF 91
Query: 85 --WKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGE 142
W ++ WL+N+++ + +L Y + +W +L RF + RI ++ L +
Sbjct: 92 GAWSRCNDIVSTWLMNSVDKKIGQSLLYIATVQGIWNNLLSRFKQDDAPRIFDIEQKLSK 151
Query: 143 CKQGKGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAEDYLHHFLMG 202
+QG ++S Y+ L +W+E YV+ P C C C CD + + FL
Sbjct: 152 IEQGS-MDISTYYTALLTLWEEHRNYVELPVCTCGRCECDAAVKWEHLQQRSRVTKFLKE 210
Query: 203 LDGAYATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGE----------YSIQQNRDNVMA 252
L+ + R ++L P+P I +A+ V QDER R + + ++ A
Sbjct: 211 LNEGFDQTRRHILMLKPIPTIKEAFNMVTQDERQRNVKPLTRVDSVAFQNTSMINEDENA 270
Query: 253 FKVAPDTRGKSKLVDNSDKFCTHCNREGHDERTCFQIHGFP 293
+ A +T + N CTHC + GH + C+++HG+P
Sbjct: 271 YVAAYNT-----VRPNQKPICTHCGKVGHTIQKCYKVHGYP 306
>At4g22040 LTR retrotransposon like protein
Length = 1109
Score = 723 bits (1867), Expect = 0.0
Identities = 348/531 (65%), Positives = 430/531 (80%)
Query: 968 VSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVMEKLPPG 1027
+SC RF+ H+ F+AAVTAG EP + EA+ D WR+AM EI++L N+T+ + LPPG
Sbjct: 578 MSCNRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRVNQTFSIVNLPPG 637
Query: 1028 KKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVA 1087
K+ALG+KWVYKIK+ SDG+IER KARLVV G+ Q EG+DYDETFAPVAKM TVR FL VA
Sbjct: 638 KRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVAKMSTVRLFLGVA 697
Query: 1088 AIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGFKNTDPNLVCRLKKSLYGLKQAPRCWFA 1147
A + W VHQMDVHNAFLHGDL+EEVYMK+P GF+ DP+ VCRL KSLYGLKQAPRCWF+
Sbjct: 698 AARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSLYGLKQAPRCWFS 757
Query: 1148 KLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIAALKIFKAYLGVCF 1207
KL +ALK+YGF QS SDYSLF+ + + ++VLVYVDDLII+G+ A+ FK+YL CF
Sbjct: 758 KLSSALKQYGFTQSLSDYSLFSYNNDGVFVHVLVYVDDLIISGSCPDAVAQFKSYLESCF 817
Query: 1208 HMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAKPADFPMEQHHKLALVS 1267
HMKDLG+LKYFLG+EV+RN +G YL QRKY L+II E GLLGA+P+ FP+EQ+HKL+L +
Sbjct: 818 HMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGARPSAFPLEQNHKLSLST 877
Query: 1268 GKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEAALRVVRYLKKHP 1327
L D YRRL+GRLIYL+VTRP+L+YSVH L+QFMQ P ++HW AA+RVVRYLK +P
Sbjct: 878 SPLLSDSSRYRRLVGRLIYLAVTRPELSYSVHTLAQFMQNPRQDHWNAAIRVVRYLKSNP 937
Query: 1328 GQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAE 1387
GQGILL S S L++ GWCDSD+A+CPLTRRSLTG+ V L +P+SWKTKKQPT+SRSSAE
Sbjct: 938 GQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQLGDTPISWKTKKQPTISRSSAE 997
Query: 1388 AEYRSMAMTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVFHERTKHIEADC 1447
AEYR+MA T EL WLK++L DLGVSH Q M+++ DSKSA+ ++ NPV HERTKH+E DC
Sbjct: 998 AEYRAMAFLTQELMWLKRVLYDLGVSHVQAMRIFSDSKSAIALSVNPVQHERTKHVEVDC 1057
Query: 1448 HFVRDAVVAGIICPLYVPTSVQLADIFTKALGKAQFEFLLRKLGIRDLHAP 1498
HF+RDA++ GII +VP+ QLADI TKALG+ + + LRKLGI D+HAP
Sbjct: 1058 HFIRDAILDGIIATSFVPSHKQLADILTKALGEKEVRYFLRKLGILDVHAP 1108
Score = 468 bits (1205), Expect = e-132
Identities = 216/407 (53%), Positives = 295/407 (72%), Gaps = 1/407 (0%)
Query: 402 ASHHVTGNFSCLINGKRITNTPVGLPNGKDATAIQEGSVILDGGLRLNNVLFVPQLTCNL 461
ASHH+TGN L + ++ + L +G A+ EG+V L L L +V +V +L +L
Sbjct: 169 ASHHMTGNLELLSGMRSMSPVLIILADGNKRVAVSEGTVRLGSHLILKSVFYVKELESDL 228
Query: 462 ISVTQLIDDSNCIVQFTNALCVIQDRTTRTLIGAGERIDGLYFFRGVPKVHALMVEGDSA 521
ISV Q++D+++C+VQ + VIQDRTTR + G G+R +G + FRG+ A+ +
Sbjct: 229 ISVGQMMDENHCVVQLADHFLVIQDRTTRMVTGIGKRENGSFCFRGMENAAAVHTSVKAP 288
Query: 522 MDLWHKRLGHPSEKVLKFIPH-VSQHSRSKNNRPCDVCPRAKQHRDSFPLSENNAASLFE 580
DLWH+RLGH S+K++ +P + + CD C RAKQ RD+FPL +N + F+
Sbjct: 289 FDLWHRRLGHASDKIVNLLPRELLSSGKEILENVCDTCMRAKQTRDTFPLRDNRSMDSFQ 348
Query: 581 LVHCDLWGSYRTRSSCGAQYYLTIVNDYSRAVWVYLLCNKTEIETMFLNFVAFVDRQFDK 640
L+HCD+WG YRT S GA+Y+LTIV+DYSR VWVYL+ +K+E + +F+A V+RQFD
Sbjct: 349 LIHCDVWGPYRTPSYSGARYFLTIVDDYSRGVWVYLMTDKSETQKHLKDFMALVERQFDT 408
Query: 641 KIKKVRSDNGTEFNCLRDYFFNNGIVFETSCVGTPQQNGRVERKHQHIMNVARALRFQGH 700
+IK VRSDNGTEF C+R+YF + GI ETSCVGTP QNGRVERKH+HI+N+ARALRFQ +
Sbjct: 409 EIKTVRSDNGTEFLCMREYFLHKGIAHETSCVGTPHQNGRVERKHRHILNIARALRFQSY 468
Query: 701 LPMQFWGECVLTACYLINRTPSSVLNYKTPYEKLFGKVPKFDNMKIFGCLCYAHNQRRDG 760
LP+QFWGEC+L+A YLINRTPS +L K+PYE L+ PK+ ++++FG LCYAHNQ G
Sbjct: 469 LPIQFWGECILSAAYLINRTPSMLLQGKSPYEMLYKTAPKYSHLRVFGSLCYAHNQNHKG 528
Query: 761 DKFASRSRKCIFVGYPYGKKGWKLYDLESKEYIVSRDVKFYEHEFPF 807
DKFA+RSR+C+FVGYP+G+KGW+L+DLE +++ VSRDV F E EFP+
Sbjct: 529 DKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIFQETEFPY 575
Score = 115 bits (289), Expect = 1e-25
Identities = 55/150 (36%), Positives = 98/150 (64%), Gaps = 4/150 (2%)
Query: 21 KKVDLVFFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPT--T 78
++ L + L + DN G VI+ L+ NY+EW+ +T+ ++++K+GF++G IP+P +
Sbjct: 15 RRTILPYDLTAADNSGAVISHPILKTNNYEEWACGFKTALRSRKKFGFLDGTIPQPLDGS 74
Query: 79 PEKLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKA 138
P+ LEDW + ++L++W+ TI+ L + +S+ D A LW +++RF V NG + ++KA
Sbjct: 75 PD-LEDWLTINALLVSWMKMTIDSELLTNISHRDVARDLWEQIRKRFSVSNGPKNQKMKA 133
Query: 139 SLGECKQGKGEEVSAYFGRLSRIWDELVTY 168
L CKQ +G V Y+G+L++IWD + +Y
Sbjct: 134 DLATCKQ-EGMTVEGYYGKLNKIWDNINSY 162
>At2g24660 putative retroelement pol polyprotein
Length = 1156
Score = 668 bits (1724), Expect = 0.0
Identities = 345/620 (55%), Positives = 445/620 (71%), Gaps = 32/620 (5%)
Query: 906 EPEVVAEREAIVASEMGRGMRNKVPNIKLKDFVTH--TIRKVK------SSKSSSAQEDA 957
+P V + + A+ + R +++L+D+V + T+ + SS SS+
Sbjct: 547 QPLFVVDSPFVEATSPRQRKRQIRQSVRLQDYVLYNATVSPINPHALPDSSSQSSSMVQG 606
Query: 958 SGTPYPITYFVSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNE 1017
+ + YP++ +VS + FS H+ F+AA+TA EP +FKEAV+ W DAM E+ ALE N+
Sbjct: 607 TSSLYPLSDYVSDDCFSAGHKAFLAAITANDEPKHFKEAVRIKVWNDAMFKEVDALEINK 666
Query: 1018 TWVMEKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKM 1077
TW + LPPGK A+GS+WVYK K+++DGSIER KARLVV G+ Q+EG DY+ETFAPV KM
Sbjct: 667 TWDIVDLPPGKVAIGSQWVYKTKYNADGSIERYKARLVVQGNKQVEGEDYNETFAPVVKM 726
Query: 1078 VTVRTFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGFKNTDPNLVCRLKKSLYG 1137
TVRT L + A +WEV+QMDV+NAFLHGDL+EEVYMK+PPGF+++ P+ VCRL+KSLYG
Sbjct: 727 TTVRTLLRLVAANQWEVYQMDVNNAFLHGDLDEEVYMKLPPGFRHSHPDKVCRLRKSLYG 786
Query: 1138 LKQAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIAALK 1197
LKQAPRCWF KL AL R+GFVQ + DYS F+ R I++ VLVYVDDL+I GND L+
Sbjct: 787 LKQAPRCWFKKLSDALLRFGFVQGHEDYSFFSYTRNGIELRVLVYVDDLLICGNDGYMLQ 846
Query: 1198 IFKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAKPADFPM 1257
FK YLG CF MKDLG LKYFLG+EV+R EGI+L QRKYAL+II ++G LG +PA P+
Sbjct: 847 KFKEYLGRCFSMKDLGKLKYFLGIEVSRGSEGIFLSQRKYALDIITDSGNLGCRPALTPL 906
Query: 1258 EQHHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEAAL 1317
EQ+H LA G L D +PYRRL+GRL+YL TRP+L+YSVH+LSQFMQ P E H AA+
Sbjct: 907 EQNHHLATDDGPLLADAKPYRRLVGRLLYLLHTRPELSYSVHVLSQFMQTPREAHLAAAM 966
Query: 1318 RVVRYLKKHPGQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTKK 1377
R+VR+LK PGQGILL +D+EL+L+ +CDSD+ +CP TRRSL+ +VVLL SP+SWKTKK
Sbjct: 967 RIVRFLKGSPGQGILLNADTELQLDVYCDSDFQTCPKTRRSLSAYVVLLGGSPISWKTKK 1026
Query: 1378 QPTVSRSSAEAEYRSMAMTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVFH 1437
Q TVS SSAEAEYR+M++ E+KWL++LL +L NPVFH
Sbjct: 1027 QDTVSHSSAEAEYRAMSVALREIKWLRKLLKELA---------------------NPVFH 1065
Query: 1438 ERTKHIEADCHFVRDAVVAGIICPLYVPTSVQLADIFTKALGKAQFEFLLRKLGIRDLHA 1497
ERTKHIE+DCH VRDAV GII +V T+ QLADIFTKALG++QF +L+ KLGI+DLH
Sbjct: 1066 ERTKHIESDCHSVRDAVRDGIITTHHVRTNEQLADIFTKALGRSQFLYLMSKLGIQDLHT 1125
Query: 1498 PEGGYWDYCDIIVIYFRLMW 1517
P G C + + L+W
Sbjct: 1126 PTNGRHICCQEMEL---LLW 1142
Score = 398 bits (1023), Expect = e-110
Identities = 196/372 (52%), Positives = 253/372 (67%), Gaps = 12/372 (3%)
Query: 486 DRTTRTLIGAGERIDGLYFFRGVP--KVHALMVEGDSAMDLWHKRLGHPSEKVLKFIPHV 543
DR +RTLIG+GE DG+Y+ V K+H V D A LWH+RLGHPS VL +P +
Sbjct: 52 DRFSRTLIGSGEERDGVYYLTDVATAKIHTAKVSSDQA--LWHQRLGHPSFSVLSSLPVL 109
Query: 544 SQHSRSKNNRPCDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGSYRTRSSCGAQYYLT 603
+ S S +R CDVC RAKQ R+ FP+S N + F L+HCD+WG YR SSCGA Y+LT
Sbjct: 110 TSSSLSVGSRSCDVCFRAKQTREVFPVSTNKSIECFSLIHCDVWGPYRVPSSCGAVYFLT 169
Query: 604 IVNDYSRAVWVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTEFNCLRDYFFNN 663
IV+D+SRAVW YLL K+E+ T+ NF+ + ++QF K +K +RSDNGTEF CL YF +
Sbjct: 170 IVDDFSRAVWTYLLLAKSEVRTVLTNFLVYTEKQFGKSVKVLRSDNGTEFMCLASYFREH 229
Query: 664 GIVFETSCVGTPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLTACYLINRTPSS 723
GIV +TSCVGTPQQNGRVERKH+HI+NVARA+ FQ LP+QFWGE VLTA YLINRTP+S
Sbjct: 230 GIVHQTSCVGTPQQNGRVERKHRHILNVARAILFQASLPIQFWGEAVLTAAYLINRTPTS 289
Query: 724 VLNYKTPYEKLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIFVGYPYGKKGWK 783
+ N +PYE L P ++++++FG CY H RD DKF RSR C+F+GYP+ +KGWK
Sbjct: 290 LHNGLSPYEILHNSKPNYEHLRVFGSACYVHRASRDKDKFGERSRLCVFIGYPFAQKGWK 349
Query: 784 LYDLESKEYIVSRDVKFYEHEFPFDVQLDTTHSTPFIDSEYVTED--IGFETYDASFEGG 841
++D+E KE++VSRDV F E FP+ +T H + V ED + T D
Sbjct: 350 VFDMEKKEFLVSRDVVFREDVFPY-AATNTDHVSASPAPLLVDEDWMVSTTTLDR----- 403
Query: 842 GASMALQDNEQT 853
G+S L N+ T
Sbjct: 404 GSSAVLSANKNT 415
>At4g10690 retrotransposon like protein
Length = 1515
Score = 648 bits (1671), Expect = 0.0
Identities = 449/1471 (30%), Positives = 714/1471 (48%), Gaps = 134/1471 (9%)
Query: 67 GFVEGKIPKPTTP--------------EKLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYD 112
GFV G P+P + ++ W + ++ AW+ ++ + +
Sbjct: 40 GFVTGATPRPASTIIVTKDDIQSEEANQEFLKWTRIDQLVKAWIFGSLSEEALKVVIGLN 99
Query: 113 DAESLWTHLKQRFCVVNGARICQLKASLGECKQGKGEEVSAYFGRLSRIWDELVTYVKKP 172
A+ +W L +RF + R L+ LG C + G+ + AY + I D+L +
Sbjct: 100 SAQEVWLGLARRFNRFSTTRKYDLQKRLGTCSKA-GKTMDAYLSEVKNICDQLDS----- 153
Query: 173 TCKCEGCICDINKQVTDFSAEDYLHHFLMGLDGAY---ATIRSNLLSQDPLPNIDQAYQR 229
I VT+ ++ + L GL Y AT+ + L P P D
Sbjct: 154 ----------IGFPVTE---QEKIFGVLNGLGKEYESIATVIEHSLDVYPGPCFDDVVY- 199
Query: 230 VIQDERLRQGEYSIQQNRDNVMAFKVAPDTRGKSKLVDNSDKFCTHCNREGHDERTCFQI 289
KL DK T+ F
Sbjct: 200 ----------------------------------KLTTFDDKLSTYTANSEVTPHLAF-- 223
Query: 290 HGFPEWWGDRPRGGRGSGRGGATTGRDTTGRSGG-RGRGTYNALARANKATTSSRSIGGN 348
+ D+ RG+ GR G RGRG+Y++ R S S G+
Sbjct: 224 ------YTDKSYSSRGNNNSRG-------GRYGNFRGRGSYSSRGRGFHQQFGSGSNNGS 270
Query: 349 SGQSHAPPHSSEAAGISGI---TPAQWQQILDALNISKTKDRLHGKNDIS---WIIDTGA 402
S G S T + + + L + R+ +N S W+ D+ A
Sbjct: 271 GNGSKPTCQICRKYGHSAFKCYTRFEENYLPEDLPNAFAAMRVSDQNQASSHEWLPDSAA 330
Query: 403 SHHVTGNFSCLINGKRIT-NTPVGLPNGKDATAIQEGSVILD---GGLRLNNVLFVPQLT 458
+ H+T L N + + + V + NG G++ L+ G L L +VL P +T
Sbjct: 331 TAHITNTTDGLQNSQTYSGDDSVIVGNGDFLPITHIGTIPLNISQGTLPLEDVLVCPGIT 390
Query: 459 CNLISVTQLIDDSNCIVQFTNALCVIQDRTTRTLIGAGERIDGLYFFRGVP-KVHALMVE 517
+L+SV++L DD C F + VI+D+ T+ L+ G + GLY + VP + + +
Sbjct: 391 KSLLSVSKLTDDYPCSFTFDSDSVVIKDKRTQQLLTQGNKHKGLYVLKDVPFQTYYSTRQ 450
Query: 518 GDSAMDLWHKRLGHPSEKVLKFIPHVSQHSRSK-NNRPCDVCPRAKQHRDSFPLSENNAA 576
S ++WH+RLGHP+++VL+ + +K ++ C+ C K R F SE ++
Sbjct: 451 QSSDDEVWHQRLGHPNKEVLQHLIKTKAIVVNKTSSNMCEACQMGKVCRLPFVASEFVSS 510
Query: 577 SLFELVHCDLWGSYRTRSSCGAQYYLTIVNDYSRAVWVYLLCNKTEIETMFLNFVAFVDR 636
E +HCDLWG S+ G QYY+ +++YSR W Y L K++ ++F+ F V+
Sbjct: 511 RPLERIHCDLWGPAPVTSAQGFQYYVIFIDNYSRFTWFYPLKLKSDFFSVFVLFQQLVEN 570
Query: 637 QFDKKIKKVRSDNGTEFNCLR--DYFFNNGIVFETSCVGTPQQNGRVERKHQHIMNVARA 694
Q+ KI + D G EF + + + GI SC TPQQNG ER+H+++ + +
Sbjct: 571 QYQHKIAMFQCDGGGEFVSYKFVAHLASCGIKQLISCPHTPQQNGIAERRHRYLTELGLS 630
Query: 695 LRFQGHLPMQFWGECVLTACYLINRTPSSVL-NYKTPYEKLFGKVPKFDNMKIFGCLCYA 753
L F +P + W E T+ +L N PSS L + K+PYE L G P + +++FG CY
Sbjct: 631 LMFHSKVPHKLWVEAFFTSNFLSNLLPSSTLSDNKSPYEMLHGTPPVYTALRVFGSACYP 690
Query: 754 HNQRRDGDKFASRSRKCIFVGYPYGKKGWKLYDLESKEYIVSRDVKFYEHEFPFD---VQ 810
+ + +KF +S C+F+GY KG++ + + + R V F E +FP+ Q
Sbjct: 691 YLRPYAKNKFDPKSLLCVFLGYNNKYKGYRCLHPPTGKVYICRHVLFDERKFPYSDIYSQ 750
Query: 811 LDTTHSTPFIDSEYVTEDIGFETYDASFEGGGASMALQDNEQTQLQGGLHGDC--NGADV 868
T +P + GF + S E ++ + + C N A+
Sbjct: 751 FQTISGSPL----FTAWQKGFSSTALSRETPSTNVEDIIFPSATVSSSVPTGCAPNIAET 806
Query: 869 AANEGLSVSGIEGEVEPAHEVDGADVVVMQEDDVEPGEPEVVAEREAIVASEMGRGMRNK 928
A + V+ V P + + E+ + + E ++S M
Sbjct: 807 ATAPDVDVAAAHDMVVPPSPITSTSLPTQPEESTS-DQNHYSTDSETAISSAM------T 859
Query: 929 VPNIKLKDFVTHTIRKVKSSKSSSAQEDASGTPYPITYFVSCERFSIRHRNFVAAVTAGK 988
+I + F ++S SS+ A T +P+ ++ + I N A+ + K
Sbjct: 860 PQSINVSLFEDSDFPPLQSVISSTTA--APETSHPM---ITRAKSGITKPNPKYALFSVK 914
Query: 989 ----EPNNFKEAVKDSGWRDAMRNEIQALEDNETWVMEKLPPGKKALGSKWVYKIKHHSD 1044
EP + KEA+KD GW +AM E+ + + +TW + + LG KWV+K K +SD
Sbjct: 915 SNYPEPKSVKEALKDEGWTNAMGEEMGTMHETDTWDLVPPEMVDRLLGCKWVFKTKLNSD 974
Query: 1045 GSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVAAIKKWEVHQMDVHNAFL 1104
GS++RLKARLV G+ Q EG+DY ET++PV + TVR+ L VA I KW + Q+DV NAFL
Sbjct: 975 GSLDRLKARLVARGYEQEEGVDYVETYSPVVRSATVRSILHVATINKWSLKQLDVKNAFL 1034
Query: 1105 HGDLEEEVYMKVPPGFKN-TDPNLVCRLKKSLYGLKQAPRCWFAKLVTALKRYGFVQSYS 1163
H +L+E V+M PPGF++ + P+ VC+LKK++Y LKQAPR WF K + L +YGF+ S+S
Sbjct: 1035 HDELKETVFMTQPPGFEDPSRPDYVCKLKKAIYDLKQAPRAWFDKFSSYLLKYGFICSFS 1094
Query: 1164 DYSLFTLHRGEIQINVLVYVDDLIIAGNDIAALKIFKAYLGVCFHMKDLGVLKYFLGLEV 1223
D SLF +G + +L+YVDD+I+ GN+ L+ L F MKD+G L YFLG++
Sbjct: 1095 DPSLFVYLKGRDVMFLLLYVDDMILTGNNDVLLQQLLNILSTEFRMKDMGALHYFLGIQA 1154
Query: 1224 ARNHEGIYLCQRKYALEIIDETGLLGAKPADFPMEQHHKLALVSG--KPLEDPEPYRRLI 1281
+++G++L Q KY +++ G+ P++ L L+ G KP +P +RRL
Sbjct: 1155 HYHNDGLFLSQEKYTSDLLVNAGMSDCSSMPTPLQ----LDLLQGNNKPFPEPTYFRRLA 1210
Query: 1282 GRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEAALRVVRYLKKHPGQGILLRSDSELKL 1341
G+L YL++TRPD+ ++V+ + Q M P + R++ YLK GI L S+++ L
Sbjct: 1211 GKLQYLTLTRPDIQFAVNFVCQKMHAPTMSDFHLLKRILHYLKGTMTMGINLSSNTDSVL 1270
Query: 1342 EGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMAMTTCELK 1401
+ DSDWA C TRRS G+ L + +SW K+ PTVS+SS EAEYR+++ E+
Sbjct: 1271 RCYSDSDWAGCKDTRRSTGGFCTFLGYNIISWSAKRHPTVSKSSTEAEYRTLSFAASEVS 1330
Query: 1402 WLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVFHERTKHIEADCHFVRDAVVAGIICP 1461
W+ LL ++G+ Q ++YCD+ SA++++ NP H R+KH + D ++VR+ V G +
Sbjct: 1331 WIGFLLQEIGLPQQQIPEMYCDNLSAVYLSANPALHSRSKHFQVDYYYVRERVALGALTV 1390
Query: 1462 LYVPTSVQLADIFTKALGKAQFEFLLRKLGI 1492
++P S QLADIFTK+L +A F L KLG+
Sbjct: 1391 KHIPASQQLADIFTKSLPQAPFCDLRFKLGV 1421
>At2g06840 putative retroelement pol polyprotein
Length = 1102
Score = 621 bits (1601), Expect = e-177
Identities = 332/599 (55%), Positives = 403/599 (66%), Gaps = 48/599 (8%)
Query: 900 DDVEPGEPEVVAEREAIVASEMGRGMRNKVPNIKLKDFVTHTIRKVKSSKSSSAQEDASG 959
D PG PE+ +G+G R K P + LKD+V H KV SS +S+ +
Sbjct: 551 DSSSPGLPEL-----------LGKGHRPKHPPVYLKDYVAH---KVHSSPHTSSPGLSDS 596
Query: 960 TPYPITYFVSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETW 1019
P S H F+AA+ E N+FK+ V W DAM+ EI+ALE N TW
Sbjct: 597 NVSPTV--------SANHIAFMAAILDSNEQNHFKDDVLIKEWCDAMQKEIEALEANHTW 648
Query: 1020 VMEKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVT 1079
+ LP GKKA+ SKWVYK+K +SDG++ER KARLVV G+HQ EGID+ ETFAPVAKM T
Sbjct: 649 DVTDLPHGKKAISSKWVYKLKFNSDGTLERHKARLVVMGNHQKEGIDFKETFAPVAKMTT 708
Query: 1080 VRTFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGFKNTDPNLVCRLKKSLYGLK 1139
VR LAVAA K W+V QMDVHNAFLHGDLE+E SLYGLK
Sbjct: 709 VRLLLAVAAAKDWDVFQMDVHNAFLHGDLEQE----------------------SLYGLK 746
Query: 1140 QAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIAALKIF 1199
QAPRCWFAKL TAL++ GF QSY DYSLF+L+R I+ LVYVDD II GN++ A+ F
Sbjct: 747 QAPRCWFAKLSTALRKLGFTQSYEDYSLFSLNRDGTVIHFLVYVDDFIIVGNNLKAIDHF 806
Query: 1200 KAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAKPADFPMEQ 1259
K +L CFHMKDLG LKYFLGLEV+R +G L Q+KYAL+II+E GLLG KP+ PME
Sbjct: 807 KEHLHKCFHMKDLGKLKYFLGLEVSRGADGFCLSQQKYALDIINEAGLLGYKPSAVPMEL 866
Query: 1260 HHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEAALRV 1319
HHKL +S ++P YRRL+ R IYL++TRPDL+Y+VHILSQFMQ P E HW A LR+
Sbjct: 867 HHKLGSISSPVFDNPAQYRRLVDRFIYLTITRPDLSYAVHILSQFMQTPLEAHWHATLRL 926
Query: 1320 VRYLKKHPGQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQP 1379
VRYLK P QGILLRSD L L +CDSD+ CP TRRSL+ +V+ L +P+SWKTKKQ
Sbjct: 927 VRYLKGSPDQGILLRSDRALSLTAYCDSDYNPCPRTRRSLSAYVLYLGDTPISWKTKKQD 986
Query: 1380 TVSRSSAEAEYRSMAMTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVFHER 1439
TVS SSAEAEYR+MA T E+KWLK L+ LGV H+Q + L+CDS++A+HIA NPVFHER
Sbjct: 987 TVSSSSAEAEYRAMAYTLKEIKWLKALMTTLGVDHTQPILLFCDSQAAIHIAANPVFHER 1046
Query: 1440 TKHIEADCHFVRDAVVAGIICPLYVPTSVQLADIFTKALGKAQFEFLLRKLGIRDLHAP 1498
TKHIE DCH VRDAV +I ++ T+ D+ TKAL + FE LL LG + P
Sbjct: 1047 TKHIEKDCHQVRDAVTDKVISTPHISTT----DLLTKALPRPTFERLLSTLGTCNYDLP 1101
Score = 318 bits (816), Expect = 1e-86
Identities = 216/595 (36%), Positives = 289/595 (48%), Gaps = 128/595 (21%)
Query: 199 FLMGLD-GAYATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGEYSIQQNRDNVMAFKVAP 257
FL LD +A IRS + +DPLP+ ++ Y RVI+ ++ S + + F V
Sbjct: 6 FLFELDLPRFAPIRSKITDEDPLPSHNRVYSRVIRGQQNLDVARSKETTNSEAINFSVKT 65
Query: 258 DTRGKSKLV---DNSDKFCTHCNREGHDERTCFQIHGFPEWWGDRPRGGRGSGRGGATTG 314
+ + V D+ CTHC+R+GHD CF +HGFPEW+ ++ G R S
Sbjct: 66 PSAPQVAAVYAPKPRDRSCTHCHRQGHDVTDCFLVHGFPEWYYEQKGGSRVSSDNREVVS 125
Query: 315 R---------DTTGRSGGRGRGTYNALARANKATTSSRSIGGNSGQSHAPPHSSEAAGIS 365
R + + GRGRG N+ + AP SS G
Sbjct: 126 RLENKPAKREGRSSKGNGRGRGRVNS--------------------ARAPLSSSN--GSD 163
Query: 366 GITPAQWQQILDALNISKTKDRLHGKNDIS-WIIDTGASHHVTGNFSCLINGKRITNTPV 424
IT Q +L A T +RL G ++ IID+GASHH+TG+ S L++ I + V
Sbjct: 164 QIT--QLISLLQAQRPKSTSERLSGNTCLTDVIIDSGASHHMTGDCSILVDVFDIIPSAV 221
Query: 425 GLPNGKDATAIQEGSVILDGGLRLNNVLFVPQLTCNLISVTQLIDDSNCIVQFTNALCVI 484
P+GK + A + +++L +L +VLFVP C LISV++L+ +
Sbjct: 222 TKPDGKASCATKCVTLLLSSSYKLQDVLFVPDFDCTLISVSKLLKQTGPF---------- 271
Query: 485 QDRTTRTLIGAGERIDGLYFFRGVPKVHALMVEGDSAMD--LWHKRLGHPSEKVLKFIPH 542
+RTLIGAGE + +Y+F GV A DS LWH+RLGHPS L +P
Sbjct: 272 ----SRTLIGAGEVRERVYYFTGVLVASAHKTSSDSTSSGALWHRRLGHPSTSFLLSLPE 327
Query: 543 VSQHSRSKNNRPCDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGSYRTRSSCGAQYYL 602
Q S+ L ++ CD +C
Sbjct: 328 CHQSSKD----------------------------LGKIDSCD---------TC------ 344
Query: 603 TIVNDYSRAVWVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTEFNCLRDYFFN 662
SRA K + ++ NF A DRQF K ++ +RSDNGTEF C YF
Sbjct: 345 ------SRA--------KQTLPSLIRNFCAMADRQFRKPVRSIRSDNGTEFMCHTSYFQE 390
Query: 663 NGIVFETSCVGTPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLTACYLINRTPS 722
+GI+ ETSCV TPQQN RVERKH+HI+NVAR FQG+ P TPS
Sbjct: 391 HGILHETSCVDTPQQNARVERKHRHILNVARTCLFQGNFP-----------------TPS 433
Query: 723 SVLNYKTPYEKLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIFVGYPY 777
VL KTPYE LFGK P +D ++ FGCLCYAH + RD DKFASRSRKCIF+GYP+
Sbjct: 434 PVLKGKTPYEVLFGKQPSYDMLRTFGCLCYAHIRPRDKDKFASRSRKCIFIGYPH 488
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 540 bits (1391), Expect = e-153
Identities = 308/725 (42%), Positives = 423/725 (57%), Gaps = 33/725 (4%)
Query: 775 YPYGKKGWKLYDLESKEYIVSRDVKFYEHEFPFDVQLDTTHSTPFIDSEYVTEDIGFETY 834
YP G KG+K+ DLES ++R+V F+E +FPF S + +
Sbjct: 335 YPSGYKGYKVLDLESHSISITRNVVFHETKFPFKTSKFLKESVDMFPNSILPLPAPLHFV 394
Query: 835 DASFEGGGASMALQDNEQTQLQGGLHGDCNGADVAANEGLSVSGIEGEVEPAHEVDGADV 894
+ SM L D+ L D N A + N S S I + D
Sbjct: 395 E--------SMPLDDD--------LRADDNNASTS-NSASSASSIP-PLPSTVNTQNTDA 436
Query: 895 VVMQEDDVEPGEPEVVAEREAIVASEMGRGMRNKVPNIKLKDFVTHTIRKVKSSKSSSAQ 954
+ + + V P+ R A + + N VP + T T + + SSS
Sbjct: 437 LDIDTNSVPIARPK----RNAKAPAYLSEYHCNSVPFLSSLSPTTSTSIE---TPSSSIP 489
Query: 955 EDASGTPYPITYFVSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALE 1014
TPYP++ +S ++ + +++ A EP F +A+K W A E+ ALE
Sbjct: 490 PKKITTPYPMSTAISYDKLTPLFHSYICAYNVETEPKAFTQAMKSEKWTRAANEELHALE 549
Query: 1015 DNETWVMEKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPV 1074
N+TW++E L GK +G KWV+ IK++ DGSIER KARLV G Q EGIDY ETF+PV
Sbjct: 550 QNKTWIVESLTEGKNVVGCKWVFTIKYNPDGSIERYKARLVAQGFTQQEGIDYMETFSPV 609
Query: 1075 AKMVTVRTFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGFK-----NTDPNLVC 1129
AK +V+ L +AA W + QMDV NAFLHG+L+EE+YM +P G+ + VC
Sbjct: 610 AKFGSVKLLLGLAAATGWSLTQMDVSNAFLHGELDEEIYMSLPQGYTPPTGISLPSKPVC 669
Query: 1130 RLKKSLYGLKQAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIA 1189
RL KSLYGLKQA R W+ +L + F+QS +D ++F I VLVYVDDL+IA
Sbjct: 670 RLLKSLYGLKQASRQWYKRLSSVFLGANFIQSPADNTMFVKVSCTSIIVVLVYVDDLMIA 729
Query: 1190 GNDIAALKIFKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEIIDETGLLG 1249
ND +A++ K L F +KDLG ++FLGLE+AR+ EGI +CQRKYA ++++ GL G
Sbjct: 730 SNDSSAVENLKELLRSEFKIKDLGPARFFLGLEIARSSEGISVCQRKYAQNLLEDVGLSG 789
Query: 1250 AKPADFPMEQHHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPC 1309
KP+ PM+ + L G L + YR L+GRL+YL +TRPD+ ++VH LSQF+ P
Sbjct: 790 CKPSSIPMDPNLHLTKEMGTLLPNATSYRELVGRLLYLCITRPDITFAVHTLSQFLSAPT 849
Query: 1310 EEHWEAALRVVRYLKKHPGQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGWVVLLDLS 1369
+ H +AA +V+RYLK +PGQG++ + SEL L G+ D+DW +C +RRS+TG+ + L S
Sbjct: 850 DIHMQAAHKVLRYLKGNPGQGLMYSASSELCLNGFSDADWGTCKDSRRSVTGFCIYLGTS 909
Query: 1370 PVSWKTKKQPTVSRSSAEAEYRSMAMTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSALH 1429
++WK+KKQ VSRSS E+EYRS+A TCE+ WL+QLL DL V+ + +L+CD+KSALH
Sbjct: 910 LITWKSKKQSVVSRSSTESEYRSLAQATCEIIWLQQLLKDLHVTMTCPAKLFCDNKSALH 969
Query: 1430 IAQNPVFHERTKHIEADCHFVRDAVVAGIICPLYVPTSVQLADIFTKALGKAQ---FEFL 1486
+A NPVFHERTKHIE DCH VRD + AG + L+VPT QLADI TK L Q F L
Sbjct: 970 LATNPVFHERTKHIEIDCHTVRDQIKAGKLKTLHVPTGNQLADILTKPLHPVQSPIFSLL 1029
Query: 1487 LRKLG 1491
R G
Sbjct: 1030 FRFTG 1034
Score = 72.8 bits (177), Expect = 1e-12
Identities = 46/144 (31%), Positives = 65/144 (44%), Gaps = 8/144 (5%)
Query: 395 SWIIDTGASHHVTGNFSCLINGKRITNTPVGLPNGKDATAIQEGSVILDGGLRLNNVLFV 454
+WIID+GAS HV + + ++ V LPNG G++ + L L+NVL V
Sbjct: 98 AWIIDSGASSHVCSDLTMFRELIHVSGVTVTLPNGTRVAITHTGTICITSTLILHNVLLV 157
Query: 455 PQLTCNLISVTQLIDDSNCIVQFTNALCVIQDRTTRTLIGAGERIDGLY--------FFR 506
P NLISV L+ + F C IQ+ T +IG G+ + LY F
Sbjct: 158 PDFKFNLISVCCLVKTLSYSAHFFADCCYIQELTRGLMIGRGKTYNNLYILETQRTSFSP 217
Query: 507 GVPKVHALMVEGDSAMDLWHKRLG 530
+P + LWH+RLG
Sbjct: 218 SLPAASSFTGTVQDDCLLWHQRLG 241
Score = 72.8 bits (177), Expect = 1e-12
Identities = 35/84 (41%), Positives = 52/84 (61%), Gaps = 1/84 (1%)
Query: 600 YYLTIVNDYSRAVWVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTEFNCLRDY 659
Y+LT+V+D +R WVY++ NK+E+ +F FV + Q++ KIK +RSDN E +
Sbjct: 252 YFLTLVDDCTRTTWVYMMKNKSEVSNIFPVFVKLIFTQYNAKIKAIRSDNVKEL-AFTKF 310
Query: 660 FFNNGIVFETSCVGTPQQNGRVER 683
G++ + SC TPQQN VER
Sbjct: 311 VKEQGMIHQFSCAYTPQQNSVVER 334
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 499 bits (1286), Expect = e-141
Identities = 260/539 (48%), Positives = 359/539 (66%), Gaps = 6/539 (1%)
Query: 962 YPITYFVSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVM 1021
+PI+ +S + S H ++ ++ P ++ EA W A+ EI A+E +TW +
Sbjct: 919 HPISSSLSYSQISPSHMLYINNISKIPIPQSYHEAKDSKEWCGAIDQEIGAMERTDTWEI 978
Query: 1022 EKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVR 1081
LPPGKKA+G KWV+ +K H+DGS+ER KAR+V G+ Q EG+DY ETF+PVAKM TV+
Sbjct: 979 TSLPPGKKAVGCKWVFTVKFHADGSLERFKARIVAKGYTQKEGLDYTETFSPVAKMATVK 1038
Query: 1082 TFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGF---KNTD--PNLVCRLKKSLY 1136
L V+A KKW ++Q+D+ NAFL+GDLEE +YMK+P G+ K T PN+VCRLKKS+Y
Sbjct: 1039 LLLKVSASKKWYLNQLDISNAFLNGDLEETIYMKLPDGYADIKGTSLPPNVVCRLKKSIY 1098
Query: 1137 GLKQAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIAAL 1196
GLKQA R WF K +L GF + + D++LF G I +LVYVDD++IA A
Sbjct: 1099 GLKQASRQWFLKFSNSLLALGFEKQHGDHTLFVRCIGSEFIVLLVYVDDIVIASTTEQAA 1158
Query: 1197 KIFKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAKPADFP 1256
+ L F +++LG LKYFLGLEVAR EGI L QRKYALE++ +L KP+ P
Sbjct: 1159 QSLTEALKASFKLRELGPLKYFLGLEVARTSEGISLSQRKYALELLTSADMLDCKPSSIP 1218
Query: 1257 MEQHHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEAA 1316
M + +L+ G LED E YRRL+G+L+YL++TRPD+ ++V+ L QF P H A
Sbjct: 1219 MTPNIRLSKNDGLLLEDKEMYRRLVGKLMYLTITRPDITFAVNKLCQFSSAPRTAHLAAV 1278
Query: 1317 LRVVRYLKKHPGQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTK 1376
+V++Y+K GQG+ ++ +L L+G+ D+DW +CP +RRS TG+ + + S +SW++K
Sbjct: 1279 YKVLQYIKGTVGQGLFYSAEDDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSK 1338
Query: 1377 KQPTVSRSSAEAEYRSMAMTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVF 1436
KQPTVSRSSAEAEYR++A+ +CE+ WL LL L V HS LY DS +A++IA NPVF
Sbjct: 1339 KQPTVSRSSAEAEYRALALASCEMAWLSTLLLALRV-HSGVPILYSDSTAAVYIATNPVF 1397
Query: 1437 HERTKHIEADCHFVRDAVVAGIICPLYVPTSVQLADIFTKALGKAQFEFLLRKLGIRDL 1495
HERTKHIE DCH VR+ + G + L+V T Q+ADI TK L QF LL K+ I+++
Sbjct: 1398 HERTKHIEIDCHTVREKLDNGQLKLLHVKTKDQVADILTKPLFPYQFAHLLSKMSIQNI 1456
Score = 458 bits (1179), Expect = e-129
Identities = 283/827 (34%), Positives = 434/827 (52%), Gaps = 38/827 (4%)
Query: 16 SSNSEKKVDLVFFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPK 75
SS + FFL S D+PG I +L Y +WS A+R S AK K GFV+G +P+
Sbjct: 55 SSGFDDPTQSPFFLHSADHPGLSIISHRLDETTYGDWSVAMRISLDAKNKLGFVDGSLPR 114
Query: 76 PTTPE-KLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARIC 134
P + W SM+ +WLLN++ P + ++ +DA +W L RF + N R
Sbjct: 115 PLESDPNFRLWSRCNSMVKSWLLNSVSPQIYRSILRLNDATDIWRDLFDRFNLTNLPRTY 174
Query: 135 QLKASLGECKQGKGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAED 194
L + + +QG +S Y+ L +WD+L + C C ++ + +
Sbjct: 175 NLTQEIQDLRQGT-MSLSEYYTLLKTLWDQL----DSTEALDDPCTCGKAVRLYQKAEKA 229
Query: 195 YLHHFLMGLDGAYATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGEYSIQQNRDNVMAFK 254
+ FL GL+ +YA +R ++++ LP++ + Y + QD ++G +++ +
Sbjct: 230 KIMKFLAGLNESYAIVRRQIIAKKALPSLAEVYHILDQDNS-QKGFFNVVAPPAAFQVSE 288
Query: 255 VA--PDTRGKSKLVDNSDK----FCTHCNREGHDERTCFQIHGFPEWWGDRPRGGRGSGR 308
V+ P T + V + C+ CNR GH C++ HGFP + + + +
Sbjct: 289 VSHSPITSPEIMYVQSGPNKGRPTCSFCNRVGHIAERCYKKHGFPPGFTPKGKSSDKPPK 348
Query: 309 GGATTGRDT------TGR----SGGRGRGTYNALARANKATTSSRSIGGNSGQSHAPPHS 358
A + T TG+ +G L + + + + S S
Sbjct: 349 PQAVAAQVTLSPDKMTGQLETLAGNFSPDQIQNLIALFSSQLQPQIVSPQTASSQHEASS 408
Query: 359 SEAAGISGI--TPAQWQQILDALNISKTKDRLHGKNDISWIIDTGASHHVTGNFSCLING 416
S++ SGI +P+ + + L +S + + +W+ID+GA+HHV+ +
Sbjct: 409 SQSVAPSGILFSPSTYC-FIGILAVSH-----NSLSSDTWVIDSGATHHVSHDRKLFQTL 462
Query: 417 KRITNTPVGLPNGKDATAIQEGSVILDGGLRLNNVLFVPQLTCNLISVTQLIDDSNCIVQ 476
+ V LP G + G+V+++ + L NVLF+P+ NLIS++ L D V
Sbjct: 463 DTSIVSFVNLPTGPNVRISGVGTVLINKDIILQNVLFIPEFRLNLISISSLTTDLGTRVI 522
Query: 477 FTNALCVIQDRTTRTLIGAGERIDGLYFFRGVPKVHALMVEGDSAMDLWHKRLGHPSEKV 536
F + C IQD T +G G+RI LY + A+ V + +WHKRLGHPS
Sbjct: 523 FDPSCCQIQDLTKGLTLGEGKRIGNLYVLD--TQSPAISVNAVVDVSVWHKRLGHPSFSR 580
Query: 537 LKFIPHVSQHSRSKNNRP--CDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGSYRTRS 594
L + V +R KN + C VC AKQ + SFP + N S FEL+H D+WG + +
Sbjct: 581 LDSLSEVLGTTRHKNKKSAYCHVCHLAKQKKLSFPSANNICNSTFELLHIDVWGPFSVET 640
Query: 595 SCGAQYYLTIVNDYSRAVWVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTEFN 654
G +Y+LTIV+D+SRA W+YLL +K+++ T+F F+ V+ Q+D ++K VRSDN E
Sbjct: 641 VEGYKYFLTIVDDHSRATWIYLLKSKSDVLTVFPAFIDLVENQYDTRVKSVRSDNAKEL- 699
Query: 655 CLRDYFFNNGIVFETSCVGTPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLTAC 714
+++ GIV SC TP+QN VERKHQHI+NVARAL FQ ++ + +WG+CVLTA
Sbjct: 700 AFTEFYKAKGIVSFHSCPETPEQNSVVERKHQHILNVARALMFQSNMSLPYWGDCVLTAV 759
Query: 715 YLINRTPSSVLNYKTPYEKLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIFVG 774
+LINRTPS++L+ KTP+E L GK+P + +K FGCLCY+ + KF RSR C+F+G
Sbjct: 760 FLINRTPSALLSNKTPFEVLTGKLPDYSQLKTFGCLCYSSTSSKQRHKFLPRSRACVFLG 819
Query: 775 YPYGKKGWKLYDLESKEYIVSRDVKFYEHEFPF--DVQLDTTHSTPF 819
YP+G KG+KL DLES +SR+V+F+E FP Q TT S F
Sbjct: 820 YPFGFKGYKLLDLESNVVHISRNVEFHEELFPLASSQQSATTASDVF 866
>At4g17450 retrotransposon like protein
Length = 1433
Score = 499 bits (1285), Expect = e-141
Identities = 249/524 (47%), Positives = 344/524 (65%), Gaps = 5/524 (0%)
Query: 980 FVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVMEKLPPGKKALGSKWVYKI 1039
F+ +T P + EA W DAM+ EI A+ TW + LPP KKA+G KWV+ I
Sbjct: 907 FINNITNAVIPQRYSEAKDFKAWCDAMKEEIGAMVRTNTWSVVSLPPNKKAIGCKWVFTI 966
Query: 1040 KHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVAAIKKWEVHQMDV 1099
KH++DGSIER KARLV G+ Q EG+DY+ETF+PVAK+ +VR L +AA KW VHQ+D+
Sbjct: 967 KHNADGSIERYKARLVAKGYTQEEGLDYEETFSPVAKLTSVRMMLLLAAKMKWSVHQLDI 1026
Query: 1100 HNAFLHGDLEEEVYMKVPPGFKNT-----DPNLVCRLKKSLYGLKQAPRCWFAKLVTALK 1154
NAFL+GDL+EE+YMK+PPG+ + P+ +CRL KS+YGLKQA R W+ KL LK
Sbjct: 1027 SNAFLNGDLDEEIYMKIPPGYADLVGEALPPHAICRLHKSIYGLKQASRQWYLKLSNTLK 1086
Query: 1155 RYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIAALKIFKAYLGVCFHMKDLGV 1214
GF +S +D++LF + + + VLVYVDD++I N A+ F A L F ++DLG
Sbjct: 1087 GMGFQKSNADHTLFIKYANGVLMGVLVYVDDIMIVSNSDDAVAQFTAELKSYFKLRDLGA 1146
Query: 1215 LKYFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAKPADFPMEQHHKLALVSGKPLEDP 1274
KYFLG+E+AR+ +GI +CQRKY LE++ TG LG+KP+ P++ KL G PL D
Sbjct: 1147 AKYFLGIEIARSEKGISICQRKYILELLSTTGFLGSKPSSIPLDPSVKLNKEDGVPLTDS 1206
Query: 1275 EPYRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEAALRVVRYLKKHPGQGILLR 1334
YR+L+G+L+YL +TRPD+AY+V+ L QF P H A +V+RYLK GQG+
Sbjct: 1207 TSYRKLVGKLMYLQITRPDIAYAVNTLCQFSHAPTSVHLSAVHKVLRYLKGTVGQGLFYS 1266
Query: 1335 SDSELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMA 1394
+D + L G+ DSD+ SC +RR + + + + VSWK+KKQ TVS S+AEAE+R+M+
Sbjct: 1267 ADDKFDLRGYTDSDFGSCTDSRRCVAAYCMFIGDYLVSWKSKKQDTVSMSTAEAEFRAMS 1326
Query: 1395 MTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVFHERTKHIEADCHFVRDAV 1454
T E+ WL +L D V LYCD+ +ALHI N VFHERTK +E DC+ R+AV
Sbjct: 1327 QGTKEMIWLSRLFDDFKVPFIPPAYLYCDNTAALHIVNNSVFHERTKFVELDCYKTREAV 1386
Query: 1455 VAGIICPLYVPTSVQLADIFTKALGKAQFEFLLRKLGIRDLHAP 1498
+G + ++V T Q+AD TKA+ AQF L+ K+G+ ++ AP
Sbjct: 1387 ESGFLKTMFVETGEQVADPLTKAIHPAQFHKLIGKMGVCNIFAP 1430
Score = 399 bits (1024), Expect = e-111
Identities = 270/855 (31%), Positives = 406/855 (46%), Gaps = 116/855 (13%)
Query: 27 FFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPTTPEKL-EDW 85
+FL S+D+PG I L G NY+ WS A+R S AK K FV+G +P+P +++ + W
Sbjct: 68 YFLHSSDHPGLNIVSHILDGTNYNNWSIAMRMSLDAKNKLSFVDGSLPRPDVSDRMFKIW 127
Query: 86 KAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQ 145
SM+ WLLN + +W L RF V N R QL+ S+ KQ
Sbjct: 128 SRCNSMVKTWLLNVVT--------------EMWNDLFSRFRVSNLPRKYQLEQSIHTLKQ 173
Query: 146 GKGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAEDYLHHFLMGLDG 205
G ++S Y+ + +W++L KC C+ K++ + + + FLMGL+
Sbjct: 174 GN-LDLSTYYTKKKTLWEQLANTRVLTVRKCN---CEHVKELLEEAETSRIIQFLMGLND 229
Query: 206 AYATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGEYSIQQNRDNVMAFKVAPDTRGKSKL 265
+A IR +L+ P P + + Y + QDE R N + +P + +
Sbjct: 230 NFAHIRGQILNMKPRPGLTEIYNMLDQDESQRLVGNPTLSNPTAAFQVQASPIIDSQVNM 289
Query: 266 VDNSDKF--CTHCNREGHDERTCFQIHGFPEWWGDRPRGGRGSGRGG-ATTGRDTTGRSG 322
S K C++CN+ GH C++ HG+P G + G+ G A+T +
Sbjct: 290 AQGSYKKPKCSYCNKLGHLVDKCYKKHGYPP--GSKWTKGQTIGSTNLASTQLQPVNETP 347
Query: 323 GRGRGTYNALAR-------------------ANKATTSSRSIGGNSGQSHAPPHSSEAAG 363
+Y + + TTSS SI S + P S+ +G
Sbjct: 348 NEKTDSYEEFSTDQIQTMISYLSTKLHIASASPMPTTSSASISA----SPSVPMISQISG 403
Query: 364 --ISGITPAQWQQILDALNISKTKDRLHGKNDISWIIDTGASHHVTGNFSCLINGKRITN 421
+S + A + ++ +++ + W+ID+GA+HHVT N +N + + N
Sbjct: 404 TFLSLFSNAYYDMLISSVSQEPAV------SPRGWVIDSGATHHVTHNRDLYLNFRSLEN 457
Query: 422 TPVGLPNGKDATAIQEGSVILDGGLRLNNVLFVPQLTCNLISVTQLIDDSNCIVQFTNAL 481
T V LPN G + L + L+NVL++P+ NLIS
Sbjct: 458 TFVRLPNDCTVKIAGIGFIQLSDAISLHNVLYIPEFKFNLIS------------------ 499
Query: 482 CVIQDRTTRTLIGAGERIDGLYFFRGVPKVHALMVEGDSAMD--------------LWHK 527
+ T +IG G ++ LY H + ++G ++M WHK
Sbjct: 500 ----ELTKELMIGRGSQVGNLYVLDFNENNHTVSLKGTTSMCPEFSVCSSVVVDSVTWHK 555
Query: 528 RLGHPSEKVLKFIPHVSQHSRSKNNRP-------CDVCPRAKQHRDSFPLSENNAASLFE 580
RLGHP+ + + V K N+ C VC +KQ SF +N ++ F+
Sbjct: 556 RLGHPAYSKIDLLSDVLNLKVKKINKEHSPVCHVCHVCHLSKQKHLSFQSRQNMCSAAFD 615
Query: 581 LVHCDLWGSYRTRSSCGAQYYLTIVNDYSRAVWVYLLCNKTEIETMFLNFVAFVDRQFDK 640
LVH D WG + + ND A W+YLL NK+++ +F F+ V Q+
Sbjct: 616 LVHIDTWGPFSVPT-----------ND---ATWIYLLKNKSDVLHVFPAFINMVHTQYQT 661
Query: 641 KIKKVRSDNGTEFNCLRDYFFNNGIVFETSCVGTPQQNGRVERKHQHIMNVARALRFQGH 700
K+K VRSDN E D F +GIV SC TP+QN VERKHQHI+NVARAL FQ +
Sbjct: 662 KLKSVRSDNAHELK-FTDLFAAHGIVAYHSCPETPEQNSVVERKHQHILNVARALLFQSN 720
Query: 701 LPMQFWGECVLTACYLINRTPSSVLNYKTPYEKLFGKVPKFDNMKIFGCLCYAHNQRRDG 760
+P++FWG+CVLTA +LINR P+ VLN K+PYEKL P ++++K FGCLCY+ +
Sbjct: 721 IPLEFWGDCVLTAVFLINRLPTPVLNNKSPYEKLKNIPPAYESLKTFGCLCYSSTSPKQR 780
Query: 761 DKFASRSRKCIFVGYPYGKKGWKLYDLESKEYIVSRDVKFYEHEFPF---DVQLDTTHST 817
KF R+R C+F+GYP G KG+KL D+E+ +SR V F+E FPF ++ D
Sbjct: 781 HKFEPRARACVFLGYPLGYKGYKLLDIETHAVSISRHVIFHEDIFPFISSTIKDDIKDFF 840
Query: 818 PFIDSEYVTEDIGFE 832
P + T+D+ E
Sbjct: 841 PLLQFPARTDDLPLE 855
>At4g23160 putative protein
Length = 1240
Score = 489 bits (1260), Expect = e-138
Identities = 247/504 (49%), Positives = 346/504 (68%), Gaps = 5/504 (0%)
Query: 957 ASGTPYPITYFVSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDN 1016
AS T + I+ F+S E+ S + +F+ + KEP+ + EA + W AM +EI A+E
Sbjct: 53 ASLTIHDISQFLSYEKVSPLYHSFLVCIAKAKEPSTYNEAKEFLVWCGAMDDEIGAMETT 112
Query: 1017 ETWVMEKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAK 1076
TW + LPP KK +G KWVYKIK++SDG+IER KARLV G+ Q EGID+ ETF+PV K
Sbjct: 113 HTWEICTLPPNKKPIGCKWVYKIKYNSDGTIERYKARLVAKGYTQQEGIDFIETFSPVCK 172
Query: 1077 MVTVRTFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGFK-----NTDPNLVCRL 1131
+ +V+ LA++AI + +HQ+D+ NAFL+GDL+EE+YMK+PPG+ + PN VC L
Sbjct: 173 LTSVKLILAISAIYNFTLHQLDISNAFLNGDLDEEIYMKLPPGYAARQGDSLPPNAVCYL 232
Query: 1132 KKSLYGLKQAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGN 1191
KKS+YGLKQA R WF K L +GFVQS+SD++ F + + VLVYVDD+II N
Sbjct: 233 KKSIYGLKQASRQWFLKFSVTLIGFGFVQSHSDHTYFLKITATLFLCVLVYVDDIIICSN 292
Query: 1192 DIAALKIFKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAK 1251
+ AA+ K+ L CF ++DLG LKYFLGLE+AR+ GI +CQRKYAL+++DETGLLG K
Sbjct: 293 NDAAVDELKSQLKSCFKLRDLGPLKYFLGLEIARSAAGINICQRKYALDLLDETGLLGCK 352
Query: 1252 PADFPMEQHHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPCEE 1311
P+ PM+ + SG D + YRRLIGRL+YL +TR D++++V+ LSQF + P
Sbjct: 353 PSSVPMDPSVTFSAHSGGDFVDAKAYRRLIGRLMYLQITRLDISFAVNKLSQFSEAPRLA 412
Query: 1312 HWEAALRVVRYLKKHPGQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPV 1371
H +A ++++ Y+K GQG+ S +E++L+ + D+ + SC TRRS G+ + L S +
Sbjct: 413 HQQAVMKILHYIKGTVGQGLFYSSQAEMQLQVFSDASFQSCKDTRRSTNGYCMFLGTSLI 472
Query: 1372 SWKTKKQPTVSRSSAEAEYRSMAMTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIA 1431
SWK+KKQ VS+SSAEAEYR+++ T E+ WL Q +L + S+ L+CD+ +A+HIA
Sbjct: 473 SWKSKKQQVVSKSSAEAEYRALSFATDEMMWLAQFFRELQLPLSKPTLLFCDNTAAIHIA 532
Query: 1432 QNPVFHERTKHIEADCHFVRDAVV 1455
N VFHERTKHIE+DCH VR+ V
Sbjct: 533 TNAVFHERTKHIESDCHSVRERSV 556
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 483 bits (1243), Expect = e-136
Identities = 258/540 (47%), Positives = 355/540 (64%), Gaps = 8/540 (1%)
Query: 962 YPITYFVSCERFSIRHRNFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVM 1021
YPI+ +S + S H ++ +T P N+ EA W +A+ EI A+E TW +
Sbjct: 912 YPISSTISYSKISPSHMCYINNITKIPIPTNYAEAQDTKEWCEAVDAEIGAMEKTNTWEI 971
Query: 1022 EKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVR 1081
LP GKKA+G KWV+ +K +DG++ER KARLV G+ Q EG+DY +TF+PVAKM T++
Sbjct: 972 TTLPKGKKAVGCKWVFTLKFLADGNLERYKARLVAKGYTQKEGLDYTDTFSPVAKMTTIK 1031
Query: 1082 TFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGFKNTD-----PNLVCRLKKSLY 1136
L V+A KKW + Q+DV NAFL+G+LEEE++MK+P G+ N+V RLK+S+Y
Sbjct: 1032 LLLKVSASKKWFLKQLDVSNAFLNGELEEEIFMKIPEGYAERKGIVLPSNVVLRLKRSIY 1091
Query: 1137 GLKQAPRCWFAKLVTALKRYGFVQSYSDYSLFT-LHRGEIQINVLVYVDDLIIAGNDIAA 1195
GLKQA R WF K ++L GF +++ D++LF ++ GE I VLVYVDD++IA AA
Sbjct: 1092 GLKQASRQWFKKFSSSLLSLGFKKTHGDHTLFLKMYDGEFVI-VLVYVDDIVIASTSEAA 1150
Query: 1196 LKIFKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAKPADF 1255
L F ++DLG LKYFLGLEVAR GI +CQRKYALE++ TG+L KP
Sbjct: 1151 AAQLTEELDQRFKLRDLGDLKYFLGLEVARTTAGISICQRKYALELLQSTGMLACKPVSV 1210
Query: 1256 PMEQHHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEA 1315
PM + K+ G +ED E YRR++G+L+YL++TRPD+ ++V+ L QF P H A
Sbjct: 1211 PMIPNLKMRKDDGDLIEDIEQYRRIVGKLMYLTITRPDITFAVNKLCQFSSAPRTTHLTA 1270
Query: 1316 ALRVVRYLKKHPGQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKT 1375
A RV++Y+K GQG+ + S+L L+G+ DSDWASC +RRS T + + + S +SW++
Sbjct: 1271 AYRVLQYIKGTVGQGLFYSASSDLTLKGFADSDWASCQDSRRSTTSFTMFVGDSLISWRS 1330
Query: 1376 KKQPTVSRSSAEAEYRSMAMTTCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPV 1435
KKQ TVSRSSAEAEYR++A+ TCE+ WL LL L S + LY DS +A++IA NPV
Sbjct: 1331 KKQHTVSRSSAEAEYRALALATCEMVWLFTLLVSLQASPPVPI-LYSDSTAAIYIATNPV 1389
Query: 1436 FHERTKHIEADCHFVRDAVVAGIICPLYVPTSVQLADIFTKALGKAQFEFLLRKLGIRDL 1495
FHERTKHI+ DCH VR+ + G + L+V T Q+ADI TK L QFE L K+ I ++
Sbjct: 1390 FHERTKHIKLDCHTVRERLDNGELKLLHVRTEDQVADILTKPLFPYQFEHLKSKMSILNI 1449
Score = 451 bits (1160), Expect = e-126
Identities = 282/814 (34%), Positives = 414/814 (50%), Gaps = 47/814 (5%)
Query: 16 SSNSEKKVDLVFFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPK 75
SS S FFL S D+PG I +L NY +WS A+ S AK K GF++G + +
Sbjct: 51 SSESGDPTQSPFFLHSADHPGLNIISHRLDETNYGDWSVAMLISLDAKNKTGFIDGTLSR 110
Query: 76 PTTPE-KLEDWKAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARIC 134
P + W SM+ +WLLN++ P + ++ +DA +W L RF V N R
Sbjct: 111 PLESDLNFRLWSRCNSMVKSWLLNSVSPQIYRSILRMNDASDIWRDLNSRFNVTNLPRTY 170
Query: 135 QLKASLGECKQGKGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAED 194
L + + +QG +S Y+ RL +WD+L E C C ++ + +
Sbjct: 171 NLTQEIQDFRQGT-LSLSEYYTRLKTLWDQL----DSTEALDEPCTCGKAMRLQQKAEQA 225
Query: 195 YLHHFLMGLDGAYATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGEYSIQQNRDNVMA-- 252
+ FL GL+ +YA +R ++++ LP++ + Y + QD QQ+ NV+A
Sbjct: 226 KIVKFLAGLNESYAIVRRQIIAKKALPSLGEVYHILDQDNS--------QQSFSNVVAPP 277
Query: 253 --FKVAPDTRGKSK-----LVDNSDK----FCTHCNREGHDERTCFQIHGFPEWWGDRPR 301
F+V+ T+ S V N C+ NR GH C++ HGFP G P+
Sbjct: 278 AAFQVSEITQSPSMDPTVCYVQNGPNKGRPICSFYNRVGHIAERCYKKHGFPP--GFTPK 335
Query: 302 GGRGSGRGGATTGRDTTGRSGGRGRGTYNALARANKATTSSRSIGGNSGQSHAPPHSSEA 361
G G S + + +K +S + PP +
Sbjct: 336 GKAGEKLQKPKPLAANVAESSEVNTSLESMVGNLSKEQLQQFIAMFSSQLQNTPPSTYAT 395
Query: 362 AGIS-----GI--TPAQWQQILDALNISKTKDRLHGKNDISWIIDTGASHHVTGNFSCLI 414
A S GI +P+ + + L +++ H + +W+ID+GA+HHV+ + S
Sbjct: 396 ASTSQSDNLGICFSPSTYS-FIGILTVAR-----HTLSSATWVIDSGATHHVSHDRSLFS 449
Query: 415 NGKRITNTPVGLPNGKDATAIQEGSVILDGGLRLNNVLFVPQLTCNLISVTQLIDDSNCI 474
+ + V LP G G++ L+ + L NVLF+P+ NLIS++ L DD
Sbjct: 450 SLDTSVLSAVNLPTGPTVKISGVGTLKLNDDILLKNVLFIPEFRLNLISISSLTDDIGSR 509
Query: 475 VQFTNALCVIQDRTTRTLIGAGERIDGLYFFRGVPKVHALMVEGDSAMDLWHKRLGHPSE 534
V F C IQD ++G G R+ LY + ++ V + +WH+RLGH S
Sbjct: 510 VIFDKNSCEIQDLIKGRMLGQGRRVANLYLLDVGDQ--SISVNAVVDISMWHRRLGHASL 567
Query: 535 KVLKFIPHVSQHSRSKNNRP--CDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGSYRT 592
+ L I +R KN C VC AKQ + SFP S +F+L+H D+WG +
Sbjct: 568 QRLDAISDSLGTTRHKNKGSDFCHVCHLAKQRKLSFPTSNKVCKEIFDLLHIDVWGPFSV 627
Query: 593 RSSCGAQYYLTIVNDYSRAVWVYLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTE 652
+ G +Y+LTIV+D+SRA W+YLL K+E+ T+F F+ V+ Q+ K+K VRSDN E
Sbjct: 628 ETVEGYKYFLTIVDDHSRATWMYLLKTKSEVLTVFPAFIQQVENQYKVKVKAVRSDNAPE 687
Query: 653 FNCLRDYFFNNGIVFETSCVGTPQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLT 712
++ GIV SC TP+QN VERKHQHI+NVARAL FQ +P+ WG+CVLT
Sbjct: 688 LK-FTSFYAEKGIVSFHSCPETPEQNSVVERKHQHILNVARALMFQSQVPLSLWGDCVLT 746
Query: 713 ACYLINRTPSSVLNYKTPYEKLFGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIF 772
A +LINRTPS +L KTPYE L G P ++ ++ FGCLCY+ + KF RSR C+F
Sbjct: 747 AVFLINRTPSQLLMNKTPYEILTGTAPVYEQLRTFGCLCYSSTSPKQRHKFQPRSRACLF 806
Query: 773 VGYPYGKKGWKLYDLESKEYIVSRDVKFYEHEFP 806
+GYP G KG+KL DLES +SR+V+F+E FP
Sbjct: 807 LGYPSGYKGYKLMDLESNTVFISRNVQFHEEVFP 840
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 457 bits (1176), Expect = e-128
Identities = 239/525 (45%), Positives = 325/525 (61%), Gaps = 2/525 (0%)
Query: 979 NFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETW-VMEKLPPGKKALGSKWVY 1037
++ ++ A EP +A+KD WR AM +EI A N TW ++ PP +G +W++
Sbjct: 928 SYATSLAANSEPRTAIQAMKDDRWRQAMGSEINAQIGNHTWDLVPPPPPSVTIVGCRWIF 987
Query: 1038 KIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVAAIKKWEVHQM 1097
K +SDGS+ R KARLV G++Q G+DY ETF+PV K ++R L VA + W + Q+
Sbjct: 988 TKKFNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQL 1047
Query: 1098 DVHNAFLHGDLEEEVYMKVPPGFKNTD-PNLVCRLKKSLYGLKQAPRCWFAKLVTALKRY 1156
DV+NAFL G L +EVYM PPGF + D P+ VCRL+K++YGLKQAPR W+ +L T L
Sbjct: 1048 DVNNAFLQGTLTDEVYMSQPPGFVDKDRPDYVCRLRKAIYGLKQAPRAWYVELRTYLLTV 1107
Query: 1157 GFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIAALKIFKAYLGVCFHMKDLGVLK 1216
GFV S SD SLF L RG I +LVYVDD++I GND LK L F +K+ L
Sbjct: 1108 GFVNSISDTSLFVLQRGRSIIYMLVYVDDILITGNDTVLLKHTLDALSQRFSVKEHEDLH 1167
Query: 1217 YFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAKPADFPMEQHHKLALVSGKPLEDPEP 1276
YFLG+E R +G++L QR+Y L+++ T +L AKP PM KL L SG L DP
Sbjct: 1168 YFLGIEAKRVPQGLHLSQRRYTLDLLARTNMLTAKPVATPMATSPKLTLHSGTKLPDPTE 1227
Query: 1277 YRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEAALRVVRYLKKHPGQGILLRSD 1336
YR ++G L YL+ TRPDL+Y+V+ LSQ+M P ++HW A RV+RYL P GI L+
Sbjct: 1228 YRGIVGSLQYLAFTRPDLSYAVNRLSQYMHMPTDDHWNALKRVLRYLAGTPDHGIFLKKG 1287
Query: 1337 SELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMAMT 1396
+ L L + D+DWA S G++V L P+SW +KKQ V RSS EAEYRS+A T
Sbjct: 1288 NTLSLHAYSDADWAGDTDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSVANT 1347
Query: 1397 TCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVFHERTKHIEADCHFVRDAVVA 1456
+ EL+W+ LL +LG+ S +YCD+ A ++ NPVFH R KHI D HF+R+ V +
Sbjct: 1348 SSELQWICSLLTELGIQLSHPPVIYCDNVGATYLCANPVFHSRMKHIALDYHFIRNQVQS 1407
Query: 1457 GIICPLYVPTSVQLADIFTKALGKAQFEFLLRKLGIRDLHAPEGG 1501
G + ++V T QLAD TK L + F+ RK+G+ + GG
Sbjct: 1408 GALRVVHVSTHDQLADTLTKPLSRVAFQNFSRKIGVIKVPPSCGG 1452
Score = 229 bits (585), Expect = 7e-60
Identities = 196/793 (24%), Positives = 325/793 (40%), Gaps = 103/793 (12%)
Query: 37 NVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKP-------TTPEKLED---WK 86
N+ +L NY WSR + F GF++G P P P D W+
Sbjct: 19 NMSNVTKLTSTNYLMWSRQVHALFDGYELAGFLDGSTPMPPATIGTDAVPRVNPDYTRWR 78
Query: 87 AVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQG 146
++ + +L I S++ +S A +W L++ + + + QL
Sbjct: 79 RQDKLIYSAILGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQL---------- 128
Query: 147 KGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAEDYLHHFLMGLDGA 206
R +D+L K ++ + L L
Sbjct: 129 ----------RFITRFDQLALLGKP------------------MDHDEQVERVLENLPDD 160
Query: 207 YATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGEYSIQQNRDNVMAFKVAPDTRGKSKLV 266
Y + + ++D P++ + ++R+I E ++A A + +V
Sbjct: 161 YKPVIDQIAAKDTPPSLTEIHERLINRE-------------SKLLALNSAEVVPITANVV 207
Query: 267 DNSDKFCTHCNREGHDERTCFQIHGFPEWWGDRPRGGRGSGRGGATTGRDTTGRSGGRGR 326
+ + T+ NR ++ GD + R + + RS R
Sbjct: 208 THRN---TNTNRNQNNR-------------GDNRNYNNNNNRSNSWQPSSSGSRSDNRQP 251
Query: 327 GTYNALARANKATTSSRSIGGNSGQSHAPPHSSEAAGISGITPAQWQQILDALNISKTKD 386
Y L R + S +++ S TP WQ N++
Sbjct: 252 KPY--LGRCQICSVQGHSAKRCPQLHQFQSTTNQQQSTSPFTP--WQP---RANLAVNSP 304
Query: 387 RLHGKNDISWIIDTGASHHVTGNFSCLINGKRITN-TPVGLPNGKDATAIQEGSVILDGG 445
N +W++D+GA+HH+T +F+ L + T V + +G GS L
Sbjct: 305 Y----NANNWLLDSGATHHITSDFNNLSFHQPYTGGDDVMIADGSTIPITHTGSASLPTS 360
Query: 446 LR---LNNVLFVPQLTCNLISVTQLIDDSNCIVQFTNALCVIQDRTTRTLIGAGERIDGL 502
R LN VL+VP + NLISV +L + + V+F A ++D T + G+ D L
Sbjct: 361 SRSLDLNKVLYVPNIHKNLISVYRLCNTNRVSVEFFPASFQVKDLNTGVPLLQGKTKDEL 420
Query: 503 YFFRGVPKVHALMVEGDSAM---DLWHKRLGHPSEKVLKFIPHVSQHSRSKNNRP----- 554
Y + M + WH RLGHPS +L + +S HS N
Sbjct: 421 YEWPIASSQAVSMFASPCSKATHSSWHSRLGHPSLAILNSV--ISNHSLPVLNPSHKLLS 478
Query: 555 CDVCPRAKQHRDSFPLSENNAASLFELVHCDLWGSYRTRSSCGAQYYLTIVNDYSRAVWV 614
C C K H+ F S ++ E ++ D+W S S +YY+ V+ ++R W+
Sbjct: 479 CSDCFINKSHKVPFSNSTITSSKPLEYIYSDVWSS-PILSIDNYRYYVIFVDHFTRYTWL 537
Query: 615 YLLCNKTEIETMFLNFVAFVDRQFDKKIKKVRSDNGTEFNCLRDYFFNNGIVFETSCVGT 674
Y L K++++ F+ F + V+ +F +I + SDNG EF LRDY +GI TS T
Sbjct: 538 YPLKQKSQVKDTFIIFKSLVENRFQTRIGTLYSDNGGEFVVLRDYLSQHGISHFTSPPHT 597
Query: 675 PQQNGRVERKHQHIMNVARALRFQGHLPMQFWGECVLTACYLINRTPSSVLNYKTPYEKL 734
P+ NG ERKH+HI+ + L +P +W A YLINR P+ +L ++P++KL
Sbjct: 598 PEHNGLSERKHRHIVEMGLTLLSHASVPKTYWPYAFSVAVYLINRLPTPLLQLQSPFQKL 657
Query: 735 FGKVPKFDNMKIFGCLCYAHNQRRDGDKFASRSRKCIFVGYPYGKKGWKLYDLESKEYIV 794
FG+ P ++ +K+FGC CY + + K +S++C F+GY + + + +
Sbjct: 658 FGQPPNYEKLKVFGCACYPWLRPYNRHKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYT 717
Query: 795 SRDVKFYEHEFPF 807
SR V+F E FPF
Sbjct: 718 SRHVQFDERCFPF 730
>At1g47650 hypothetical protein
Length = 1409
Score = 447 bits (1149), Expect = e-125
Identities = 231/467 (49%), Positives = 314/467 (66%), Gaps = 6/467 (1%)
Query: 1008 NEIQALEDNETWVMEKLPPGKKALGSKWVYKIKHHSDGSIERLKARLVVFGHHQIEGIDY 1067
+EI A+E TW + LP GKKA+G KWV+ +K +DGS+ER KARLV G+ Q EG+DY
Sbjct: 749 SEIGAMETTHTWEITTLPAGKKAVGCKWVFSLKFLADGSLERYKARLVAKGYTQKEGLDY 808
Query: 1068 DETFAPVAKMVTVRTFLAVAAIKKWEVHQMDVHNAFLHGDLEEEVYMKVPPGFK-----N 1122
+TF+PVAKM T++ L ++A KKW + Q+DV NAFL+G+LEEE+YMK+P G+
Sbjct: 809 TDTFSPVAKMTTIKLLLKISASKKWFLKQLDVSNAFLNGELEEEIYMKLPEGYAARKGIT 868
Query: 1123 TDPNLVCRLKKSLYGLKQAPRCWFAKLVTALKRYGFVQSYSDYSLFTLHRGEIQINVLVY 1182
PN VCRLK+S+YGLKQA R WF K +L GF +++ D++LF + VLVY
Sbjct: 869 LPPNSVCRLKRSIYGLKQASRQWFKKFSASLLDLGFKKTHGDHTLFIKEYDGEFVIVLVY 928
Query: 1183 VDDLIIAGNDIAALKIFKAYLGVCFHMKDLGVLKYFLGLEVARNHEGIYLCQRKYALEII 1242
VDD+ IA A L F ++DLG LKYFLGLE+AR GI +CQRKYALE++
Sbjct: 929 VDDIAIASTSEGAAIQLTQDLQQRFKLRDLGDLKYFLGLEIARTEAGISICQRKYALELL 988
Query: 1243 DETGLLGAKPADFPMEQHHKLALVSGKPLEDPEPYRRLIGRLIYLSVTRPDLAYSVHILS 1302
TG++ KP PM + K+ G L+D E YRR++G+L+YL++TR D+ ++V+ L
Sbjct: 989 ASTGMVNCKPVSVPMIPNVKMMKTDGDLLDDREQYRRIVGKLMYLTITRLDITFAVNKLC 1048
Query: 1303 QFMQKPCEEHWEAALRVVRYLKKHPGQGILLRSDSELKLEGWCDSDWASCPLTRRSLTGW 1362
QF P H +AA RV++Y+K GQG+ + ++L L+G+ DSDWASCP +RRS TG+
Sbjct: 1049 QFSSAPRTSHLQAAYRVLQYIKGSVGQGLFYSASADLTLKGFADSDWASCPDSRRSTTGF 1108
Query: 1363 VVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMAMTTCELKWLKQLLGDLGVSHSQGMQLYC 1422
+ + S +SW++KKQ VSRSSAEAEYR++A+ TCEL WL LL L S+ + LY
Sbjct: 1109 SMFVGDSLISWRSKKQHVVSRSSAEAEYRALALVTCELVWLHTLLMSLKASYLVPV-LYS 1167
Query: 1423 DSKSALHIAQNPVFHERTKHIEADCHFVRDAVVAGIICPLYVPTSVQ 1469
DS +A++IA NPVFHERTKHIE DCH VR+ + G + L+V T Q
Sbjct: 1168 DSTTAIYIATNPVFHERTKHIELDCHTVREKLDNGELKLLHVRTEDQ 1214
Score = 305 bits (782), Expect = 1e-82
Identities = 180/446 (40%), Positives = 249/446 (55%), Gaps = 30/446 (6%)
Query: 393 DISWIIDTGASHHVTGNFSCLINGKRITNTPVGLPNGKDATAIQEGSVILDGGLRLNNVL 452
D S +ID+GA HHV+ + S +N + V LP G G++ L+ + L N+L
Sbjct: 329 DKSQVIDSGAIHHVSHDKSLFLNLDTSVVSAVNLPAGPTVRISGVGTLRLNDDILLKNIL 388
Query: 453 FVPQLTCNLISVTQLIDDSNCIVQFTNALCVIQDRTTRTLIGAGERIDGLYFFRGVPKVH 512
F+P+ NLIS++ L DD V F C IQD ++G G RI LY V +
Sbjct: 389 FIPEFRLNLISISSLTDDIGSRVIFDKTSCEIQDPIKARMLGQGRRIANLY----VLDIE 444
Query: 513 ALMVEGDSAMDL--WHKRLGHPSEKVLKFIPHVSQHSRSKNNRP--CDVCPRAKQHRDSF 568
+V ++ +D+ WH+RLGH S + L I ++ KN C VC AKQ + F
Sbjct: 445 DPIVSVNAVVDISMWHRRLGHASLQRLDVISDSLGTTKPKNKGSDYCHVCHLAKQRKLPF 504
Query: 569 PLSENNAASLFELVHCDLWGSYRTRSSCGAQYYLTIVNDYSRAVWVYLLCNKTEIETMFL 628
P +F+L+H D+WG + + G +Y+LTIV+D+SRA W+YLL NK+E+ T+F
Sbjct: 505 PSQNKVCNEIFDLLHIDIWGPFSVETVDGYKYFLTIVDDHSRATWIYLLRNKSEVLTVFP 564
Query: 629 NFVAFVDRQFDKKIKKVRSDNGTEFNCLRDYFFNNGIVFETSCVGTPQQNGRVERKHQHI 688
F+ V+ Q+ ++K VRSDN E + GIV SC TP+QN VERKHQHI
Sbjct: 565 AFIEQVENQYKVRVKAVRSDNAPELK-FTSLYQQKGIVSFHSCPETPEQNLVVERKHQHI 623
Query: 689 MNVARALRFQGHLPMQFWGECVLTACYLINRTPSSVLNYKTPYEKLFGKVPKFDNMKIFG 748
+NV+RAL FQ +P+ WG+CVLTA +LINRTPS +L+ KTPYE L G VP + K
Sbjct: 624 LNVSRALMFQSQVPLSLWGDCVLTAVFLINRTPSQLLSNKTPYEILTGTVPFYFAKK--- 680
Query: 749 CLCYAHNQRRDGDKFASRSRKCIFVGYPYGKKGWKLYDLESKEYIVSRDVKFYEHEFPFD 808
KF RS+ C+F+GYP G KG+KL DLES +SR+V F+E FP
Sbjct: 681 -----------RHKFQPRSKACVFLGYPAGYKGYKLMDLESHTVFISRNVVFHEEVFPLA 729
Query: 809 VQLDTTHS----TPFIDSEYVTEDIG 830
V + S TP + V+ +IG
Sbjct: 730 VDPKSESSLKLFTPMVP---VSSEIG 752
Score = 127 bits (320), Expect = 4e-29
Identities = 82/278 (29%), Positives = 138/278 (49%), Gaps = 15/278 (5%)
Query: 27 FFLGSNDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPTTPEK-LEDW 85
FFL S D+PG I +L NY +W+ A+ S AK K GF++G +P+P +K W
Sbjct: 65 FFLHSADHPGLNIISHRLDETNYGDWNVAMLISLDAKNKSGFIDGTLPRPLESDKNFRLW 124
Query: 86 KAVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQ 145
+SM+ +WLLN++ P + ++ +DA +W L RF V N R L + + +Q
Sbjct: 125 SRCKSMVKSWLLNSVSPQIYRSILRMNDASDIWRDLHSRFNVTNLPRTYNLTQEIQDFRQ 184
Query: 146 GKGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAEDYLHHFLMGLDG 205
G +S Y+ RL +WD+L + + + CIC ++ + + + L GL+
Sbjct: 185 GT-LSLSEYYTRLKTLWDQLESTEELD----DPCICGKAMRLQQKAEQAKIVKLLAGLND 239
Query: 206 AYATIRSNLLSQDPLPNIDQAYQRVIQDERLRQGEYSIQQNRDNVMAFKVAPDTRGKSKL 265
+YA IR ++++ LP++ + Y + QD + QQ NV+A A ++ +
Sbjct: 240 SYAIIRRQIIAKKALPSLAEVYHILDQD--------NSQQGFSNVVAPPAAFQV-SEAMM 290
Query: 266 VDNSDKFCTHCNREGHDERTCFQIHGFPEWWGDRPRGG 303
N+D GH C++ HGFP + + +GG
Sbjct: 291 ATNNDVNMLCSEWVGHIAERCYKKHGFPLGFTPKGKGG 328
>At1g59265 polyprotein, putative
Length = 1466
Score = 446 bits (1148), Expect = e-125
Identities = 231/516 (44%), Positives = 321/516 (61%), Gaps = 2/516 (0%)
Query: 979 NFVAAVTAGKEPNNFKEAVKDSGWRDAMRNEIQALEDNETWVMEKLPPGKKAL-GSKWVY 1037
+ ++ A EP +A+KD WR+AM +EI A N TW + PP + G +W++
Sbjct: 945 SLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHTWDLVPPPPSHVTIVGCRWIF 1004
Query: 1038 KIKHHSDGSIERLKARLVVFGHHQIEGIDYDETFAPVAKMVTVRTFLAVAAIKKWEVHQM 1097
K++SDGS+ R KARLV G++Q G+DY ETF+PV K ++R L VA + W + Q+
Sbjct: 1005 TKKYNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQL 1064
Query: 1098 DVHNAFLHGDLEEEVYMKVPPGFKNTD-PNLVCRLKKSLYGLKQAPRCWFAKLVTALKRY 1156
DV+NAFL G L ++VYM PPGF + D PN VC+L+K+LYGLKQAPR W+ +L L
Sbjct: 1065 DVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALYGLKQAPRAWYVELRNYLLTI 1124
Query: 1157 GFVQSYSDYSLFTLHRGEIQINVLVYVDDLIIAGNDIAALKIFKAYLGVCFHMKDLGVLK 1216
GFV S SD SLF L RG+ + +LVYVDD++I GND L L F +KD L
Sbjct: 1125 GFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGNDPTLLHNTLDNLSQRFSVKDHEELH 1184
Query: 1217 YFLGLEVARNHEGIYLCQRKYALEIIDETGLLGAKPADFPMEQHHKLALVSGKPLEDPEP 1276
YFLG+E R G++L QR+Y L+++ T ++ AKP PM KL+L SG L DP
Sbjct: 1185 YFLGIEAKRVPTGLHLSQRRYILDLLARTNMITAKPVTTPMAPSPKLSLYSGTKLTDPTE 1244
Query: 1277 YRRLIGRLIYLSVTRPDLAYSVHILSQFMQKPCEEHWEAALRVVRYLKKHPGQGILLRSD 1336
YR ++G L YL+ TRPD++Y+V+ LSQFM P EEH +A R++RYL P GI L+
Sbjct: 1245 YRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEHLQALKRILRYLAGTPNHGIFLKKG 1304
Query: 1337 SELKLEGWCDSDWASCPLTRRSLTGWVVLLDLSPVSWKTKKQPTVSRSSAEAEYRSMAMT 1396
+ L L + D+DWA S G++V L P+SW +KKQ V RSS EAEYRS+A T
Sbjct: 1305 NTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSVANT 1364
Query: 1397 TCELKWLKQLLGDLGVSHSQGMQLYCDSKSALHIAQNPVFHERTKHIEADCHFVRDAVVA 1456
+ E++W+ LL +LG+ ++ +YCD+ A ++ NPVFH R KHI D HF+R+ V +
Sbjct: 1365 SSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCANPVFHSRMKHIAIDYHFIRNQVQS 1424
Query: 1457 GIICPLYVPTSVQLADIFTKALGKAQFEFLLRKLGI 1492
G + ++V T QLAD TK L + F+ K+G+
Sbjct: 1425 GALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGV 1460
Score = 223 bits (568), Expect = 7e-58
Identities = 136/431 (31%), Positives = 218/431 (50%), Gaps = 15/431 (3%)
Query: 395 SWIIDTGASHHVTGNFSCLINGKRITN-TPVGLPNGKDATAIQEGSVILDGG---LRLNN 450
+W++D+GA+HH+T +F+ L + T V + +G GS L L L+N
Sbjct: 330 NWLLDSGATHHITSDFNNLSLHQPYTGGDDVMVADGSTIPISHTGSTSLSTKSRPLNLHN 389
Query: 451 VLFVPQLTCNLISVTQLIDDSNCIVQFTNALCVIQDRTTRTLIGAGERIDGLYFFRGVPK 510
+L+VP + NLISV +L + + V+F A ++D T + G+ D LY +
Sbjct: 390 ILYVPNIHKNLISVYRLCNANGVSVEFFPASFQVKDLNTGVPLLQGKTKDELYEWPIASS 449
Query: 511 VHALMVEGDSAM---DLWHKRLGHPSEKVLKFIPHVSQHSRSKNNRP-----CDVCPRAK 562
+ S+ WH RLGHP+ +L + +S +S S N C C K
Sbjct: 450 QPVSLFASPSSKATHSSWHARLGHPAPSILNSV--ISNYSLSVLNPSHKFLSCSDCLINK 507
Query: 563 QHRDSFPLSENNAASLFELVHCDLWGSYRTRSSCGAQYYLTIVNDYSRAVWVYLLCNKTE 622
++ F S N+ E ++ D+W S S +YY+ V+ ++R W+Y L K++
Sbjct: 508 SNKVPFSQSTINSTRPLEYIYSDVWSS-PILSHDNYRYYVIFVDHFTRYTWLYPLKQKSQ 566
Query: 623 IETMFLNFVAFVDRQFDKKIKKVRSDNGTEFNCLRDYFFNNGIVFETSCVGTPQQNGRVE 682
++ F+ F ++ +F +I SDNG EF L +YF +GI TS TP+ NG E
Sbjct: 567 VKETFITFKNLLENRFQTRIGTFYSDNGGEFVALWEYFSQHGISHLTSPPHTPEHNGLSE 626
Query: 683 RKHQHIMNVARALRFQGHLPMQFWGECVLTACYLINRTPSSVLNYKTPYEKLFGKVPKFD 742
RKH+HI+ L +P +W A YLINR P+ +L ++P++KLFG P +D
Sbjct: 627 RKHRHIVETGLTLLSHASIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQKLFGTSPNYD 686
Query: 743 NMKIFGCLCYAHNQRRDGDKFASRSRKCIFVGYPYGKKGWKLYDLESKEYIVSRDVKFYE 802
+++FGC CY + + K +SR+C+F+GY + + L++ +SR V+F E
Sbjct: 687 KLRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFDE 746
Query: 803 HEFPFDVQLDT 813
+ FPF L T
Sbjct: 747 NCFPFSNYLAT 757
Score = 43.5 bits (101), Expect = 0.001
Identities = 39/208 (18%), Positives = 77/208 (36%), Gaps = 29/208 (13%)
Query: 37 NVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKP-------TTPEKLED---WK 86
N+ +L NY WSR + F GF++G P P D WK
Sbjct: 19 NMSNVTKLTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATIGTDAAPRVNPDYTRWK 78
Query: 87 AVQSMLIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQG 146
++ + +L I S++ +S A +W L++ + + + QL+ L + +G
Sbjct: 79 RQDKLIYSAVLGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLRTQLKQWTKG 138
Query: 147 KGEEVSAYFGRLSRIWDELVTYVKKPTCKCEGCICDINKQVTDFSAEDYLHHFLMGLDGA 206
+ + Y L +D+L K ++ + L L
Sbjct: 139 T-KTIDDYMQGLVTRFDQLALLGK------------------PMDHDEQVERVLENLPEE 179
Query: 207 YATIRSNLLSQDPLPNIDQAYQRVIQDE 234
Y + + ++D P + + ++R++ E
Sbjct: 180 YKPVIDQIAAKDTPPTLTEIHERLLNHE 207
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,338,991
Number of Sequences: 26719
Number of extensions: 1664408
Number of successful extensions: 6347
Number of sequences better than 10.0: 179
Number of HSP's better than 10.0 without gapping: 136
Number of HSP's successfully gapped in prelim test: 47
Number of HSP's that attempted gapping in prelim test: 4993
Number of HSP's gapped (non-prelim): 631
length of query: 1519
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1407
effective length of database: 8,326,068
effective search space: 11714777676
effective search space used: 11714777676
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC137510.5


