

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137079.8 + phase: 0
(518 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
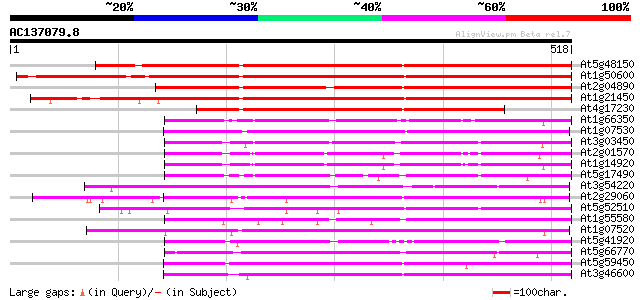
Score E
Sequences producing significant alignments: (bits) Value
At5g48150 SCARECROW gene regulator-like 554 e-158
At1g50600 scarecrow-like 5 (SCL5) 552 e-157
At2g04890 putative SCARECROW gene regulator 480 e-136
At1g21450 scarecrow-like 1 (SCL1) 392 e-109
At4g17230 scarecrow-like 13 (SCL13) 351 5e-97
At1g66350 gibberellin regulatory protein like 223 3e-58
At1g07530 transcription factor scarecrow-like 14, putative 219 4e-57
At3g03450 RGA1-like protein 214 9e-56
At2g01570 putative RGA1, giberellin repsonse modulation protein 214 1e-55
At1g14920 signal response protein (GAI) 213 3e-55
At5g17490 RGA-like protein 206 3e-53
At3g54220 SCARECROW1 205 6e-53
At2g29060 putative SCARECROW gene regulator 201 6e-52
At5g52510 scarecrow-like 8 (SCL8) 197 1e-50
At1g55580 unknown protein 196 3e-50
At1g07520 transcription factor scarecrow-like 14, putative 192 4e-49
At5g41920 SCARECROW gene regulator-like protein 186 3e-47
At5g66770 SCARECROW gene regulator 185 5e-47
At5g59450 putative SCARECROW transcriptional regulatory protein ... 185 5e-47
At3g46600 scarecrow-like protein 184 8e-47
>At5g48150 SCARECROW gene regulator-like
Length = 490
Score = 554 bits (1428), Expect = e-158
Identities = 276/440 (62%), Positives = 343/440 (77%), Gaps = 10/440 (2%)
Query: 80 SCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDPLLLEAEKWNKMIEMI 139
SC+T +D HKIRE+E M+G D+ D+L D+ E W +E I
Sbjct: 60 SCVTDELNDFKHKIREIETVMMGPDSLDLLVDCTDSF-----DSTASQEINGWRSTLEAI 114
Query: 140 SRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASS 199
SR DL+ L +CAKA+SEND+ A +M +L +MVSVSG PIQRLGAY+LE LVA++ASS
Sbjct: 115 SRRDLRADLVSCAKAMSENDLMMAHSMMEKLRQMVSVSGEPIQRLGAYLLEGLVAQLASS 174
Query: 200 GSIIYKSL-KCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHII 258
GS IYK+L +C EP S ELLS+MH+LYE+CPY KFGYMSANG IAEA+K+E+ +HII
Sbjct: 175 GSSIYKALNRCPEP---ASTELLSYMHILYEVCPYFKFGYMSANGAIAEAMKEENRVHII 231
Query: 259 DFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYN 318
DFQI QG QW++LIQA A +PGGPP+IRITG DD TSAYARGGGL IVG RL+KLA+ +N
Sbjct: 232 DFQIGQGSQWVTLIQAFAARPGGPPRIRITGIDDMTSAYARGGGLSIVGNRLAKLAKQFN 291
Query: 319 VAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKC 378
V FEF+++ VS SEV+ ++L +R GEA+AVNFA +LHH+PDE V +NHRDRL+R+ K
Sbjct: 292 VPFEFNSVSVSVSEVKPKNLGVRPGEALAVNFAFVLHHMPDESV-STENHRDRLLRMVKS 350
Query: 379 LSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLARE 438
LSPKVVTLVEQESNTN FF RF+ETMNYY A+FESIDV LPR+H++RINVEQHCLAR+
Sbjct: 351 LSPKVVTLVEQESNTNTAAFFPRFMETMNYYAAMFESIDVTLPRDHKQRINVEQHCLARD 410
Query: 439 VVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHYTLQEK 498
VVN++ACEGA+RVERHE+L KWRS F MAGFTPYPLS +N +I++LL NY Y L+E+
Sbjct: 411 VVNIIACEGADRVERHELLGKWRSRFGMAGFTPYPLSPLVNSTIKSLLRNYSDKYRLEER 470
Query: 499 DGALYLGWMNQPLITSSAWR 518
DGALYLGWM++ L+ S AW+
Sbjct: 471 DGALYLGWMHRDLVASCAWK 490
>At1g50600 scarecrow-like 5 (SCL5)
Length = 597
Score = 552 bits (1423), Expect = e-157
Identities = 292/514 (56%), Positives = 366/514 (70%), Gaps = 19/514 (3%)
Query: 7 IRNLDSYCFLQNENLENYSSSDNGSHSTYPSFQALEQNLE-SSNNSPVSKLQSKSYTFTS 65
++ DSYC L+ SSS SH + SSN SP+S+ + + + +
Sbjct: 101 VQTFDSYCTLE-------SSSGTKSHPCLNNKNNSSSTTSFSSNESPISQANNNNLSRFN 153
Query: 66 QNSLEIINDS-LENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDP 124
+S E N+S L S N+ +L +++LE AM+ D + YN+ Q+
Sbjct: 154 NHSPEENNNSPLSGSSATNTNETELSLMLKDLETAMMEPDVDNS---YNNQGGFGQQHGV 210
Query: 125 LLLEAEKWNKMIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRL 184
+ + + +EMISRGDLK +L+ CAKA+ D+E +WL+S+L +MVSVSG P+QRL
Sbjct: 211 V---SSAMYRSMEMISRGDLKGVLYECAKAVENYDLEMTDWLISQLQQMVSVSGEPVQRL 267
Query: 185 GAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGV 244
GAYMLE LVAR+ASSGS IYK+L+CK+P T ELL++MH+LYE CPY KFGY SANG
Sbjct: 268 GAYMLEGLVARLASSGSSIYKALRCKDP---TGPELLTYMHILYEACPYFKFGYESANGA 324
Query: 245 IAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLG 304
IAEA+K+ES +HIIDFQI+QG QW+SLI+AL +PGGPP +RITG DD S++AR GGL
Sbjct: 325 IAEAVKNESFVHIIDFQISQGGQWVSLIRALGARPGGPPNVRITGIDDPRSSFARQGGLE 384
Query: 305 IVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHG 364
+VG+RL KLAE V FEFH + +EV +E L +R GEA+AVNF ++LHH+PDE V
Sbjct: 385 LVGQRLGKLAEMCGVPFEFHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTV 444
Query: 365 GKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREH 424
+NHRDRL+RL K LSP VVTLVEQE+NTN PF RFVETMN+Y AVFESIDV L R+H
Sbjct: 445 -ENHRDRLLRLVKHLSPNVVTLVEQEANTNTAPFLPRFVETMNHYLAVFESIDVKLARDH 503
Query: 425 RERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQN 484
+ERINVEQHCLAREVVNL+ACEG ER ERHE L KWRS F MAGF PYPLSSY+N +I+
Sbjct: 504 KERINVEQHCLAREVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKG 563
Query: 485 LLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
LLE+Y YTL+E+DGALYLGW NQPLITS AWR
Sbjct: 564 LLESYSEKYTLEERDGALYLGWKNQPLITSCAWR 597
>At2g04890 putative SCARECROW gene regulator
Length = 413
Score = 480 bits (1236), Expect = e-136
Identities = 238/384 (61%), Positives = 299/384 (76%), Gaps = 11/384 (2%)
Query: 135 MIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVA 194
++E ISRGDLK +L CAKA+SEN++ A W M EL MVS+SG PIQRLGAYMLE LVA
Sbjct: 41 IVEAISRGDLKLVLVACAKAVSENNLLMARWCMGELRGMVSISGEPIQRLGAYMLEGLVA 100
Query: 195 RIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESE 254
R+A+SGS IYKSL+ +EP S E LS+++VL+E+CPY KFGYMSANG IAEA+KDE
Sbjct: 101 RLAASGSSIYKSLQSREP---ESYEFLSYVYVLHEVCPYFKFGYMSANGAIAEAMKDEER 157
Query: 255 IHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLA 314
IHIIDFQI QG QW++LIQA A +PGG P IRITG D G L V +RL KLA
Sbjct: 158 IHIIDFQIGQGSQWIALIQAFAARPGGAPNIRITGVGD-------GSVLVTVKKRLEKLA 210
Query: 315 ESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVR 374
+ ++V F F+A+ EV +E+L++R GEA+ VNFA MLHH+PDE V +NHRDRL+R
Sbjct: 211 KKFDVPFRFNAVSRPSCEVEVENLDVRDGEALGVNFAYMLHHLPDESV-SMENHRDRLLR 269
Query: 375 LAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHC 434
+ K LSPKVVTLVEQE NTN PF RF+ET++YY A+FESIDV LPR H+ERIN+EQHC
Sbjct: 270 MVKSLSPKVVTLVEQECNTNTSPFLPRFLETLSYYTAMFESIDVMLPRNHKERINIEQHC 329
Query: 435 LAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHYT 494
+AR+VVN++ACEGAER+ERHE+L KW+S F+MAGF PYPLSS I+ +I+ LL +Y Y
Sbjct: 330 MARDVVNIIACEGAERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALLRDYSNGYA 389
Query: 495 LQEKDGALYLGWMNQPLITSSAWR 518
++E+DGALYLGWM++ L++S AW+
Sbjct: 390 IEERDGALYLGWMDRILVSSCAWK 413
>At1g21450 scarecrow-like 1 (SCL1)
Length = 593
Score = 392 bits (1007), Expect = e-109
Identities = 219/519 (42%), Positives = 321/519 (61%), Gaps = 37/519 (7%)
Query: 20 NLENYSSSDNGSHSTYP------SFQALEQNLESSNNSPVSKLQSKSYTFTSQNSLEIIN 73
++ ++ S D+ + + P FQ + +S+ + Q+ SY+ S++++
Sbjct: 92 SVSSFGSLDSFPYQSRPVLGCSMEFQLPLDSTSTSSTRLLGDYQAVSYS----PSMDVVE 147
Query: 74 DSLENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVII----------QQESD 123
+ + + + KI+ELE A+LG + M+ I N I Q +
Sbjct: 148 E---------FDDEQMRSKIQELERALLGDEDDKMVGIDNLMEIDSEWSYQNESEQHQDS 198
Query: 124 PLLLEAEKWNKMI---EMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNP 180
P + N + E++S+ K+IL +CA+A+SE +E A +++EL ++VS+ G+P
Sbjct: 199 PKESSSADSNSHVSSKEVVSQATPKQILISCARALSEGKLEEALSMVNELRQIVSIQGDP 258
Query: 181 IQRLGAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMS 240
QR+ AYM+E L AR+A+SG IY++LKCKEP S E L+ M VL+E+CP KFG+++
Sbjct: 259 SQRIAAYMVEGLAARMAASGKFIYRALKCKEP---PSDERLAAMQVLFEVCPCFKFGFLA 315
Query: 241 ANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARG 300
ANG I EA+K E E+HIIDF INQG Q+M+LI+++A PG P++R+TG DD S
Sbjct: 316 ANGAILEAIKGEEEVHIIDFDINQGNQYMTLIRSIAELPGKRPRLRLTGIDDPESVQRSI 375
Query: 301 GGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDE 360
GGL I+G RL +LAE V+F+F A+ S V L + GE + VNFA LHH+PDE
Sbjct: 376 GGLRIIGLRLEQLAEDNGVSFKFKAMPSKTSIVSPSTLGCKPGETLIVNFAFQLHHMPDE 435
Query: 361 DVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVAL 420
V N RD L+ + K L+PK+VT+VEQ+ NTN PFF RF+E YY AVFES+D+ L
Sbjct: 436 SVTT-VNQRDELLHMVKSLNPKLVTVVEQDVNTNTSPFFPRFIEAYEYYSAVFESLDMTL 494
Query: 421 PREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINY 480
PRE +ER+NVE+ CLAR++VN+VACEG ER+ER+E KWR+ MAGF P P+S+ +
Sbjct: 495 PRESQERMNVERQCLARDIVNIVACEGEERIERYEAAGKWRARMMMAGFNPKPMSAKVTN 554
Query: 481 SIQNLL-ENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
+IQNL+ + Y Y L+E+ G L+ W + LI +SAWR
Sbjct: 555 NIQNLIKQQYCNKYKLKEEMGELHFCWEEKSLIVASAWR 593
>At4g17230 scarecrow-like 13 (SCL13)
Length = 287
Score = 351 bits (901), Expect = 5e-97
Identities = 180/285 (63%), Positives = 218/285 (76%), Gaps = 4/285 (1%)
Query: 173 MVSVSGNPIQRLGAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICP 232
MVSVSG+PIQRLG YM E L AR+ SGS IYKSLKC EP T +EL+S+M VLYEICP
Sbjct: 1 MVSVSGSPIQRLGTYMAEGLRARLEGSGSNIYKSLKCNEP---TGRELMSYMSVLYEICP 57
Query: 233 YLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDD 292
Y KF Y +AN I EA+ E+ +HIIDFQI QG Q+M LIQ LA PGGPP +R+TG DD
Sbjct: 58 YWKFAYTTANVEILEAIAGETRVHIIDFQIAQGSQYMFLIQELAKHPGGPPLLRVTGVDD 117
Query: 293 STSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAM 352
S S YARGGGL +VGERL+ LA+S V FEFH +S +V+ E L L G A+ VNF
Sbjct: 118 SQSTYARGGGLSLVGERLATLAQSCGVPFEFHDAIMSGCKVQREHLGLEPGFAVVVNFPY 177
Query: 353 MLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAV 412
+LHH+PDE V +NHRDRL+ L K LSPK+VTLVEQESNTN PF +RFVET++YY A+
Sbjct: 178 VLHHMPDESV-SVENHRDRLLHLIKSLSPKLVTLVEQESNTNTSPFLSRFVETLDYYTAM 236
Query: 413 FESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVL 457
FESID A PR+ ++RI+ EQHC+AR++VN++ACE +ERVERHEVL
Sbjct: 237 FESIDAARPRDDKQRISAEQHCVARDIVNMIACEESERVERHEVL 281
>At1g66350 gibberellin regulatory protein like
Length = 511
Score = 223 bits (567), Expect = 3e-58
Identities = 135/379 (35%), Positives = 211/379 (55%), Gaps = 28/379 (7%)
Query: 144 LKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSII 203
L L CA+A+ +N+++ A+ L+ + + S ++++ Y E L RI I
Sbjct: 152 LVHALLACAEAVQQNNLKLADALVKHVGLLASSQAGAMRKVATYFAEGLARRIYR----I 207
Query: 204 YKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQIN 263
Y ++ + +S +H YE CPYLKF + +AN I E ++H+ID +N
Sbjct: 208 YP----RDDVALSSFSDTLQIH-FYESCPYLKFAHFTANQAILEVFATAEKVHVIDLGLN 262
Query: 264 QGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEF 323
G+QW +LIQALA +P GPP R+TG S + + VG +L +LA + V FEF
Sbjct: 263 HGLQWPALIQALALRPNGPPDFRLTGIGYSLT------DIQEVGWKLGQLASTIGVNFEF 316
Query: 324 HAIGVSP-SEVRLEDLELRRG-EAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSP 381
+I ++ S+++ E L++R G E++AVN LH + H G D+ + K + P
Sbjct: 317 KSIALNNLSDLKPEMLDIRPGLESVAVNSVFELHRLL---AHPGS--IDKFLSTIKSIRP 371
Query: 382 KVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVN 441
++T+VEQE+N N F RF E+++YY ++F+S++ P + R + + L R+++N
Sbjct: 372 DIMTVVEQEANHNGTVFLDRFTESLHYYSSLFDSLE-GPPSQDRV---MSELFLGRQILN 427
Query: 442 LVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQG--HYTLQEKD 499
LVACEG +RVERHE L +WR+ F + GF P + S LL Y G Y ++E +
Sbjct: 428 LVACEGEDRVERHETLNQWRNRFGLGGFKPVSIGSNAYKQASMLLALYAGADGYNVEENE 487
Query: 500 GALYLGWMNQPLITSSAWR 518
G L LGW +PLI +SAWR
Sbjct: 488 GCLLLGWQTRPLIATSAWR 506
>At1g07530 transcription factor scarecrow-like 14, putative
Length = 769
Score = 219 bits (557), Expect = 4e-57
Identities = 128/377 (33%), Positives = 203/377 (52%), Gaps = 6/377 (1%)
Query: 143 DLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSI 202
DL+ +L CA+A+S +D TA ++ ++ + S GN +RL Y +L AR+A +G+
Sbjct: 392 DLRTLLVLCAQAVSVDDRRTANEMLRQIREHSSPLGNGSERLAHYFANSLEARLAGTGTQ 451
Query: 203 IYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQI 262
IY +L K+ A ++L +CP+ K + AN + + + IHIIDF I
Sbjct: 452 IYTALSSKKTSAA---DMLKAYQTYMSVCPFKKAAIIFANHSMMRFTANANTIHIIDFGI 508
Query: 263 NQGIQWMSLIQALA-GKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAF 321
+ G QW +LI L+ +PGG PK+RITG + + G+ G RL++ + +NV F
Sbjct: 509 SYGFQWPALIHRLSLSRPGGSPKLRITGIELPQRGFRPAEGVQETGHRLARYCQRHNVPF 568
Query: 322 EFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSP 381
E++AI +++EDL+LR+GE + VN ++ DE V + RD +++L + ++P
Sbjct: 569 EYNAIAQKWETIQVEDLKLRQGEYVVVNSLFRFRNLLDETVLVN-SPRDAVLKLIRKINP 627
Query: 382 KVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVN 441
V N N F RF E + +Y AVF+ D L RE R+ E+ RE+VN
Sbjct: 628 NVFIPAILSGNYNAPFFVTRFREALFHYSAVFDMCDSKLAREDEMRLMYEKEFYGREIVN 687
Query: 442 LVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLEN-YQGHYTLQEKDG 500
+VACEG ERVER E K+W++ AGF PL + +++ +EN Y ++ + +
Sbjct: 688 VVACEGTERVERPETYKQWQARLIRAGFRQLPLEKELMQNLKLKIENGYDKNFDVDQNGN 747
Query: 501 ALYLGWMNQPLITSSAW 517
L GW + + SS W
Sbjct: 748 WLLQGWKGRIVYASSLW 764
>At3g03450 RGA1-like protein
Length = 547
Score = 214 bits (545), Expect = 9e-56
Identities = 130/382 (34%), Positives = 202/382 (52%), Gaps = 23/382 (6%)
Query: 144 LKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSII 203
L L CA+AI + ++ A+ L+ + + + ++ Y +AL RI
Sbjct: 180 LVHALVACAEAIHQENLNLADALVKRVGTLAGSQAGAMGKVATYFAQALARRI------- 232
Query: 204 YKSLKCKEPITAT---SKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDF 260
Y+ + + A S E + MH YE CPYLKF + +AN I EA+ +H+ID
Sbjct: 233 YRDYTAETDVCAAVNPSFEEVLEMH-FYESCPYLKFAHFTANQAILEAVTTARRVHVIDL 291
Query: 261 QINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVA 320
+NQG+QW +L+QALA +PGGPP R+TG + L +G +L++ A++ V
Sbjct: 292 GLNQGMQWPALMQALALRPGGPPSFRLTGIGPPQT--ENSDSLQQLGWKLAQFAQNMGVE 349
Query: 321 FEFHAIGV-SPSEVRLEDLELR-RGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKC 378
FEF + S S++ E E R E + VN LH + ++L+ K
Sbjct: 350 FEFKGLAAESLSDLEPEMFETRPESETLVVNSVFELHR-----LLARSGSIEKLLNTVKA 404
Query: 379 LSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLARE 438
+ P +VT+VEQE+N N + F RF E ++YY ++F+S++ + ++R+ E + L R+
Sbjct: 405 IKPSIVTVVEQEANHNGIVFLDRFNEALHYYSSLFDSLEDSYSLPSQDRVMSEVY-LGRQ 463
Query: 439 VVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQ--GHYTLQ 496
++N+VA EG++RVERHE +WR AGF P L S LL Y Y ++
Sbjct: 464 ILNVVAAEGSDRVERHETAAQWRIRMKSAGFDPIHLGSSAFKQASMLLSLYATGDGYRVE 523
Query: 497 EKDGALYLGWMNQPLITSSAWR 518
E DG L +GW +PLIT+SAW+
Sbjct: 524 ENDGCLMIGWQTRPLITTSAWK 545
>At2g01570 putative RGA1, giberellin repsonse modulation protein
Length = 587
Score = 214 bits (544), Expect = 1e-55
Identities = 137/380 (36%), Positives = 210/380 (55%), Gaps = 24/380 (6%)
Query: 144 LKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSII 203
L L CA+AI +N++ AE L+ ++ + ++++ Y EAL RI
Sbjct: 221 LVHALMACAEAIQQNNLTLAEALVKQIGCLAVSQAGAMRKVATYFAEALARRI------- 273
Query: 204 YKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQIN 263
Y+ + I + L MH YE CPYLKF + +AN I EA + + +H+IDF +N
Sbjct: 274 YRLSPPQNQIDHCLSDTLQ-MH-FYETCPYLKFAHFTANQAILEAFEGKKRVHVIDFSMN 331
Query: 264 QGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEF 323
QG+QW +L+QALA + GGPP R+TG A L VG +L++LAE+ +V FE+
Sbjct: 332 QGLQWPALMQALALREGGPPTFRLTGI--GPPAPDNSDHLHEVGCKLAQLAEAIHVEFEY 389
Query: 324 HA-IGVSPSEVRLEDLELRRG--EAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLS 380
+ S +++ LELR EA+AVN LH + G ++++ + K +
Sbjct: 390 RGFVANSLADLDASMLELRPSDTEAVAVNSVFELH-----KLLGRPGGIEKVLGVVKQIK 444
Query: 381 PKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVV 440
P + T+VEQESN N F RF E+++YY +F+S++ +P ++++ E + L +++
Sbjct: 445 PVIFTVVEQESNHNGPVFLDRFTESLHYYSTLFDSLE-GVPNS-QDKVMSEVY-LGKQIC 501
Query: 441 NLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLE--NYQGHYTLQEK 498
NLVACEG +RVERHE L +W + F +G P L S LL N Y ++E
Sbjct: 502 NLVACEGPDRVERHETLSQWGNRFGSSGLAPAHLGSNAFKQASMLLSVFNSGQGYRVEES 561
Query: 499 DGALYLGWMNQPLITSSAWR 518
+G L LGW +PLIT+SAW+
Sbjct: 562 NGCLMLGWHTRPLITTSAWK 581
>At1g14920 signal response protein (GAI)
Length = 533
Score = 213 bits (541), Expect = 3e-55
Identities = 133/380 (35%), Positives = 212/380 (55%), Gaps = 24/380 (6%)
Query: 144 LKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSII 203
L L CA+A+ + ++ AE L+ ++ + ++++ Y EAL RI
Sbjct: 169 LVHALLACAEAVQKENLTVAEALVKQIGFLAVSQIGAMRKVATYFAEALARRI------- 221
Query: 204 YKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQIN 263
Y+ + PI + + L MH YE CPYLKF + +AN I EA + + +H+IDF ++
Sbjct: 222 YRLSPSQSPIDHSLSDTLQ-MH-FYETCPYLKFAHFTANQAILEAFQGKKRVHVIDFSMS 279
Query: 264 QGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEF 323
QG+QW +L+QALA +PGGPP R+TG A L VG +L+ LAE+ +V FE+
Sbjct: 280 QGLQWPALMQALALRPGGPPVFRLTGI--GPPAPDNFDYLHEVGCKLAHLAEAIHVEFEY 337
Query: 324 HA-IGVSPSEVRLEDLELRRG--EAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLS 380
+ + +++ LELR E++AVN LH + G D+++ + +
Sbjct: 338 RGFVANTLADLDASMLELRPSEIESVAVNSVFELH-----KLLGRPGAIDKVLGVVNQIK 392
Query: 381 PKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVV 440
P++ T+VEQESN N F RF E+++YY +F+S++ +P ++++ E + L +++
Sbjct: 393 PEIFTVVEQESNHNSPIFLDRFTESLHYYSTLFDSLE-GVP-SGQDKVMSEVY-LGKQIC 449
Query: 441 NLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQG--HYTLQEK 498
N+VAC+G +RVERHE L +WR+ F AGF + S LL + G Y ++E
Sbjct: 450 NVVACDGPDRVERHETLSQWRNRFGSAGFAAAHIGSNAFKQASMLLALFNGGEGYRVEES 509
Query: 499 DGALYLGWMNQPLITSSAWR 518
DG L LGW +PLI +SAW+
Sbjct: 510 DGCLMLGWHTRPLIATSAWK 529
>At5g17490 RGA-like protein
Length = 523
Score = 206 bits (523), Expect = 3e-53
Identities = 129/383 (33%), Positives = 208/383 (53%), Gaps = 31/383 (8%)
Query: 144 LKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSII 203
L + L CA+A+ ++ A+ L+ + + + + ++ Y EAL RI I
Sbjct: 157 LVQALVACAEAVQLENLSLADALVKRVGLLAASQAGAMGKVATYFAEALARRIYR----I 212
Query: 204 YKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQIN 263
+ S +P S E + M+ Y+ CPYLKF + +AN I EA+ +H+ID +N
Sbjct: 213 HPSAAAIDP----SFEEILQMN-FYDSCPYLKFAHFTANQAILEAVTTSRVVHVIDLGLN 267
Query: 264 QGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEF 323
QG+QW +L+QALA +PGGPP R+TG + ++ G+ +G +L++LA++ V F+F
Sbjct: 268 QGMQWPALMQALALRPGGPPSFRLTGVGNPSNR----EGIQELGWKLAQLAQAIGVEFKF 323
Query: 324 HAIGVSPSEVRLEDLE------LRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAK 377
+ + + RL DLE E + VN LH V + ++L+ K
Sbjct: 324 NGL----TTERLSDLEPDMFETRTESETLVVNSVFELHPVLSQ-----PGSIEKLLATVK 374
Query: 378 CLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAR 437
+ P +VT+VEQE+N N F RF E ++YY ++F+S++ + ++R+ E + L R
Sbjct: 375 AVKPGLVTVVEQEANHNGDVFLDRFNEALHYYSSLFDSLEDGVVIPSQDRVMSEVY-LGR 433
Query: 438 EVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSS--YINYSIQNLLENYQGHYTL 495
+++NLVA EG++R+ERHE L +WR AGF P L S + S+ L Y +
Sbjct: 434 QILNLVATEGSDRIERHETLAQWRKRMGSAGFDPVNLGSDAFKQASLLLALSGGGDGYRV 493
Query: 496 QEKDGALYLGWMNQPLITSSAWR 518
+E DG+L L W +PLI +SAW+
Sbjct: 494 EENDGSLMLAWQTKPLIAASAWK 516
>At3g54220 SCARECROW1
Length = 653
Score = 205 bits (521), Expect = 6e-53
Identities = 148/457 (32%), Positives = 225/457 (48%), Gaps = 25/457 (5%)
Query: 70 EIINDSLENESCLTHNQDDLWHK-----IRELENAMLGQDAADMLDIYNDTV-IIQQESD 123
+I N+ + H Q HK I++ E DA + TV +Q +
Sbjct: 209 QISNNPSPPQQQQQHQQQQQQHKPPPPPIQQQERENSSTDAPPQPETVTATVPAVQTNTA 268
Query: 124 PLLLEAEKWNKMIEMISRG-DLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQ 182
L E ++ K + G L +L CA+A+S +++E A L+ E+S++ + G Q
Sbjct: 269 EALRERKEEIKRQKQDEEGLHLLTLLLQCAEAVSADNLEEANKLLLEISQLSTPYGTSAQ 328
Query: 183 RLGAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSAN 242
R+ AY EA+ AR+ +S IY +L + S +++S V I P +KF + +AN
Sbjct: 329 RVAAYFSEAMSARLLNSCLGIYAALPSRWMPQTHSLKMVSAFQVFNGISPLVKFSHFTAN 388
Query: 243 GVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGG 302
I EA + E +HIID I QG+QW L LA +PGGPP +R+TG S A
Sbjct: 389 QAIQEAFEKEDSVHIIDLDIMQGLQWPGLFHILASRPGGPPHVRLTGLGTSMEA------ 442
Query: 303 LGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAM-MLHHVPDED 361
L G+RLS A+ + FEF + + E L +R+ EA+AV++ L+ V D
Sbjct: 443 LQATGKRLSDFADKLGLPFEFCPLAEKVGNLDTERLNVRKREAVAVHWLQHSLYDVTGSD 502
Query: 362 VHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALP 421
H + L + L+PKVVT+VEQ+ ++ F RFVE ++YY A+F+S+ +
Sbjct: 503 AH--------TLWLLQRLAPKVVTVVEQDL-SHAGSFLGRFVEAIHYYSALFDSLGASYG 553
Query: 422 REHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYS 481
E ER VEQ L++E+ N++A G R + WR GF L+
Sbjct: 554 EESEERHVVEQQLLSKEIRNVLAVGGPSR-SGEVKFESWREKMQQCGFKGISLAGNAATQ 612
Query: 482 IQNLLENYQGH-YTLQEKDGALYLGWMNQPLITSSAW 517
LL + YTL + +G L LGW + L+T+SAW
Sbjct: 613 ATLLLGMFPSDGYTLVDDNGTLKLGWKDLSLLTASAW 649
>At2g29060 putative SCARECROW gene regulator
Length = 1336
Score = 201 bits (512), Expect = 6e-52
Identities = 126/379 (33%), Positives = 200/379 (52%), Gaps = 8/379 (2%)
Query: 143 DLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSI 202
DL+ +L +CA+A+S ND TA+ L+S + + S G+ +RL Y +L AR+A G+
Sbjct: 317 DLRTMLVSCAQAVSINDRRTADELLSRIRQHSSSYGDGTERLAHYFANSLEARLAGIGTQ 376
Query: 203 IYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESE--IHIIDF 260
+Y +L K+ T+TS ++L +CP+ K + AN I + IHIIDF
Sbjct: 377 VYTALSSKK--TSTS-DMLKAYQTYISVCPFKKIAIIFANHSIMRLASSANAKTIHIIDF 433
Query: 261 QINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVA 320
I+ G QW SLI LA + G K+RITG + + G+ G RL+K + +N+
Sbjct: 434 GISDGFQWPSLIHRLAWRRGSSCKLRITGIELPQRGFRPAEGVIETGRRLAKYCQKFNIP 493
Query: 321 FEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLS 380
FE++AI ++LEDL+L+ GE +AVN ++ DE V + RD +++L + +
Sbjct: 494 FEYNAIAQKWESIKLEDLKLKEGEFVAVNSLFRFRNLLDETV-AVHSPRDTVLKLIRKIK 552
Query: 381 PKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVV 440
P V + N F RF E + +Y ++F+ D L RE R+ E+ RE++
Sbjct: 553 PDVFIPGILSGSYNAPFFVTRFREVLFHYSSLFDMCDTNLTREDPMRVMFEKEFYGREIM 612
Query: 441 NLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLEN--YQGHYTLQEK 498
N+VACEG ERVER E K+W++ AGF PL + ++ ++E+ + + +
Sbjct: 613 NVVACEGTERVERPESYKQWQARAMRAGFRQIPLEKELVQKLKLMVESGYKPKEFDVDQD 672
Query: 499 DGALYLGWMNQPLITSSAW 517
L GW + + SS W
Sbjct: 673 CHWLLQGWKGRIVYGSSIW 691
Score = 186 bits (473), Expect = 2e-47
Identities = 141/552 (25%), Positives = 263/552 (47%), Gaps = 61/552 (11%)
Query: 22 ENYSSSDNGSHSTYPSFQALEQNLESSNNSPVSKLQSKSYTFTSQNSLE----------- 70
+++S +D+ +++ + +++++ S++ PV+++ KS ++++L+
Sbjct: 787 QSFSPADSLITNSWDASGSIDESAYSADPQPVNEIMVKSMFSDAESALQFKKGVEEASKF 846
Query: 71 -------IIN---------DSLENESCLTH-----NQDDLWHKIRELENAMLGQDAADML 109
+IN DS++ E L N + + ++R + + + +
Sbjct: 847 LPNSDQWVINLDIERSERRDSVKEEMGLDQLRVKKNHERDFEEVRSSKQFASNVEDSKVT 906
Query: 110 DIYNDTVIIQQESDP-LLLEAE----------------KWNKMIEMISRGDLKEILFTCA 152
D+++ +++ E DP LL++E K K +++ D + +L CA
Sbjct: 907 DMFDKVLLLDGECDPQTLLDSEIQAIRSSKNIGEKGKKKKKKKSQVV---DFRTLLTHCA 963
Query: 153 KAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARI-ASSGSII---YKSLK 208
+AIS D TA + ++ + S G+ QRL AL AR+ S+G +I Y +L
Sbjct: 964 QAISTGDKTTALEFLLQIRQQSSPLGDAGQRLAHCFANALEARLQGSTGPMIQTYYNALT 1023
Query: 209 CKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQW 268
+ T+ + + V P++ Y + +I + KD +HI+DF I G QW
Sbjct: 1024 SS--LKDTAADTIRAYRVYLSSSPFVTLMYFFSIWMILDVAKDAPVLHIVDFGILYGFQW 1081
Query: 269 MSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGV 328
IQ+++ + P K+RITG + + + G RL++ + +NV FE+ AI
Sbjct: 1082 PMFIQSISDRKDVPRKLRITGIELPQCGFRPAERIEETGRRLAEYCKRFNVPFEYKAIAS 1141
Query: 329 SPSE-VRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLV 387
E +R+EDL++R E +AVN + L ++ DE RD +++L + ++P V
Sbjct: 1142 QNWETIRIEDLDIRPNEVLAVNAGLRLKNLQDETGSEENCPRDAVLKLIRNMNPDVFIHA 1201
Query: 388 EQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEG 447
+ N F +RF E + +Y A+F+ D LPR+++ERI E+ RE +N++ACE
Sbjct: 1202 IVNGSFNAPFFISRFKEAVYHYSALFDMFDSTLPRDNKERIRFEREFYGREAMNVIACEE 1261
Query: 448 AERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGH--YTLQEKDGALYLG 505
A+RVER E ++W+ AGF + + + L+ ++ H + + E L G
Sbjct: 1262 ADRVERPETYRQWQVRMVRAGFKQKTIKPELVELFRGKLKKWRYHKDFVVDENSKWLLQG 1321
Query: 506 WMNQPLITSSAW 517
W + L SS W
Sbjct: 1322 WKGRTLYASSCW 1333
>At5g52510 scarecrow-like 8 (SCL8)
Length = 525
Score = 197 bits (501), Expect = 1e-50
Identities = 144/468 (30%), Positives = 234/468 (49%), Gaps = 47/468 (10%)
Query: 84 HNQDDLWHKIRELENAML-------GQDAADML-----DIYNDTVIIQQESDPLLLEAEK 131
H +++ + +RELE +L G D ++ D + V +P+L +
Sbjct: 72 HESENMLNSLRELEKQLLDDDDESGGDDDVSVITNSNSDWIQNLVTPNPNPNPVLSFSPS 131
Query: 132 WNKMIEMISRGDL------KEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLG 185
+ S ++ + A AI+E E A +++ +S+ ++ N ++L
Sbjct: 132 SSSSSSSPSTASTTTSVCSRQTVMEIATAIAEGKTEIATEILARVSQTPNLERNSEEKLV 191
Query: 186 AYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVI 245
+M+ AL +RIAS + +Y KE L +LYE+ P K G+ +AN I
Sbjct: 192 DFMVAALRSRIASPVTELY------------GKEHLISTQLLYELSPCFKLGFEAANLAI 239
Query: 246 AEALKDESE----IHIIDFQINQGIQWMSLIQALAGKPGGP------PKIRITGFDDST- 294
+A + H+IDF I +G Q+++L++ L+ + G P ++IT ++
Sbjct: 240 LDAADNNDGGMMIPHVIDFDIGEGGQYVNLLRTLSTRRNGKSQSQNSPVVKITAVANNVY 299
Query: 295 SAYARGGG---LGIVGERLSKLAESYNVAFEFHAI-GVSPSEVRLEDLELRRGEAIAVNF 350
GG L VG+ LS+L + ++ F+ + + ++ E L E +AVN
Sbjct: 300 GCLVDDGGEERLKAVGDLLSQLGDRLGISVSFNVVTSLRLGDLNRESLGCDPDETLAVNL 359
Query: 351 AMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYF 410
A L+ VPDE V +N RD L+R K L P+VVTLVEQE N+N PF R E+ Y
Sbjct: 360 AFKLYRVPDESVCT-ENPRDELLRRVKGLKPRVVTLVEQEMNSNTAPFLGRVSESCACYG 418
Query: 411 AVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFT 470
A+ ES++ +P + +R VE+ + R++VN VACEG +R+ER EV KWR +MAGF
Sbjct: 419 ALLESVESTVPSTNSDRAKVEEG-IGRKLVNAVACEGIDRIERCEVFGKWRMRMSMAGFE 477
Query: 471 PYPLSSYINYSIQNLLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
PLS I S+++ +T++E +G + GWM + L +SAWR
Sbjct: 478 LMPLSEKIAESMKSRGNRVHPGFTVKEDNGGVCFGWMGRALTVASAWR 525
>At1g55580 unknown protein
Length = 445
Score = 196 bits (498), Expect = 3e-50
Identities = 127/415 (30%), Positives = 209/415 (49%), Gaps = 50/415 (12%)
Query: 144 LKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARI------- 196
L+ +LFT A +S+++ A+ L+S LS S G+ +RL +AL RI
Sbjct: 41 LRRLLFTAANFVSQSNFTAAQNLLSILSLNSSPHGDSTERLVHLFTKALSVRINRQQQDQ 100
Query: 197 -------------ASSGSIIYKSLKCKEPITATSKELLSHMHVLY-----EICPYLKFGY 238
S S ++ S CKE +K S Y ++ P+++FG+
Sbjct: 101 TAETVATWTTNEMTMSNSTVFTSSVCKEQFLFRTKNNNSDFESCYYLWLNQLTPFIRFGH 160
Query: 239 MSANGVIAEALK--DESEIHIIDFQINQGIQWMSLIQALAGKPGGP----PKIRITGFDD 292
++AN I +A + D +HI+D I+QG+QW L+QALA + P P +RITG
Sbjct: 161 LTANQAILDATETNDNGALHILDLDISQGLQWPPLMQALAERSSNPSSPPPSLRITGCGR 220
Query: 293 STSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSE-------VRLEDLELRRGEA 345
+ GL G+RL++ A+S + F+FH + + + +RL L +GE
Sbjct: 221 DVT------GLNRTGDRLTRFADSLGLQFQFHTLVIVEEDLAGLLLQIRLLALSAVQGET 274
Query: 346 IAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVET 405
IAVN LH + ++D + + K L+ ++VT+ E+E+N + F RF E
Sbjct: 275 IAVNCVHFLHKIFNDD----GDMIGHFLSAIKSLNSRIVTMAEREANHGDHSFLNRFSEA 330
Query: 406 MNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFT 465
+++Y A+F+S++ LP RER+ +EQ +E++++VA E ER +RH + W
Sbjct: 331 VDHYMAIFDSLEATLPPNSRERLTLEQRWFGKEILDVVAAEETERKQRHRRFEIWEEMMK 390
Query: 466 MAGFTPYPLSSYINYSIQNLLE-NYQGH-YTLQEKDGALYLGWMNQPLITSSAWR 518
GF P+ S+ + LL +Y Y LQ + +L+LGW N+PL + S+W+
Sbjct: 391 RFGFVNVPIGSFALSQAKLLLRLHYPSEGYNLQFLNNSLFLGWQNRPLFSVSSWK 445
>At1g07520 transcription factor scarecrow-like 14, putative
Length = 695
Score = 192 bits (488), Expect = 4e-49
Identities = 127/462 (27%), Positives = 225/462 (48%), Gaps = 19/462 (4%)
Query: 72 INDSLENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDPLLLEAEK 131
I+ + +N +DDL R + + ++ + ++++ +++ E DP ++E +
Sbjct: 234 ISKTRKNHHEREEEEDDLEEARRRSKQFAVNEEDGKLTEMFDKVLLLDGECDPQIIEDGE 293
Query: 132 WNKMIEMISRG---------DLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQ 182
++ +G D + +L CA+++S D TA+ L+ ++ K S G+ Q
Sbjct: 294 NGSSKALVKKGRAKKKSRAVDFRTLLTLCAQSVSAGDKITADDLLRQIRKQCSPVGDASQ 353
Query: 183 RLGAYMLEALVARI-ASSGSII---YKSLKCKEPITATSKELLSHMHVLYEICPYLKFGY 238
RL + AL AR+ S+G++I Y S+ K+ T+ ++L V P++ Y
Sbjct: 354 RLAHFFANALEARLEGSTGTMIQSYYDSISSKK---RTAAQILKSYSVFLSASPFMTLIY 410
Query: 239 MSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYA 298
+N +I +A KD S +HI+DF I G QW IQ L+ G K+RITG +
Sbjct: 411 FFSNKMILDAAKDASVLHIVDFGILYGFQWPMFIQHLSKSNPGLRKLRITGIEIPQHGLR 470
Query: 299 RGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSE-VRLEDLELRRGEAIAVNFAMMLHHV 357
+ G RL++ + + V FE++AI E +++E+ ++R E +AVN + ++
Sbjct: 471 PTERIQDTGRRLTEYCKRFGVPFEYNAIASKNWETIKMEEFKIRPNEVLAVNAVLRFKNL 530
Query: 358 PDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESID 417
D RD ++L + ++P V + N F RF E + +Y A+F+
Sbjct: 531 RDVIPGEEDCPRDGFLKLIRDMNPNVFLSSTVNGSFNAPFFTTRFKEALFHYSALFDLFG 590
Query: 418 VALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSY 477
L +E+ ERI+ E REV+N++ACEG +RVER E K+W+ AGF P+ +
Sbjct: 591 ATLSKENPERIHFEGEFYGREVMNVIACEGVDRVERPETYKQWQVRMIRAGFKQKPVEAE 650
Query: 478 INYSIQNLLENYQGH--YTLQEKDGALYLGWMNQPLITSSAW 517
+ + ++ + H + L E GW + L +SS W
Sbjct: 651 LVQLFREKMKKWGYHKDFVLDEDSNWFLQGWKGRILFSSSCW 692
>At5g41920 SCARECROW gene regulator-like protein
Length = 405
Score = 186 bits (472), Expect = 3e-47
Identities = 133/383 (34%), Positives = 201/383 (51%), Gaps = 30/383 (7%)
Query: 144 LKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSII 203
L +L CA+ ++ + + A L+SE+S++ S G+ +R+ AY +AL R+ SS
Sbjct: 40 LLSLLLQCAEYVATDHLREASTLLSEISEICSPFGSSPERVVAYFAQALQTRVISS---- 95
Query: 204 YKSLKC----KEPITAT-SKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHII 258
Y S C ++P+T S+++ S + + P +KF + +AN I +AL E +HII
Sbjct: 96 YLSGACSPLSEKPLTVVQSQKIFSALQTYNSVSPLIKFSHFTANQAIFQALDGEDSVHII 155
Query: 259 DFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYN 318
D + QG+QW +L LA +P IRITGF S+ L G RL+ A S N
Sbjct: 156 DLDVMQGLQWPALFHILASRPRKLRSIRITGFGSSSDL------LASTGRRLADFASSLN 209
Query: 319 VAFEFHAI-GVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAK 377
+ FEFH I G+ + + L R+GEA+ V++ M H + DV G N+ + L + +
Sbjct: 210 LPFEFHPIEGIIGNLIDPSQLATRQGEAVVVHW--MQHRL--YDVTG--NNLETL-EILR 262
Query: 378 CLSPKVVTLVEQE-SNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLA 436
L P ++T+VEQE S + F RFVE ++YY A+F+++ L E ER VEQ L
Sbjct: 263 RLKPNLITVVEQELSYDDGGSFLGRFVEALHYYSALFDALGDGLGEESGERFTVEQIVLG 322
Query: 437 REVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGH-YTL 495
E+ N+VA G R KW+ + GF P L LL + YTL
Sbjct: 323 TEIRNIVAHGGGRRKR-----MKWKEELSRVGFRPVSLRGNPATQAGLLLGMLPWNGYTL 377
Query: 496 QEKDGALYLGWMNQPLITSSAWR 518
E++G L LGW + L+T+SAW+
Sbjct: 378 VEENGTLRLGWKDLSLLTASAWK 400
>At5g66770 SCARECROW gene regulator
Length = 584
Score = 185 bits (470), Expect = 5e-47
Identities = 127/381 (33%), Positives = 200/381 (52%), Gaps = 19/381 (4%)
Query: 144 LKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSII 203
L + ++ CA+ IS++D A + ++ + VS G+P +R+ Y EAL R++ +
Sbjct: 217 LLKAIYDCAR-ISDSDPNEASKTLLQIRESVSELGDPTERVAFYFTEALSNRLSPNSPAT 275
Query: 204 YKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQIN 263
S ++++++L+ L + CPY KF +++AN I EA + ++IHI+DF I
Sbjct: 276 SSS-------SSSTEDLILSYKTLNDACPYSKFAHLTANQAILEATEKSNKIHIVDFGIV 328
Query: 264 QGIQWMSLIQALAGKPGG-PPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFE 322
QGIQW +L+QALA + G P +IR++G + + L G RL A+ ++ F+
Sbjct: 329 QGIQWPALLQALATRTSGKPTQIRVSGIPAPSLGESPEPSLIATGNRLRDFAKVLDLNFD 388
Query: 323 FHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPK 382
F I + + E +AVNF + L+ + DE D +RLAK L+P+
Sbjct: 389 FIPILTPIHLLNGSSFRVDPDEVLAVNFMLQLYKLLDET----PTIVDTALRLAKSLNPR 444
Query: 383 VVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNL 442
VVTL E E + N + F R + +Y AVFES++ L R+ ER+ VE+ R + L
Sbjct: 445 VVTLGEYEVSLNRVGFANRVKNALQFYSAVFESLEPNLGRDSEERVRVERELFGRRISGL 504
Query: 443 VACE--GAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLL--ENYQGHYTLQE- 497
+ E G R ER E ++WR AGF LS+Y + LL NY Y++ E
Sbjct: 505 IGPEKTGIHR-ERMEEKEQWRVLMENAGFESVKLSNYAVSQAKILLWNYNYSNLYSIVES 563
Query: 498 KDGALYLGWMNQPLITSSAWR 518
K G + L W + PL+T S+WR
Sbjct: 564 KPGFISLAWNDLPLLTLSSWR 584
>At5g59450 putative SCARECROW transcriptional regulatory protein
(F2O15.13)
Length = 610
Score = 185 bits (470), Expect = 5e-47
Identities = 111/380 (29%), Positives = 188/380 (49%), Gaps = 8/380 (2%)
Query: 143 DLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSI 202
DL+ +L CA+A++ D A + E+ S +G+ QRL Y EAL ARI +
Sbjct: 223 DLRSLLTQCAQAVASFDQRRATDKLKEIRAHSSSNGDGTQRLAFYFAEALEARITGN--- 279
Query: 203 IYKSLKCKEPITATSK-ELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQ 261
I + P + TS ++L + CP Y +AN I E +++HI+DF
Sbjct: 280 ISPPVSNPFPSSTTSMVDILKAYKLFVHTCPIYVTDYFAANKSIYELAMKATKLHIVDFG 339
Query: 262 INQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAF 321
+ G QW L++AL+ +PGGPP +R+TG + + + + G RL + + +NV F
Sbjct: 340 VLYGFQWPCLLRALSKRPGGPPMLRVTGIELPQAGFRPSDRVEETGRRLKRFCDQFNVPF 399
Query: 322 EFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSP 381
EF+ I + L++L + GE VN L + PDE V + RD +++L + ++P
Sbjct: 400 EFNFIAKKWETITLDELMINPGETTVVNCIHRLQYTPDETV-SLDSPRDTVLKLFRDINP 458
Query: 382 KVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVAL--PREHRERINVEQHCLAREV 439
+ E N F RF E + +Y ++F+ D + E++ R +E+ L R+
Sbjct: 459 DLFVFAEINGMYNSPFFMTRFREALFHYSSLFDMFDTTIHAEDEYKNRSLLERELLVRDA 518
Query: 440 VNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLL-ENYQGHYTLQEK 498
+++++CEGAER R E K+WR AGF P +S I + ++ + Y + +
Sbjct: 519 MSVISCEGAERFARPETYKQWRVRILRAGFKPATISKQIMKEAKEIVRKRYHRDFVIDSD 578
Query: 499 DGALYLGWMNQPLITSSAWR 518
+ + GW + + S W+
Sbjct: 579 NNWMLQGWKGRVIYAFSCWK 598
>At3g46600 scarecrow-like protein
Length = 583
Score = 184 bits (468), Expect = 8e-47
Identities = 115/382 (30%), Positives = 186/382 (48%), Gaps = 16/382 (4%)
Query: 143 DLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSI 202
D++ +L CA+A++ D A + E+ + S G+ QRLG + EAL ARI + +
Sbjct: 208 DMRNLLMQCAQAVASFDQRRAFEKLKEIREHSSRHGDATQRLGYHFAEALEARITGTMTT 267
Query: 203 IYKSLKCKEPITATSK-----ELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHI 257
PI+ATS ++L + CP L Y +AN I E + +HI
Sbjct: 268 ---------PISATSSRTSMVDILKAYKGFVQACPTLIMCYFTANRTINELASKATTLHI 318
Query: 258 IDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESY 317
IDF I G QW LIQAL+ + GPP +R+TG + S + + G RL + + +
Sbjct: 319 IDFGILYGFQWPCLIQALSKRDIGPPLLRVTGIELPQSGFRPSERVEETGRRLKRFCDKF 378
Query: 318 NVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAK 377
NV FE+ I + + L+DL + GE VN + L + PDE V + RD ++L +
Sbjct: 379 NVPFEYSFIAKNWENITLDDLVINSGETTVVNCILRLQYTPDETV-SLNSPRDTALKLFR 437
Query: 378 CLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAR 437
++P + E N F RF E + + ++F+ + L + R VE+ + R
Sbjct: 438 DINPDLFVFAEINGTYNSPFFLTRFREALFHCSSLFDMYETTLSEDDNCRTLVERELIIR 497
Query: 438 EVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLL-ENYQGHYTLQ 496
+ ++++ACEG+ER R E K+W+ AGF P LS I + ++ E Y + +
Sbjct: 498 DAMSVIACEGSERFARPETYKQWQVRILRAGFRPAKLSKQIVKDGKEIVKERYHKDFVID 557
Query: 497 EKDGALYLGWMNQPLITSSAWR 518
+ ++ GW + L S W+
Sbjct: 558 NDNHWMFQGWKGRVLYAVSCWK 579
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,697,372
Number of Sequences: 26719
Number of extensions: 497531
Number of successful extensions: 1581
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1419
Number of HSP's gapped (non-prelim): 51
length of query: 518
length of database: 11,318,596
effective HSP length: 104
effective length of query: 414
effective length of database: 8,539,820
effective search space: 3535485480
effective search space used: 3535485480
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC137079.8


