

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137077.8 - phase: 0
(1091 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
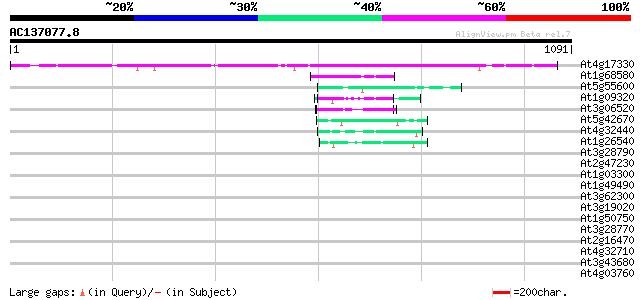
Score E
Sequences producing significant alignments: (bits) Value
At4g17330 G2484-1 protein 747 0.0
At1g68580 unknown protein 61 4e-09
At5g55600 unknown protein 59 1e-08
At1g09320 unknown protein 56 1e-07
At3g06520 hypothetical protein 54 5e-07
At5g42670 unknown protein 53 1e-06
At4g32440 unknown protein 44 4e-04
At1g26540 unknown protein 44 4e-04
At3g28790 unknown protein 42 0.001
At2g47230 unknown protein 42 0.002
At1g03300 unknown protein 42 0.002
At1g49490 hypothetical protein 41 0.004
At3g62300 putative protein 40 0.009
At3g19020 hypothetical protein 40 0.009
At1g50750 hypothetical protein 40 0.009
At3g28770 hypothetical protein 39 0.012
At2g16470 unknown protein 39 0.012
At4g32710 putative protein kinase 39 0.016
At3g43680 putative protein 39 0.021
At4g03760 hypothetical protein 38 0.028
>At4g17330 G2484-1 protein
Length = 2037
Score = 747 bits (1929), Expect = 0.0
Identities = 467/1108 (42%), Positives = 657/1108 (59%), Gaps = 101/1108 (9%)
Query: 1 MISAYGGTGTCGRMSGVSAWKGSVV---KNLILIPQKHLCSQDLAARTSDSTVKQSVLQG 57
MISA+GG G+ S +W+ VV K+L+ P+ L S+ G
Sbjct: 980 MISAFGGADG-GKGSWEKSWRTCVVRAQKSLVATPETPLQSRP----------------G 1022
Query: 58 KGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPL 117
K + G +SK + NP+IPLSSPLWSL T S D+LQSS++ RGS + L+
Sbjct: 1023 KTETPSAGHTNSKESSG-TNPMIPLSSPLWSLST-SVDTLQSSSVQRGSAATHQPLLSAS 1080
Query: 118 HPYQSPSPRNFLGHSTSWISQAPLRGPWIGSP-TPAPDNNTHLSASPSSDTIKLASVKGS 176
H +Q+P +N +GH+T W+S P R W+ S T D + P +D +KL +K S
Sbjct: 1081 HAHQTPPTQNIVGHNTPWMSPLPFRNAWLASQQTSGFDVGSRFPVYPITDPVKLTPMKES 1140
Query: 177 LPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSE 236
S K V G ++ S + T L+ + V PAQ S+ K++KRKK +S
Sbjct: 1141 SMTLSGAKHVQSGTSSNVSKVTPT-------LEPTSTVVAPAQHSTRVKSRKRKKMPVSV 1193
Query: 237 DHGQKLLQSL----------TPAVASRASTSVSAATPVGNVPMSSVEKSVVSV------- 279
+ G +L SL P + A+ +A T V M++V +VS
Sbjct: 1194 ESGPNILNSLKQTELAASPLVPFTPTPANLGYNAGTLPSVVSMTAVPMDLVSTFPGKKIK 1253
Query: 280 ----SPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDK 335
SP+ + ++ +LS++++ K+KEA++HAE+ASAL+ AAV+HS +W Q+++
Sbjct: 1254 SSFPSPIFGGNLVREVKQRSVLSEDTIEKLKEAKMHAEDASALATAAVSHSEYVWKQIEQ 1313
Query: 336 HKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYEN 395
++G + + +LASAAVAIAAAAAVAKAAAAAANVA+NAA QAKLMA+EA + +
Sbjct: 1314 QSHAGLQPETQDRLASAAVAIAAAAAVAKAAAAAANVAANAALQAKLMAEEASLPNA--- 1370
Query: 396 TSQGNNTFLPEGTSNL--GQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKR 453
+ QG LP+ ++ GQ TPASILKG + S ++AA+EA ++RVEAA+AATKR
Sbjct: 1371 SDQG----LPKSYDSILPGQGTPASILKGEGAVVNSSSVLIAAREASKKRVEAATAATKR 1426
Query: 454 AENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRESSREVGLLKD 513
AENMD+I+KAAELA+EAVSQAG +V+MG P L +L+EAGP W+ ++ES +EV K
Sbjct: 1427 AENMDSIVKAAELASEAVSQAGILVSMGHPPLLNKLVEAGPSNYWRQAQES-QEVQPCKT 1485
Query: 514 MTRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEK---------NTRGQQARTVSD 564
+ + + E + A + I+ +E S+ + +GQ+ +D
Sbjct: 1486 VVLEKETVST----SEGTFAGPK-IVQTEFGGSVNTADGVSGPVPATGKLKGQEGDKYAD 1540
Query: 565 LVKPVDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILS 624
L K D+V E ++ S + + + KEGS VEVFK+E G + AW+ N+LS
Sbjct: 1541 LAKNNDVVFEPEVGSKF-SIDAQQTIKATKNEDIKEGSNVEVFKEEPGLRTAWYSANVLS 1599
Query: 625 LKDGKVYVCYTSLVAVEGP--LKEWVSLECEGDKPPRIRTARPLTSLQHEGTRKRRRAAM 682
L+D K YV ++ L +G LKEWV+L+ EGD+ P+IR AR +T+L +EGTRKRRRAA+
Sbjct: 1600 LEDDKAYVLFSDLSVEQGTDKLKEWVALKGEGDQAPKIRPARSVTALPYEGTRKRRRAAL 1659
Query: 683 GDYAWSVGDRVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWK 742
GD+ W +GDRVD+W+ +SW EGVITEKNKKDE T+TVH PA ET ++AWNLRPSL+WK
Sbjct: 1660 GDHIWKIGDRVDSWVHDSWLEGVITEKNKKDENTVTVHFPAEEETLTIKAWNLRPSLVWK 1719
Query: 743 DGQWLDFSKVGANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRS 802
DG+W++ S G SS+H+GDTP EKRP+LG+ A+ + KD K +D + P +
Sbjct: 1720 DGKWIECSSSGETISSSHEGDTPKEKRPRLGTPALVAEVKDTSMKIVDDPDLGKPPQTGV 1779
Query: 803 LNLTENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYV 862
LNL +E FNIGKS+ E+K DP R R+GLQK+GSKVIFGVPKPGKKRKFM+VSKHYV
Sbjct: 1780 LNLGVSENTFNIGKSTREENKPDPLRMKRTGLQKQGSKVIFGVPKPGKKRKFMDVSKHYV 1839
Query: 863 AHGSSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKT-----KFGKP 917
+ S+K ++ + VK +PQ S + W+ SK S EK S+PKT K +
Sbjct: 1840 SEASTKTQERKEPVKPVRSIVPQNSGIGSWKMPSKTISIEKQTTISRPKTFKPAPKPKEK 1899
Query: 918 PGVLGRVNPPRNTSVSNTEMNKDSSNHTKNASQS-ESRVERAPYSTTDGATQVPIVFSSQ 976
PG R+ P KDS N T + +S ES R P S G + V
Sbjct: 1900 PGATARIIP-----------RKDSRNTTASDMESDESAENRGPGS---GVSFKGTVEEQT 1945
Query: 977 ATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRSNRR 1036
+S++ +K + + +KG++AP + +L K KAL + S+ + +EPRRS RR
Sbjct: 1946 TSSSHDTGSKNSSSLSTNKGRVAPTAGRLAKIEEDKALAE---NSSKTSEGMEPRRSIRR 2002
Query: 1037 IQPTSRLLEGLQSSLMVSKIPSVSHNRN 1064
IQPTSRLLEGLQ+S+M SKIPSVSH+++
Sbjct: 2003 IQPTSRLLEGLQTSMMTSKIPSVSHSKS 2030
>At1g68580 unknown protein
Length = 648
Score = 60.8 bits (146), Expect = 4e-09
Identities = 44/170 (25%), Positives = 76/170 (43%), Gaps = 13/170 (7%)
Query: 585 TVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEG-- 642
++ G E+ + K+GSL+EV ++ G + WF +L KV V Y + +
Sbjct: 350 SIFKGDEDGSSHHIKKGSLIEVLSEDSGIRGCWFKALVLKKHKDKVKVQYQDIQDADDES 409
Query: 643 -PLKEWV--SLECEGDKPPRIR-TARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQ 698
L+EW+ S GD +R R + + +++ +G VG VD W
Sbjct: 410 KKLEEWILTSRVAAGDHLGDLRIKGRKVVRPMLKPSKENDVCVIG-----VGMPVDVWWC 464
Query: 699 ESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLD 748
+ W EG++ + + E V++P + S +LR S W D +WL+
Sbjct: 465 DGWWEGIVVQ--EVSEEKFEVYLPGEKKMSAFHRNDLRQSREWLDDEWLN 512
>At5g55600 unknown protein
Length = 663
Score = 59.3 bits (142), Expect = 1e-08
Identities = 61/287 (21%), Positives = 105/287 (36%), Gaps = 36/287 (12%)
Query: 599 KEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEG--PLKEWVSLECEGDK 656
K + +E + G + WF +L + +V + Y + +G L+EWV
Sbjct: 382 KTDAKIEFLCQDSGIRGCWFRCTVLDVSRKQVKLQYDDIEDEDGYGNLEEWV-------- 433
Query: 657 PPRIRTARPLTSLQHEGTRKRRRAAMGD-----YAWSVGDRVDAWIQESWREGVITEKNK 711
P ++A P R R A D + ++G+ VDAW + W EGV+ K
Sbjct: 434 -PAFKSAMPDKLGIRLSNRPTIRPAPRDVKTAYFDLTIGEAVDAWWNDGWWEGVVIATGK 492
Query: 712 KDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTPHEKRPK 771
D L ++IP + ++R S W W+D + + K
Sbjct: 493 PDTEDLKIYIPGENLCLTVLRKDIRISRDWVGDSWVDIDPKPEILAIVSSDSSSESKLSM 552
Query: 772 LGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTENEIVFNIGKSSTNESKQDPQRQVR 831
L + + + K K NI E A P+ ++ N +G+ + +
Sbjct: 553 LSTLSKDTKAKPSAMPNI--VEEAEPEGEKAYNSL-------LGEQNKEHKDVVVKEDDE 603
Query: 832 SGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDKNDSVKI 878
+ L KE K + +K YV H ++ + K D V +
Sbjct: 604 NKLSKEEDKEVGS-----------NETKTYVNHENTVEDHKEDDVTL 639
>At1g09320 unknown protein
Length = 517
Score = 55.8 bits (133), Expect = 1e-07
Identities = 51/157 (32%), Positives = 76/157 (47%), Gaps = 29/157 (18%)
Query: 599 KEGSLVEVFKDEEGHKAAWFMGNILSL-----KDG-KVYVCYTSL-VAVEG--PLKEWVS 649
K GS VE+ DE G + +W+MG ++++ KD K V YT+L EG PLKE V
Sbjct: 40 KPGSAVEISSDEIGFRGSWYMGKVITIPSSSDKDSVKCQVEYTTLFFDKEGTKPLKEVVD 99
Query: 650 LECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEK 709
+ +PP A P++ ++ K+++ VG+ VDA+ + W EG +TE
Sbjct: 100 M--SQLRPP----APPMSEIE-----KKKKIV-------VGEEVDAFYNDGWWEGDVTE- 140
Query: 710 NKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQW 746
D+ +V +S E R LR W DG W
Sbjct: 141 -VLDDGKFSVFFRSSKEQIRFRKDELRFHREWVDGAW 176
Score = 52.4 bits (124), Expect = 1e-06
Identities = 50/210 (23%), Positives = 77/210 (35%), Gaps = 36/210 (17%)
Query: 593 LEENTFKEGSLVEVFKDEEGHKAAWFMGNILS-LKDGKVYVCYTSLVAVEG--PLKEWVS 649
+ + F G++VEV DEEG + WF ++ + + K V Y L +G PLKE
Sbjct: 216 IAKQMFSSGTVVEVSSDEEGFQGCWFAAKVVEPVGEDKFLVEYRDLREKDGIEPLKEETD 275
Query: 650 LECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEK 709
PPR D ++VGD+++A+ + W GV+ +
Sbjct: 276 FLHIRPPPPRDE----------------------DIDFAVGDKINAFYNDGWWVGVVIDG 313
Query: 710 NKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTPHEKR 769
K T+ ++ S E LR W DG W G EK
Sbjct: 314 MK--HGTVGIYFRQSQEKMRFGRQGLRLHKDWVDGTW---------QLPLKGGKIKREKT 362
Query: 770 PKLGSNAVEVKGKDKMSKNIDAAESANPDE 799
N K +K + +I +P+E
Sbjct: 363 VSCNRNVRPKKATEKQAFSIGTPIEVSPEE 392
Score = 41.2 bits (95), Expect = 0.003
Identities = 42/157 (26%), Positives = 68/157 (42%), Gaps = 26/157 (16%)
Query: 594 EENTFKEGSLVEVFKDEEGHKAAWFMGNILSLK-DGKVYVCYTSLVAVEG--PLKEWVSL 650
E+ F G+ +EV +EEG + +WF+ ++ + K V Y +L A +G PL+E V++
Sbjct: 376 EKQAFSIGTPIEVSPEEEGFEDSWFLAKLIEYRGKDKCLVEYDNLKAEDGKEPLREEVNV 435
Query: 651 ECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKN 710
RIR PL S+ + D+V+A + W GVI +
Sbjct: 436 S-------RIRPL-PLESVMVSPFERH-------------DKVNALYNDGWWVGVIRKVL 474
Query: 711 KKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWL 747
K ++ V + E LR W DG+W+
Sbjct: 475 AK--SSYLVLFKNTQELLKFHHSQLRLHQEWIDGKWI 509
>At3g06520 hypothetical protein
Length = 466
Score = 53.9 bits (128), Expect = 5e-07
Identities = 44/153 (28%), Positives = 70/153 (44%), Gaps = 20/153 (13%)
Query: 595 ENTFKEGSLVEVFKDEEGHKAAWFMGNILS-LKDGKVYVCYTSLVAVEGPLKEWVSLECE 653
E F G VEV DE G++A+WF I+S L + + V Y +L + +E + E
Sbjct: 330 EKVFNNGMEVEVRSDEPGYEASWFSAKIVSYLGENRYTVEYQTLKTDDE--RELLKEEAR 387
Query: 654 GDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKD 713
G IR P L +G Y + + + VDAW E W G + + N +
Sbjct: 388 GSD---IRPPPP--PLIPKG-----------YRYELYELVDAWYNEGWWSGRVYKIN-NN 430
Query: 714 ETTLTVHIPASGETSVLRAWNLRPSLIWKDGQW 746
+T V+ + E+ +LRP +W++G+W
Sbjct: 431 KTRYGVYFQTTDESLEFAYNDLRPCQVWRNGKW 463
Score = 52.4 bits (124), Expect = 1e-06
Identities = 42/159 (26%), Positives = 71/159 (44%), Gaps = 24/159 (15%)
Query: 598 FKEGSLVEVFKDEEGHKAAWFMGNILSLK-DGKVYVCYTSLVAVEG---PLKEWVSLECE 653
+++G+LVEV +E+ +K +W+ IL L D K V + +G PL++ V E +
Sbjct: 169 YEKGALVEVRSEEKAYKGSWYCARILCLLGDDKYIVEHLKFSRDDGESIPLRDVV--EAK 226
Query: 654 GDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKD 713
+P P+ + G VDAW + W +++
Sbjct: 227 DIRPVPPSELSPVV------------------CYEPGVIVDAWFNKRWWTSRVSKVLGGG 268
Query: 714 ETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKV 752
+V I ++GE + + +NLRP W +GQW+ SKV
Sbjct: 269 SNKYSVFIISTGEETTILNFNLRPHKDWINGQWVIPSKV 307
>At5g42670 unknown protein
Length = 294
Score = 52.8 bits (125), Expect = 1e-06
Identities = 58/233 (24%), Positives = 90/233 (37%), Gaps = 32/233 (13%)
Query: 598 FKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEGP----LKEWVSLECE 653
F+ G VE+ E G + AWF K K + G K + LE
Sbjct: 32 FEVGQTVELKSFELGFRGAWFR-----CKKRKALFFDVEYIDYPGEGIHSTKVFQQLEGG 86
Query: 654 GDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKD 713
+K IR P ++E A + +W VGD VD W + G + E + +
Sbjct: 87 NEKCLMIRPVYPPQCRENEVDGGLEEALVVHGSWKVGDLVDWWEAYCYWSGTVLEVKENE 146
Query: 714 ETTLTVHIPASGETSVLRAW--NLRPSLIW--KDGQWLDFSKVG-----------ANDSS 758
+ + P GE S A +LRPSL W +DG + FSK G N+
Sbjct: 147 SVQIELLAPPYGEGSSYEAQSKDLRPSLEWSLEDGWTVPFSKDGEKRQCAKLMKQLNEDQ 206
Query: 759 THKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTENEIV 811
+ ++ E++ + E K ++K E DE LN+ ++E +
Sbjct: 207 VSEAESMEEEKQR----GAEPKEEEKQR----PTEKLKSDEALQLNIMDSESI 251
>At4g32440 unknown protein
Length = 393
Score = 44.3 bits (103), Expect = 4e-04
Identities = 55/220 (25%), Positives = 86/220 (39%), Gaps = 41/220 (18%)
Query: 599 KEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEGPLKEWVSLECEGDKPP 658
++GS VEVF ++E AW I+S +G Y +E E +K P
Sbjct: 4 RKGSRVEVFSNKEAPYGAWRCAEIVS-GNGHTYNVRFYSFQIEHE-------EAVMEKVP 55
Query: 659 R--IRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKDETT 716
R IR PL ++ W G+ V+ SW+ + E +
Sbjct: 56 RKIIRPCPPLVDVER---------------WDTGELVEVLDNFSWKAATVRE--ELSGHY 98
Query: 717 LTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKV-GANDSSTHKGDTPHEKRPKLGSN 775
V + + E NLR W+D +W+ K+ G+ SST G H+K + N
Sbjct: 99 YVVRLLGTPEELTFHKVNLRARKSWQDERWVAIGKISGSLKSSTLTGSDVHQKL-QPHRN 157
Query: 776 AVEVKGKDKMSKNI----------DAAES--ANPDEMRSL 803
++ + +S + + AES NP +MRSL
Sbjct: 158 SMPLHEPSVVSARLLKRPSPYNWSECAESCTGNPKKMRSL 197
>At1g26540 unknown protein
Length = 695
Score = 44.3 bits (103), Expect = 4e-04
Identities = 57/224 (25%), Positives = 87/224 (38%), Gaps = 37/224 (16%)
Query: 603 LVEVFKDEEGHKAAWFMGNILSLKDG-----KVYVCYTSLVAVEG--PLKEWVSLECEGD 655
+VEV +EEG + AWF +L G K+ V Y++L+ ++G PL E +
Sbjct: 13 VVEVSSEEEGFEGAWFRA-VLEENPGNSSRRKLRVRYSTLLDMDGSSPLIEHIEQRFIRP 71
Query: 656 KPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKDET 715
PP E +K D G VDA ++ W GV+ +K + D
Sbjct: 72 VPP------------EENQQK-------DVVLEEGLLVDADHKDGWWTGVVVKKMEDDNY 112
Query: 716 TLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTPH--EKRPKLG 773
+ +P + LR LIW G W+ +N S G + +
Sbjct: 113 LVYFDLPP--DIIQFERKQLRTHLIWTGGTWIQPEIEESNKSMFSPGTMVEVFSAKEAVW 170
Query: 774 SNAVEVKGKDK------MSKNIDAAESANPDEMRSLNLTENEIV 811
S A+ VK D + K+ + S N DE R N+ + V
Sbjct: 171 SPAMVVKETDVDDKKKFIVKDCNRYLSCNGDEARPTNIVNSRRV 214
Score = 40.8 bits (94), Expect = 0.004
Identities = 39/161 (24%), Positives = 67/161 (41%), Gaps = 29/161 (18%)
Query: 591 ENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEGPLKEWVS- 649
E ++ F G++VEVF +E A W ++ D V+ K V
Sbjct: 147 EESNKSMFSPGTMVEVFSAKE---AVWSPAMVVKETD------------VDDKKKFIVKD 191
Query: 650 ----LECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGV 705
L C GD+ ARP + R + D +++ + V+ + W +G
Sbjct: 192 CNRYLSCNGDE------ARPTNIVNSRRVRPIPPPSSVD-KYALLESVETFSGLGWHKGQ 244
Query: 706 ITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQW 746
+ + E TV + A+ + S +R +LRP ++W+DG W
Sbjct: 245 V--RKILSENRYTVRLEATQQESTIRHSDLRPFMVWEDGVW 283
>At3g28790 unknown protein
Length = 608
Score = 42.4 bits (98), Expect = 0.001
Identities = 56/247 (22%), Positives = 100/247 (39%), Gaps = 19/247 (7%)
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S SSD ++ K S S + T G +++G T G+ S A V
Sbjct: 202 SSSSDNESSSNTKSQGTSSKSGSESTAGSIETNTG-SKTEAGSKSSSSAKTKEV------ 254
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
SG + KD G S TP ++ ++ + +TP + P S + +P
Sbjct: 255 SGGSSGNTYKDTTGSSSGASPSGSPTPTPSTPTPSTPTPSTPTPSTPTPS---TPTPSTP 311
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALS-AAAVNHSLELWNQLDKHK--- 337
P +T EK ES KE+ +E SA S + + + D +K
Sbjct: 312 APSTPAAGKTSEK---GSESASMKKESNSKSESESAASGSVSKTKETNKGSSGDTYKDTT 368
Query: 338 --NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYEN 395
+SG S + + + + A+ +A+A+A +++A+ A A+E+ S E+
Sbjct: 369 GTSSGSPSGSPSGSPTPSTSTDGKASSKGSASASAGASASASAGASASAEESAASQKKES 428
Query: 396 TSQGNNT 402
S+ +++
Sbjct: 429 NSKSSSS 435
Score = 35.8 bits (81), Expect = 0.14
Identities = 42/189 (22%), Positives = 70/189 (36%), Gaps = 23/189 (12%)
Query: 147 GSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDS 206
GSPTP P T + +PS+ T S P S+ TP P ++G S +
Sbjct: 277 GSPTPTPSTPTPSTPTPSTPT-------PSTPTPSTPTPSTPAPSTPAAGKTSEKGSESA 329
Query: 207 QLDANNVTVPPAQQSSGPKAKKRK-----------KDVLSEDHGQ---KLLQSLTPAVAS 252
+ + + ++ ++ K K KD G S TP+ ++
Sbjct: 330 SMKKESNSKSESESAASGSVSKTKETNKGSSGDTYKDTTGTSSGSPSGSPSGSPTPSTST 389
Query: 253 --RASTSVSAATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARV 310
+AS+ SA+ G +S S + A Q K + S + +K E +
Sbjct: 390 DGKASSKGSASASAGASASASAGASASAEESAASQKKESNSKSSSSSSSTTSVKEVETQT 449
Query: 311 HAEEASALS 319
+E S +S
Sbjct: 450 SSEVNSFIS 458
>At2g47230 unknown protein
Length = 701
Score = 42.0 bits (97), Expect = 0.002
Identities = 51/228 (22%), Positives = 93/228 (40%), Gaps = 39/228 (17%)
Query: 586 VRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEGPLK 645
+R + L+++ F G++ EV + + AWF I+ + V+G K
Sbjct: 138 LRPDIQELDKSMFSSGTMAEVSTIVDKAEVAWFPAMIIKE------------IEVDGE-K 184
Query: 646 EWVSLECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGV 705
+++ +C AR +++ R + + + DRV+ + WR+G+
Sbjct: 185 KFIVKDCNKHLSFSGDEARTNSTIDSSRVRPTPPPFPVE-KYELMDRVEVFRGSVWRQGL 243
Query: 706 ITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTP 765
+ + D V + + E V++ +LRP +W+DG W D K TP
Sbjct: 244 V--RGVLDHNCYMVCLVVTKEEPVVKHSDLRPCKVWEDGVWQDGPK-----------QTP 290
Query: 766 HEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDE-----MRSLNLTEN 808
+ P SN + K K ++ A+S P RSLNL ++
Sbjct: 291 VIETP---SNVM----KTKPMRSCSGAKSMTPKRTTKHARRSLNLEKS 331
Score = 37.4 bits (85), Expect = 0.047
Identities = 44/157 (28%), Positives = 64/157 (40%), Gaps = 24/157 (15%)
Query: 595 ENTFKEGSLVEVFKDEEGHKAAWFMGNILS--LKDG--KVYVCYTSLVAVEGPLKEWVSL 650
E T ++GS VEV EEG AWF G + K G K+ V Y +L+ + +S
Sbjct: 2 EETIRKGSEVEVSSTEEGFADAWFRGILQENPTKSGRKKLRVRYLTLLNDDA-----LSP 56
Query: 651 ECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKN 710
E +P IR P ++ G G VDA ++ W GVI +K
Sbjct: 57 LIENIEPRFIRPVPP--ENEYNG-----------IVLEEGTVVDADHKDGWWTGVIIKKL 103
Query: 711 KKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWL 747
+ + V+ + + LRP L W +WL
Sbjct: 104 ENGK--FWVYYDSPPDIIEFERNQLRPHLRWSGWKWL 138
>At1g03300 unknown protein
Length = 670
Score = 41.6 bits (96), Expect = 0.002
Identities = 37/154 (24%), Positives = 67/154 (43%), Gaps = 16/154 (10%)
Query: 593 LEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEGPLKEWVSLEC 652
L ++ F G+LVEV + + +W I+ + E K+++ C
Sbjct: 145 LSKSMFSPGTLVEVSCVIDKVEVSWVTAMIVKEIE-------------ESGEKKFIVKVC 191
Query: 653 EGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKK 712
R+ A+P ++ R R + + + D V+ + SWR+GV+ K
Sbjct: 192 NKHLSCRVDEAKPNMTVDSCCVRPRPPLFFVE-EYDLRDCVEVFHGSSWRQGVV--KGVH 248
Query: 713 DETTLTVHIPASGETSVLRAWNLRPSLIWKDGQW 746
E TV + A+ + V++ +LRP +W+DG W
Sbjct: 249 IEKQYTVTLEATKDKLVVKHSDLRPFKVWEDGVW 282
>At1g49490 hypothetical protein
Length = 847
Score = 40.8 bits (94), Expect = 0.004
Identities = 46/170 (27%), Positives = 68/170 (39%), Gaps = 20/170 (11%)
Query: 63 PLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPL-HPYQ 121
P+G + PT SS +PT S++S QS L+ Q+ TP P Q
Sbjct: 649 PMGAPTPTQAPTP-------SSETTQVPTPSSESDQSQILSPVQAPTPVQSSTPSSEPTQ 701
Query: 122 SPSPRNFLGHSTSWISQA----PLRGPWIGS-----PTPAPDNNTHLSASPSSDTIKLAS 172
P+P + + +S P++ P S PTP+ ++N S +P T L
Sbjct: 702 VPTPSSSESYQAPNLSPVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQAP---TPILEP 758
Query: 173 VKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSS 222
V P S ++ TP SS QS V N +VP + S+
Sbjct: 759 VHAPTPNSKPVQSPTPSSEPVSSPEQSEEVEAPEPTPVNPSSVPSSSPST 808
>At3g62300 putative protein
Length = 708
Score = 39.7 bits (91), Expect = 0.009
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 2/70 (2%)
Query: 691 DRVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFS 750
+ VDA ++ W GV+++ V + + E+ LRPS+ WKDG W
Sbjct: 234 ENVDALVESGWCPGVVSKVLAGKR--YAVDLGPNRESKEFSRLQLRPSIEWKDGIWHRKE 291
Query: 751 KVGANDSSTH 760
KV ++ S+H
Sbjct: 292 KVSGSEESSH 301
Score = 30.8 bits (68), Expect = 4.4
Identities = 24/86 (27%), Positives = 35/86 (39%), Gaps = 4/86 (4%)
Query: 690 GDRVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDF 749
GD VDA + W G + + V++ + L +LRP +WKD +W
Sbjct: 94 GDVVDAAYKGGWCSGSVVKV--LGNRRFLVYLRFQPDVIELLRKDLRPHFVWKDEEWFRC 151
Query: 750 SK--VGANDSSTHKGDTPHEKRPKLG 773
K + +D S K K KLG
Sbjct: 152 EKQQLIESDFSAGKSVEVRTKVDKLG 177
>At3g19020 hypothetical protein
Length = 951
Score = 39.7 bits (91), Expect = 0.009
Identities = 42/147 (28%), Positives = 61/147 (40%), Gaps = 29/147 (19%)
Query: 73 PTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTP-LHPYQSPSPRNFLGH 131
P PL P +SP+ + PT S+ G + QA TP ++PS N H
Sbjct: 815 PKPVTPLPPATSPMANAPTPSSSE-------SGEISTPVQAPTPDSEDIEAPSDSN---H 864
Query: 132 STSWISQAPLRGPWIGSPTPAPDNNTHLSAS-PSSDTIKLASVKGSLPPSSSIKDVTPGP 190
S + S SP P+PD+ + A PSS+ A + PSSS P
Sbjct: 865 SPVFKS----------SPAPSPDSEPEVEAPVPSSEPEVEAPKQSEATPSSS-------P 907
Query: 191 PASSSGLQSTFVGTDSQLDANNVTVPP 217
P+S+ T ++ D +N +PP
Sbjct: 908 PSSNPSPDVTAPPSEDNDDGDNFILPP 934
>At1g50750 hypothetical protein
Length = 816
Score = 39.7 bits (91), Expect = 0.009
Identities = 51/234 (21%), Positives = 89/234 (37%), Gaps = 39/234 (16%)
Query: 775 NAVEVKGKDKMSKNIDAAESANPDEMRSLNLTENEIVFNIGKSSTNESKQDPQRQVR--- 831
+A +K +D+ S D + N +N+T++ + + +Q R
Sbjct: 382 SAETLKARDRKSLGADTSSKMNRTRKDEMNITDDSNKRRNYNKQARDKRLKYMKQAREKR 441
Query: 832 ---------SGLQKEGSKVIFGVPKPG--------KKRKFM----EVSKHYVA----HGS 866
+G + GSKV K G K+RK+M E ++ ++ H
Sbjct: 442 LQYIKQARKNGNNESGSKVN-RTSKDGMKIAEDFDKRRKYMKRVRENNESHMGQCHTHVH 500
Query: 867 SKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLG------ADSKPKTKFGKPPGV 920
S ND++D++ IA F + RNS + D+ E LG AD+ F K +
Sbjct: 501 SDNNDEDDNLSIAQFLRLNKKKYSDVRNSGR-DASEPLGKISRVEADNNDLGTFQKLASI 559
Query: 921 LGRVNPPRNTSVSNTEMNKDSSNHTKNASQS---ESRVERAPYSTTDGATQVPI 971
+ PP + N E ++ S N +S E+ P+ G + +
Sbjct: 560 RDEIVPPSEIAHRNGETDEAGSRAGTNMDESPFDENNSSEPPFCADRGVVDIVV 613
>At3g28770 hypothetical protein
Length = 2081
Score = 39.3 bits (90), Expect = 0.012
Identities = 42/216 (19%), Positives = 83/216 (37%), Gaps = 15/216 (6%)
Query: 756 DSSTHKGDTPHEKRPKLGSNA---VEVKGKDKMSKNIDAAESANPDEMRSLNLTENEIVF 812
+ H GD+ ++ + + VEVK D S+ + + N D M L E
Sbjct: 659 EKEVHVGDSTNDNNMESKEDTKSEVEVKKNDGSSEKGEEGKENNKDSMEDKKLENKESQT 718
Query: 813 NIGKSSTNESKQDPQRQVRSGLQKEGSKV-IFGVPKPGKKRKFMEVSKHYVAHGSSKV-N 870
+ + + KQ+ + Q+ G K+ V G K K+ K + +++ V + V
Sbjct: 719 DSKDDKSVDDKQE-EAQIYGGESKDDKSVEAKGKKKESKENKKTKTNENRVRNKEENVQG 777
Query: 871 DKNDSVKIANFSMPQGSELRGWR---------NSSKNDSKEKLGADSKPKTKFGKPPGVL 921
+K +S K+ + + + +++++KE+ G D+K + K +
Sbjct: 778 NKKESEKVEKGEKKESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKEDKEESKDYQSV 837
Query: 922 GRVNPPRNTSVSNTEMNKDSSNHTKNASQSESRVER 957
N V NK+ S K+ E + +
Sbjct: 838 EAKEKNENGGVDTNVGNKEDSKDLKDDRSVEVKANK 873
Score = 35.4 bits (80), Expect = 0.18
Identities = 75/379 (19%), Positives = 140/379 (36%), Gaps = 42/379 (11%)
Query: 694 DAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVG 753
D E+ + V TE+ KD V E S+ ++ G+ L ++ G
Sbjct: 1617 DGKTNENGGKEVSTEEGSKDSNI--VERNGGKEDSIKEGSEDGKTVEINGGEELS-TEEG 1673
Query: 754 ANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTE------ 807
+ D +G E K GS DK+ + ++ E++ + + + E
Sbjct: 1674 SKDGKIEEGKEGKENSTKEGSK------DDKIEEGMEGKENSTKESSKDGKINEIHGDKE 1727
Query: 808 ---NEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAH 864
E + G +ST + +D + +G++ + K K G + + V
Sbjct: 1728 ATMEEGSKDGGTNSTGKDSKDSKSVEINGVKDDSLK---DDSKNGDINEINNGKEDSVKD 1784
Query: 865 GSSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRV 924
+++ ++S+ + S P G +L ++S KN++ E G + T G
Sbjct: 1785 NVTEIQGNDNSLTNSTSSEPNGDKLDTNKDSMKNNTMEAQGGSNGDSTN--------GET 1836
Query: 925 NPPR--NTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQV-PIVFSSQATSTN 981
+ N S++N M SN +Q+ + +TTD + V S +
Sbjct: 1837 EETKESNVSMNNQNMQDVGSNENSMNNQTTGTGDDIISTTTDTESNTSKEVTSFISNLEE 1896
Query: 982 TLPTKRTFTSRASKGK-----LAPASDKLRKGGGGKALNDKPTTSTSEPDA---LEPRRS 1033
P + F S K K L P S +++ + +T DA L+ ++S
Sbjct: 1897 KSPGTQEFQSFFQKLKDYMKYLCPVSSTFEAKDSRSYMSEMISMATKLSDAMAVLQAKKS 1956
Query: 1034 NRRIQPTSRLLEGLQSSLM 1052
T+ L+G Q +M
Sbjct: 1957 GSGQMKTT--LQGYQQEVM 1973
Score = 33.1 bits (74), Expect = 0.88
Identities = 105/489 (21%), Positives = 184/489 (37%), Gaps = 108/489 (22%)
Query: 503 ESSREVGLLKDMTR-DLVNI--DMVRDIPETSHAQNRDILSSEISASIMI-NEKNTRGQQ 558
+ S EV + K+ T+ + VNI + + D + + +N++ + ++ A+ N R Q+
Sbjct: 484 QRSNEVYMNKETTKGENVNIQGESIGDSTKDNSLENKEDVKPKVDANESDGNSTKERHQE 543
Query: 559 ART---VSDLVKPVDMVLGSESETQDPSF--TVRNGSENLEENTFKEGSLVEVFKDEEGH 613
A+ VS K +D + E + D S T +G E+ +G+ E K+E
Sbjct: 544 AQVNNGVSTEDKNLDNIGADEQKKNDKSVEVTTNDGDHTKEKREETQGNNGESVKNE--- 600
Query: 614 KAAWFMGNILSLKDGKVYVCYTSLVAVEGPLKEWVSLECEGDKPPRIRTARPLTSLQHEG 673
N+ + +D K LK+ S+ + + + R T H+
Sbjct: 601 -------NLENKEDKK-------------ELKDDESVGAKTNNETSLEEKREQTQKGHDN 640
Query: 674 TRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNK---KDETTLTVHIPASGETSVL 730
+ + + G D+ ++ G T N K++T V + + +S
Sbjct: 641 SINSKIVD------NKGGNADSNKEKEVHVGDSTNDNNMESKEDTKSEVEVKKNDGSSEK 694
Query: 731 RAWNLRPSLIWKDGQWLDFSKVGANDSSTHKG-DTPHEKRPKLGSNA-----VEVKGKDK 784
+ + + L+ +K DS K D E+ G + VE KGK K
Sbjct: 695 GEEGKENNKDSMEDKKLE-NKESQTDSKDDKSVDDKQEEAQIYGGESKDDKSVEAKGKKK 753
Query: 785 MSKNIDAAESANPDEMRSLNLTENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFG 844
SK EN+ K+ TNE++ + + G +KE KV G
Sbjct: 754 ESK-------------------ENK------KTKTNENRVRNKEENVQGNKKESEKVEKG 788
Query: 845 VPKPGKKRKFMEVSKHYVAHGSSKVNDKNDSVKIANFSMPQGSELRGWRN-SSKNDSKEK 903
K K K +E +K N K S + + + E G N K +SK+
Sbjct: 789 EKKESKDAKSVE----------TKDNKKLSSTE----NRDEAKERSGEDNKEDKEESKDY 834
Query: 904 LGADSKPKTKFGKPPGVLGRVNPPRNTSVSNTEMNKD---------SSNHTKNASQSESR 954
++K K + G GV +T+V N E +KD +N ++ +
Sbjct: 835 QSVEAKEKNENG---GV--------DTNVGNKEDSKDLKDDRSVEVKANKEESMKKKREE 883
Query: 955 VERAPYSTT 963
V+R S+T
Sbjct: 884 VQRNDKSST 892
Score = 32.3 bits (72), Expect = 1.5
Identities = 53/289 (18%), Positives = 98/289 (33%), Gaps = 25/289 (8%)
Query: 755 NDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTENEIVFNI 814
++ S K E K E K + K S+ D E R + E
Sbjct: 1002 SEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQ--DKKREEKDSEERKSKKEKEESRDLK 1059
Query: 815 GKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDKND 874
K E+K+ + + +KE K K+ E KH + K DK D
Sbjct: 1060 AKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKD 1119
Query: 875 SVKIANFSMPQGSELRGWRNSS--------KNDSKEKLGADSKPKTKFGKPPGVLGRVNP 926
K+ + + + E + + S ++D KEK + K +TK +
Sbjct: 1120 MEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETK------EIESSKS 1173
Query: 927 PRNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTK 986
+N + + K ES ++ + D Q + + + T K
Sbjct: 1174 QKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNK 1233
Query: 987 ----RTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPR 1031
+ T++ S GK + + +A N + + +T++ D+ E +
Sbjct: 1234 PKDDKKNTTKQSGGKKESMESESK-----EAENQQKSQATTQADSDESK 1277
>At2g16470 unknown protein
Length = 670
Score = 39.3 bits (90), Expect = 0.012
Identities = 44/165 (26%), Positives = 68/165 (40%), Gaps = 14/165 (8%)
Query: 69 SKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTP-LHPYQSPSPRN 127
S+ +PT +P S +S+ + Q+S + VV+ + AL P +P +P P N
Sbjct: 327 SRWSPTKPSPQSANQSMNYSVA--QSGQSQTSRIDIPVVVNSAGALQPQTYPIPTPDPIN 384
Query: 128 F-LGHSTSWISQAPLRGPWI---------GSPTPAPDNNTHLSASPSSDTIKLASVKGSL 177
+ HS + S P G GS TP+ NN+ +PS + S G
Sbjct: 385 VSVNHSATLHSPTPAGGKQSWGSMQTDHGGSNTPSSQNNSTSYGTPSPSVLPSQSQPG-F 443
Query: 178 PPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSS 222
PPS S K P P + + Q ++ ++ P Q SS
Sbjct: 444 PPSDSWKVAVPSQPNAQAQAQWGMNMVNNNQNSAQPQAPANQNSS 488
>At4g32710 putative protein kinase
Length = 731
Score = 38.9 bits (89), Expect = 0.016
Identities = 41/153 (26%), Positives = 61/153 (39%), Gaps = 20/153 (13%)
Query: 76 ANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSW 135
A PL PL PL S P L + S+ A + +P P +SP P
Sbjct: 34 APPLSPLPPPLSSPPPLPSPPPLSAPTASPPPLPVESPPSP--PIESPPPP--------- 82
Query: 136 ISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSS 195
+ ++P P P+P H+SA S + K S PPSS + PP ++
Sbjct: 83 LLESPPPPPLESPSPPSP----HVSAPSGSPPLPFLPAKPSPPPSSPPSETV--PPGNTI 136
Query: 196 GLQSTFVGTDSQLDANNVTVP---PAQQSSGPK 225
+ ++S N + P P ++ SGPK
Sbjct: 137 SPPPRSLPSESTPPVNTASPPPPSPPRRRSGPK 169
Score = 36.2 bits (82), Expect = 0.10
Identities = 59/239 (24%), Positives = 87/239 (35%), Gaps = 28/239 (11%)
Query: 62 SPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQ 121
SPL S P + P PLS+P S P L +S S + S P P +
Sbjct: 38 SPLPPPLSSPPPLPSPP--PLSAPTASPPPLPVESPPSPPIE--SPPPPLLESPPPPPLE 93
Query: 122 SPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIK---LASVKGSLP 178
SPSP + H ++ PL P++ + P ++ P +TI + S P
Sbjct: 94 SPSPPS--PHVSAPSGSPPL--PFLPAKPSPPPSSPPSETVPPGNTISPPPRSLPSESTP 149
Query: 179 PSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDH 238
P ++ P PP SG + +F + N P S P+ K LS
Sbjct: 150 PVNTASPPPPSPPRRRSGPKPSFPPPINSSPPN----PSPNTPSLPETSPPPKPPLSTTP 205
Query: 239 GQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV-----------VSVSPLADQP 286
S P S A+ ++ P G +P +V + S+SP QP
Sbjct: 206 FPS--SSTPPPKKSPAAVTLPFFGPAGQLPDGTVAPPIGPVIEPKTSPAESISPGTPQP 262
>At3g43680 putative protein
Length = 539
Score = 38.5 bits (88), Expect = 0.021
Identities = 29/102 (28%), Positives = 47/102 (45%), Gaps = 9/102 (8%)
Query: 225 KAKKRKKDVLSEDHGQKLLQSLTPAVASRAST-----SVSAATPVGNVPMSSVEKSVVSV 279
K K+ K+ L+ED + ++P AS +T + A PV P S E +
Sbjct: 174 KKKEDKERRLAEDKRLAEARLISPRAASPEATQDQDVAPDVAAPVDPAPAESQEVDPTAA 233
Query: 280 SPLADQ----PKNDQTVEKRILSDESLMKVKEARVHAEEASA 317
+PL + P +D+ + KR+ +D+S K K + A A A
Sbjct: 234 APLPEAIVALPASDKAMGKRVRTDDSSCKKKSKKKKASSAEA 275
>At4g03760 hypothetical protein
Length = 723
Score = 38.1 bits (87), Expect = 0.028
Identities = 68/311 (21%), Positives = 120/311 (37%), Gaps = 36/311 (11%)
Query: 225 KAKKRKKDVLSEDHGQKLLQSLTPAVASRAST-----SVSAATPVGNVPMSSVEKSVVSV 279
K K+ K+ L+E+ + P VAS +T + A PV P + E +V
Sbjct: 399 KKKEDKERRLAEEKRLANAGLILPRVASPDATQDRNVAPEVAAPVDPTPSEAQEVDPTAV 458
Query: 280 SPLADQ----PKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAV--NHSLELWNQL 333
+PL + P +D KR+ +++S K K + A A A + + + N++
Sbjct: 459 APLPEAVVVLPASDMAAGKRVRTNDSSSKKKSKKKKASSAEAGKELPIFEDRVVASMNRM 518
Query: 334 DKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGY 393
+ +++E AA +A A A + A + A + A AK+ +E G
Sbjct: 519 VHSYDLAMRNNME-----AANRLAEADARIQVAERERDEALSQAAAAKVAKEE-----GE 568
Query: 394 ENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKR 453
+ + NL + LKG ++A +R A ++ T +
Sbjct: 569 KEAFVNKANAIKMAELNLRADSEVVRLKG----------MLAEARGLRDSEVARASQTAK 618
Query: 454 AENMDAILKAAELAAEAVSQAGKI----VTMGDPLPLIELIEAGPEGCWKASRESSREVG 509
E + + + A + VS I + + P +LI+A EG + + E +
Sbjct: 619 RETSEVFVARLKAAEQKVSLLDGINDQFIYLSQARPNAQLIKALEEG-GELATEKDQVEE 677
Query: 510 LLKDMTRDLVN 520
LKD VN
Sbjct: 678 WLKDFANAEVN 688
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.308 0.125 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,085,564
Number of Sequences: 26719
Number of extensions: 1059755
Number of successful extensions: 3668
Number of sequences better than 10.0: 167
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 147
Number of HSP's that attempted gapping in prelim test: 3424
Number of HSP's gapped (non-prelim): 319
length of query: 1091
length of database: 11,318,596
effective HSP length: 110
effective length of query: 981
effective length of database: 8,379,506
effective search space: 8220295386
effective search space used: 8220295386
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC137077.8


