

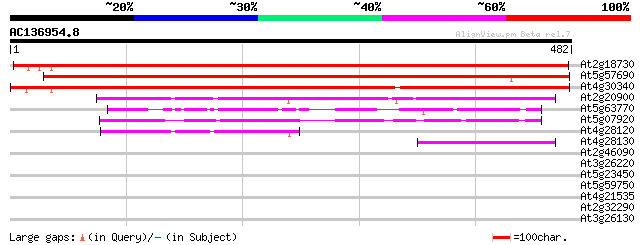
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.8 + phase: 0
(482 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g18730 putative diacylglycerol kinase 656 0.0
At5g57690 diacylglycerol kinase-like protein 639 0.0
At4g30340 unknown protein 616 e-176
At2g20900 putative diacylglycerol kinase 246 3e-65
At5g63770 diacylglycerol kinase 150 2e-36
At5g07920 diacylglycerol kinase (ATDGK1) 138 8e-33
At4g28120 putative protein 110 2e-24
At4g28130 putative diacylglycerol kinase (fragment) 104 9e-23
At2g46090 Unknown protein 38 0.011
At3g26220 cytochrome P450 monooxygenase (CYP71B3) 35 0.12
At5g23450 sphingosine kinase (AtLCBK1) 33 0.35
At5g59750 GTP cyclohydrolase II / 3,4-dihydroxy-2-butanone-4-pho... 29 5.0
At4g21535 predicted protein 29 5.0
At2g32290 beta-amylase like protein 29 6.6
At3g26130 cellulase, putative 28 8.6
>At2g18730 putative diacylglycerol kinase
Length = 488
Score = 656 bits (1693), Expect = 0.0
Identities = 313/484 (64%), Positives = 385/484 (78%), Gaps = 7/484 (1%)
Query: 4 SPSSTTEDSKK--IQVRSSLVES--IRGCGLSGMR---IDKEDLKKQLTLPQYLRFAMRD 56
SP S T+ SK+ + R S +S +RGCGL+ + +DK +L+++L +P+YLR AMRD
Sbjct: 3 SPVSKTDASKEKFVASRPSTADSKTMRGCGLANLAWVGVDKVELRQRLMMPEYLRLAMRD 62
Query: 57 SIRLQDPSAGETLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVF 116
I+ +D SA AP +PMVVFIN SGGRHGP LKERLQQLMSEEQVF
Sbjct: 63 CIKRKDSSAIPDHLLLPGGAVADMAPHAPMVVFINPNSGGRHGPVLKERLQQLMSEEQVF 122
Query: 117 DLADVKPHEFVLYGLACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQL 176
DL +VKPHEFV YGL CLE +A GD CAKE R +LR+MVAGGDGTVGWVLGCL EL +
Sbjct: 123 DLTEVKPHEFVRYGLGCLEKVAAEGDECAKECRARLRIMVAGGDGTVGWVLGCLGELNKD 182
Query: 177 GREPVPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSIS 236
G+ +PPVG++PLGTGNDLSRSF WGGSFPFAW+SA+KRTL +AS+G V RLDSW++ +S
Sbjct: 183 GKSQIPPVGVIPLGTGNDLSRSFGWGGSFPFAWRSAVKRTLHRASMGPVARLDSWKILVS 242
Query: 237 MPESTTVKPPYCLKQAEEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHR 296
MP V PPY LK AEE LDQG++ + P K+YEGV+YNY SIGMDAQVAYGFH
Sbjct: 243 MPSGEVVDPPYSLKPAEENELDQGLDAGIDAPPLAKAYEGVFYNYLSIGMDAQVAYGFHH 302
Query: 297 LRDEKPYLASGPIANKIIYSGYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWEQ 356
LR+ KPYLA GPI+NKIIYS + C+QGWF TPC +DPGLRGLRNI+++HIK+V+ S+WE+
Sbjct: 303 LRNTKPYLAQGPISNKIIYSSFGCSQGWFCTPCVNDPGLRGLRNIMKIHIKKVNCSQWEE 362
Query: 357 VAIPKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHAS 416
+A+PK+VR+IVALNLHSYGSG +PWG KP+YLEK+GFVEA DG +EIFG KQGWHAS
Sbjct: 363 IAVPKNVRSIVALNLHSYGSGSHPWGNLKPDYLEKRGFVEAHCDDGLIEIFGFKQGWHAS 422
Query: 417 FVMVDLITAKHIAQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFVEIKREPFQS 476
FVM +LI+AKHIAQAAA+R ELRGG W++A+LQMDGEPWKQP+S ++STFVEIK+ P+QS
Sbjct: 423 FVMAELISAKHIAQAAAVRFELRGGDWRDAFLQMDGEPWKQPMSTEYSTFVEIKKVPYQS 482
Query: 477 LVVD 480
L+++
Sbjct: 483 LMIN 486
>At5g57690 diacylglycerol kinase-like protein
Length = 498
Score = 639 bits (1647), Expect = 0.0
Identities = 304/463 (65%), Positives = 365/463 (78%), Gaps = 11/463 (2%)
Query: 30 LSGMRIDKEDLKKQLTLPQYLRFAMRDSIRLQDPSAGETLYRNRAEGEDSAAPTSPMVVF 89
L+ +R+ K +L++++ LPQYLR A+RD I +D S + ++ P P++VF
Sbjct: 35 LASIRVSKAELRQRVMLPQYLRIAIRDCILRKDDSFDASSSVAPPLENNALTPEVPLMVF 94
Query: 90 INARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLGDSCAKETR 149
+N +SGGR GP +KERLQ L+SEEQV+DL +VKP+EF+ YGL CLE A GD CAKE R
Sbjct: 95 VNPKSGGRQGPLIKERLQNLISEEQVYDLTEVKPNEFIRYGLGCLEAFASRGDECAKEIR 154
Query: 150 EKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFNWGGSFPFAW 209
EK+R++VAGGDGTVGWVLGCL EL R PVPPV I+PLGTGNDLSRSF WGGSFPFAW
Sbjct: 155 EKMRIVVAGGDGTVGWVLGCLGELNLQNRLPVPPVSIMPLGTGNDLSRSFGWGGSFPFAW 214
Query: 210 KSAIKRTLQKASVGSVHRLDSWRLSISMPESTTVKPPYCLKQAEEFTLDQGIEIEGELPD 269
KSAIKRTL +ASV + RLDSW + I+MP V PPY LK +E +DQ +EIEGE+P
Sbjct: 215 KSAIKRTLHRASVAPISRLDSWNILITMPSGEIVDPPYSLKATQECYIDQNLEIEGEIPP 274
Query: 270 KVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKIIYSGYSCTQGWFFTPC 329
YEGV+YNYFSIGMDAQVAYGFH LR+EKPYLA+GPIANKIIYSGY C+QGWF T C
Sbjct: 275 STNGYEGVFYNYFSIGMDAQVAYGFHHLRNEKPYLANGPIANKIIYSGYGCSQGWFLTHC 334
Query: 330 TSDPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLHSYGSGRNPWGKPKPEYL 389
+DPGLRGL+NI+ +HIK++ SSEWE+V +PKSVRA+VALNLHSYGSGRNPWG K +YL
Sbjct: 335 INDPGLRGLKNIMTLHIKKLDSSEWEKVPVPKSVRAVVALNLHSYGSGRNPWGNLKQDYL 394
Query: 390 EKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIA-----------QAAAIRLEL 438
EK+GFVEA DG LEIFGLKQGWHASFVMV+LI+AKHIA QAAAIRLE+
Sbjct: 395 EKRGFVEAQADDGLLEIFGLKQGWHASFVMVELISAKHIAQLMCKNLGFNEQAAAIRLEI 454
Query: 439 RGGGWKNAYLQMDGEPWKQPLSKDFSTFVEIKREPFQSLVVDG 481
RGG WK+A++QMDGEPWKQP+++D+STFV+IKR P QSLVV G
Sbjct: 455 RGGDWKDAFMQMDGEPWKQPMTRDYSTFVDIKRVPHQSLVVKG 497
>At4g30340 unknown protein
Length = 490
Score = 616 bits (1588), Expect = e-176
Identities = 301/493 (61%), Positives = 376/493 (76%), Gaps = 16/493 (3%)
Query: 1 MESSPSSTTEDSK------KIQVRSSLVESIRGCGLSGMR---IDKEDLKKQLTLPQYLR 51
ME +P S E S + ++ ++RGCG + + IDKE+L+ +L +P+YLR
Sbjct: 1 MEETPRSVGEASTTNFVAARPSAKTDDAVTMRGCGFANLALVGIDKEELRGRLAMPEYLR 60
Query: 52 FAMRDSIRLQDPSAGETLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMS 111
AMRD I+ +D + AP +PMVVFIN +SGGRHGP LKERLQQLM+
Sbjct: 61 IAMRDCIKRKDSTEISDHLLLPGGAAADMAPHAPMVVFINPKSGGRHGPVLKERLQQLMT 120
Query: 112 EEQVFDLADVKPHEFVLYGLACLEMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLT 171
EEQVFDL +VKPHEFV YGL CL+ LA GD CA+E REK+R+MVAGGDGTVGWVLGCL
Sbjct: 121 EEQVFDLTEVKPHEFVRYGLGCLDTLAAKGDECARECREKIRIMVAGGDGTVGWVLGCLG 180
Query: 172 ELRQLGREPVPPVGIVPLGTGNDLSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSW 231
EL + G+ +PPVG++PLGTGNDLSRSF+WGGSFPFAW+SA+KRTL +A++GS+ RLDSW
Sbjct: 181 ELHKDGKSHIPPVGVIPLGTGNDLSRSFSWGGSFPFAWRSAMKRTLHRATLGSIARLDSW 240
Query: 232 RLSISMPESTTVKPPYCLKQA-EEFTLDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQV 290
++ +SMP V PPY LK EE LDQ ++ +G++P K KSYEGV+YNYFSIGMDAQV
Sbjct: 241 KIVVSMPSGEVVDPPYSLKPTIEETALDQALDADGDVPPKAKSYEGVFYNYFSIGMDAQV 300
Query: 291 AYGFHRLRDEKPYLASGPIANKI-IYSG-YSCTQGWFFTPCTSDPGLRGLRNILRMHIKR 348
AYGFH LR+EKPYLA GP+ NK+ IY +SC F S RGLRNI+++HIK+
Sbjct: 301 AYGFHHLRNEKPYLAQGPVTNKVAIYQNLHSCLNYLFELQLYS----RGLRNIMKIHIKK 356
Query: 349 VSSSEWEQVAIPKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFG 408
+ SEWE++ +PKSVR+IV LNL++YGSGR+PWG +P+YLEK+GFVEA DG +EIFG
Sbjct: 357 ANCSEWEEIHVPKSVRSIVVLNLYNYGSGRHPWGNLRPKYLEKRGFVEAHCDDGLIEIFG 416
Query: 409 LKQGWHASFVMVDLITAKHIAQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFSTFVE 468
LKQGWHASFVM ++I+AKHIAQAAAIR ELRGG WKNA+LQMDGEPWKQP+ D+STFVE
Sbjct: 417 LKQGWHASFVMAEIISAKHIAQAAAIRFELRGGDWKNAFLQMDGEPWKQPMKSDYSTFVE 476
Query: 469 IKREPFQSLVVDG 481
IK+ PFQSL+++G
Sbjct: 477 IKKVPFQSLMING 489
>At2g20900 putative diacylglycerol kinase
Length = 509
Score = 246 bits (627), Expect = 3e-65
Identities = 143/402 (35%), Positives = 211/402 (51%), Gaps = 16/402 (3%)
Query: 75 EGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACL 134
E E P SP++VFIN++SGG+ G L + L++ QVFDL P + + L
Sbjct: 30 EEESRPTPASPVLVFINSKSGGQLGGELILTYRSLLNHNQVFDLDQETPDKVLRRIYLNL 89
Query: 135 EMLAGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGND 194
E L D A++ REKL+++VAGGDGT GW+LG + +L+ PP+ VPLGTGN+
Sbjct: 90 ERLKD--DDFARQIREKLKIIVAGGDGTAGWLLGVVCDLKL---SHPPPIATVPLGTGNN 144
Query: 195 LSRSFNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMP---ESTTVKPPYCLKQ 251
L +F WG P ++A++ L++ V ++D+W + + M E + P L+
Sbjct: 145 LPFAFGWGKKNPGTDRTAVESFLEQVLKAKVMKIDNWHILMRMKTPKEGGSCDPVAPLEL 204
Query: 252 AEEFTLDQGIEIEGEL-PDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIA 310
+ EL + ++ G ++NYFS+GMDAQ++Y FH R P +
Sbjct: 205 PHSLHAFHRVSPTDELNKEGCHTFRGGFWNYFSLGMDAQISYAFHSERKLHPEKFKNQLV 264
Query: 311 NKIIYSGYSCTQGWFFTPCTS--DPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVA 368
N+ Y CTQGWF C S P R + + ++ I + +W+ + IP S+R+IV
Sbjct: 265 NQSTYVKLGCTQGWF---CASLFHPASRNIAQLAKVKIA-TRNGQWQDLHIPHSIRSIVC 320
Query: 369 LNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHI 428
LNL S+ G NPWG P P +G V DG +E+ G + WH ++ +
Sbjct: 321 LNLPSFSGGLNPWGTPNPRKQRDRGLTPPFVDDGLIEVVGFRNAWHGLVLLAPNGHGTRL 380
Query: 429 AQAAAIRLELRGGGWKNAYLQMDGEPWKQPLSKDFST-FVEI 469
AQA IR E G + +++MDGEPWKQPL D T VEI
Sbjct: 381 AQANRIRFEFHKGATDHTFMRMDGEPWKQPLPLDDETVMVEI 422
>At5g63770 diacylglycerol kinase
Length = 712
Score = 150 bits (379), Expect = 2e-36
Identities = 121/379 (31%), Positives = 173/379 (44%), Gaps = 75/379 (19%)
Query: 85 PMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLGDSC 144
P++VFINA+SGG+ GP L RL L++ QVF+L +C AGL D C
Sbjct: 342 PLLVFINAKSGGQLGPFLHRRLNMLLNPVQVFELG------------SCQGPDAGL-DLC 388
Query: 145 AKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRSFNWG-G 203
+K + RV+V GGDGTV WVL + E R E PPV I+PLGTGNDLSR WG G
Sbjct: 389 SKV--KYFRVLVCGGDGTVAWVLDAI-EKRNF--ESPPPVAILPLGTGNDLSRVLQWGRG 443
Query: 204 SFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMPESTTVKPPYCLKQAEEFTLDQGIEI 263
+ +++ LQ +V LD W S+ + E +T K P ++ +F +
Sbjct: 444 ISVVDGQGSLRTFLQDIDHAAVTMLDRW--SVKIVEESTEKFP--AREGHKFMM------ 493
Query: 264 EGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKIIYSGYSCTQG 323
NY IG DA+VAY FH +R EKP NK+ Y+
Sbjct: 494 ----------------NYLGIGCDAKVAYEFHMMRQEKPEKFCSQFVNKLRYA------- 530
Query: 324 WFFTPCTSDPGLRGLRNILRMHIKRVSSSEW-----EQVAIPKSVRAIVALNLHSYGSGR 378
G R+I+ + W + + IPK ++ LN+ SY G
Sbjct: 531 -----------KEGARDIMDRACADLPWQVWLEVDGKDIEIPKDSEGLIVLNIGSYMGGV 579
Query: 379 NPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAAAIRLEL 438
+ W + +Y F + D LE+ ++ WH + V L A+ +AQ IR+ +
Sbjct: 580 DLW---QNDYEHDDNFSIQCMHDKTLEVVCVRGAWHLGKLQVGLSQARRLAQGKVIRIHV 636
Query: 439 RGGGWKNAYLQMDGEPWKQ 457
+Q+DGEP+ Q
Sbjct: 637 S----SPFPVQIDGEPFIQ 651
>At5g07920 diacylglycerol kinase (ATDGK1)
Length = 728
Score = 138 bits (347), Expect = 8e-33
Identities = 102/381 (26%), Positives = 161/381 (41%), Gaps = 68/381 (17%)
Query: 78 DSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEML 137
D + P++VFIN +SG + G +L++RL ++ QVF+L+ V+ E L+ +
Sbjct: 354 DLPSDARPLLVFINKKSGAQRGDSLRQRLHLHLNPVQVFELSSVQGPEVGLFLFRKV--- 410
Query: 138 AGLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSR 197
RV+V GGDGT GWVL + + + P V I+P GTGNDLSR
Sbjct: 411 ------------PHFRVLVCGGDGTAGWVLDAIEKQNFIS---PPAVAILPAGTGNDLSR 455
Query: 198 SFNWGGSF-PFAWKSAIKRTLQKASVGSVHRLDSWRLSISMPESTTVKPPYCLKQAEEFT 256
NWGG + + LQ +V LD W++SI + ++PP +
Sbjct: 456 VLNWGGGLGSVERQGGLSTVLQNIEHAAVTVLDRWKVSILNQQGKQLQPPKYMN------ 509
Query: 257 LDQGIEIEGELPDKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKIIYS 316
NY +G DA+VA H LR+E P NK++Y+
Sbjct: 510 -----------------------NYIGVGCDAKVALEIHNLREENPERFYSQFMNKVLYA 546
Query: 317 GYSCTQGWFFTPCTSDPGLRGLRNILRMHIKRVSSSEWEQVAIPKSVRAIVALNLHSYGS 376
D +R+ + V + +P+ I+ N+ SY
Sbjct: 547 REGARS-------IMDRTFEDFPWQVRVEVDGV------DIEVPEDAEGILVANIGSYMG 593
Query: 377 GRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLKQGWHASFVMVDLITAKHIAQAAAIRL 436
G + W Y + F + D +E+ + WH + V L A+ +AQ +A+++
Sbjct: 594 GVDLWQNEDETY---ENFDPQSMHDKIVEVVSISGTWHLGKLQVGLSRARRLAQGSAVKI 650
Query: 437 ELRGGGWKNAYLQMDGEPWKQ 457
+L +Q+DGEPW Q
Sbjct: 651 QL----CAPLPVQIDGEPWNQ 667
>At4g28120 putative protein
Length = 216
Score = 110 bits (274), Expect = 2e-24
Identities = 64/177 (36%), Positives = 102/177 (57%), Gaps = 11/177 (6%)
Query: 79 SAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLA 138
+ AP +P++VFIN++SGG+ G L + L++++QVFDL P + + LE L
Sbjct: 34 TTAPENPILVFINSKSGGQLGAELILTYRTLLNDKQVFDLEVETPDKVLQRIYLNLERLK 93
Query: 139 GLGDSCAKETREKLRVMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRS 198
DS A + R+KL+++VAGGDGT GW+LG +++ L PP+ VPLGTGN+L +
Sbjct: 94 D--DSLASKIRDKLKIIVAGGDGTAGWLLGVVSD---LNLSNPPPIATVPLGTGNNLPFA 148
Query: 199 FNWGGSFPFAWKSAIKRTLQKASVGSVHRLDSWRLSISMPE------STTVKPPYCL 249
F WG P +S+++ L K ++D+W++ + M T+K P+ L
Sbjct: 149 FGWGKKNPGTDRSSVESFLGKVINAKEMKIDNWKILMRMKHPKEGSCDITLKLPHSL 205
>At4g28130 putative diacylglycerol kinase (fragment)
Length = 176
Score = 104 bits (260), Expect = 9e-23
Identities = 50/120 (41%), Positives = 71/120 (58%), Gaps = 1/120 (0%)
Query: 351 SSEWEQVAIPKSVRAIVALNLHSYGSGRNPWGKPKPEYLEKKGFVEADVADGRLEIFGLK 410
+ +W + IP+S+R+IV LNL S+ G NPWG P P+ + V DG +EI G +
Sbjct: 14 NGQWNDLHIPQSIRSIVCLNLPSFSGGLNPWGTPNPKKQRDRSLTAPFVDDGLIEIVGFR 73
Query: 411 QGWHASFVMVDLITAKHIAQAAAIRLELRGGGWKNAYLQMDGEPWKQPL-SKDFSTFVEI 469
WH ++ +AQA +RLE + G K+AY+++DGEPWKQPL S D + VEI
Sbjct: 74 NAWHGLILLSPNGHGTRLAQANRVRLEFKKGAAKHAYMRIDGEPWKQPLPSNDETVMVEI 133
>At2g46090 Unknown protein
Length = 364
Score = 38.1 bits (87), Expect = 0.011
Identities = 24/65 (36%), Positives = 36/65 (54%), Gaps = 11/65 (16%)
Query: 148 TREKLR-----VMVAGGDGTVGWVL-GCLTELRQLG-----REPVPPVGIVPLGTGNDLS 196
TRE +R V+ GGDGT+ V+ G E + +G +G++PLGTG+D +
Sbjct: 103 TREAIRDGADAVIAVGGDGTLHEVVNGFFWEGKPVGYLSGEASRSTALGLIPLGTGSDFA 162
Query: 197 RSFNW 201
R+F W
Sbjct: 163 RTFGW 167
>At3g26220 cytochrome P450 monooxygenase (CYP71B3)
Length = 501
Score = 34.7 bits (78), Expect = 0.12
Identities = 35/120 (29%), Positives = 58/120 (48%), Gaps = 14/120 (11%)
Query: 13 KKIQVRSSLVESIRGC-GLSGM-RIDKEDLKKQLTLPQYLRFAMRDSIRLQDPS----AG 66
K +V E IR C G+ RI++ED+ K QYL+ +++++RL P+
Sbjct: 322 KNPRVMKKAQEEIRTCIGIKQKERIEEEDVDKL----QYLKLVIKETLRLHPPAPLLLPR 377
Query: 67 ETLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEF 126
ET+ + +G D T ++ +NA S GR+ P L E ++ E + D K + F
Sbjct: 378 ETMADIKIQGYDIPRKT---ILLVNAWSIGRN-PELWENPEEFNPERFIDCPMDYKGNSF 433
>At5g23450 sphingosine kinase (AtLCBK1)
Length = 763
Score = 33.1 bits (74), Expect = 0.35
Identities = 38/114 (33%), Positives = 52/114 (45%), Gaps = 17/114 (14%)
Query: 86 MVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEFVLYGLACLEMLAGLGDS-- 143
M+V +N RSG HG ++K + E +F LA +K LA D
Sbjct: 250 MLVILNPRSG--HGRSIKVFHNVV---EPIFKLAGIKMEVVKTTKAGHARELASTVDINL 304
Query: 144 CAKETREKLRVMVAGGDGTVGWVL-GCLTELRQLGREPVP-PVGIVPLGTGNDL 195
C+ ++ GGDG + VL G LT R +E V P+GIVP G+ N L
Sbjct: 305 CSDG------IICVGGDGIINEVLNGLLT--RSNPKEGVSIPIGIVPAGSDNSL 350
>At5g59750 GTP cyclohydrolase II /
3,4-dihydroxy-2-butanone-4-phoshate synthase - like
protein
Length = 509
Score = 29.3 bits (64), Expect = 5.0
Identities = 18/59 (30%), Positives = 30/59 (50%), Gaps = 6/59 (10%)
Query: 68 TLYRNRAEGEDSAAPTSPMVVFINARSGGRHGPALKERLQQLMSEEQVFDLADVKPHEF 126
TL E EDS+APT + ++A+SG G + +R +++ + D KP +F
Sbjct: 169 TLMSPEMEDEDSSAPT--FTITVDAKSGTSTGVSASDRAMTVLALSSL----DAKPDDF 221
>At4g21535 predicted protein
Length = 455
Score = 29.3 bits (64), Expect = 5.0
Identities = 15/45 (33%), Positives = 23/45 (50%)
Query: 154 VMVAGGDGTVGWVLGCLTELRQLGREPVPPVGIVPLGTGNDLSRS 198
++ GDG + V+ L E P+G+VP GTGN + +S
Sbjct: 174 IVCVSGDGILVEVVNGLLERADWRNALKLPIGMVPAGTGNGMIKS 218
>At2g32290 beta-amylase like protein
Length = 577
Score = 28.9 bits (63), Expect = 6.6
Identities = 24/94 (25%), Positives = 39/94 (40%), Gaps = 26/94 (27%)
Query: 269 DKVKSYEGVYYNYFSIGMDAQVAYGFHRLRDEKPYLASGPIANKIIYSGYSCTQGWFFTP 328
D +KS+ ++ S G+ + G GP A ++ Y YS TQGW F
Sbjct: 222 DYMKSFRENMEDFISSGVIIDIEVGL------------GP-AGELRYPSYSETQGWVF-- 266
Query: 329 CTSDPGL-------RGLRNILRMHIKRVSSSEWE 355
PG+ + LR+ ++R+ EW+
Sbjct: 267 ----PGIGEFQCYDKYLRSDYEEEVRRIGHPEWK 296
>At3g26130 cellulase, putative
Length = 551
Score = 28.5 bits (62), Expect = 8.6
Identities = 14/42 (33%), Positives = 24/42 (56%)
Query: 213 IKRTLQKASVGSVHRLDSWRLSISMPESTTVKPPYCLKQAEE 254
++++L + +GS +R +SWRLS S + CLK E+
Sbjct: 408 VRKSLFQLKLGSCNRSESWRLSSHRVLSLAEEQILCLKAYEK 449
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,349,095
Number of Sequences: 26719
Number of extensions: 500761
Number of successful extensions: 1108
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1074
Number of HSP's gapped (non-prelim): 16
length of query: 482
length of database: 11,318,596
effective HSP length: 103
effective length of query: 379
effective length of database: 8,566,539
effective search space: 3246718281
effective search space used: 3246718281
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC136954.8


