

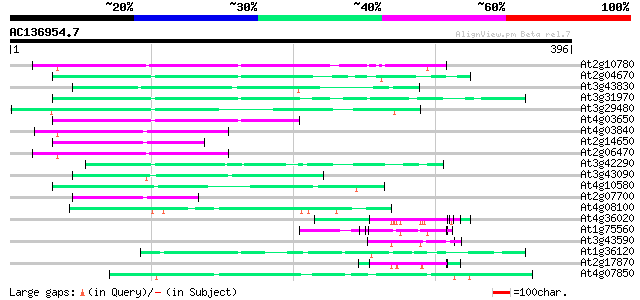
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.7 - phase: 0
(396 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 107 1e-23
At2g04670 putative retroelement pol polyprotein 86 3e-17
At3g43830 putative protein 86 4e-17
At3g31970 hypothetical protein 85 6e-17
At3g29480 hypothetical protein 84 1e-16
At4g03650 putative reverse transcriptase 79 4e-15
At4g03840 putative transposon protein 68 8e-12
At2g14650 putative retroelement pol polyprotein 68 8e-12
At2g06470 putative retroelement pol polyprotein 68 1e-11
At3g42290 putative protein 66 3e-11
At3g43090 putative protein 64 2e-10
At4g10580 putative reverse-transcriptase -like protein 60 3e-09
At2g07700 putative retroelement pol polyprotein 60 3e-09
At4g08100 putative polyprotein 55 5e-08
At4g36020 glycine-rich protein 55 7e-08
At1g75560 DNA-binding protein 55 9e-08
At3g43590 putative protein 54 1e-07
At1g36120 putative reverse transcriptase gb|AAD22339.1 53 3e-07
At2g17870 putative glycine-rich, zinc-finger DNA-binding protein 52 7e-07
At4g07850 putative polyprotein 51 1e-06
>At2g10780 pseudogene
Length = 1611
Score = 107 bits (267), Expect = 1e-23
Identities = 84/308 (27%), Positives = 133/308 (42%), Gaps = 29/308 (9%)
Query: 17 AQAVQQQPQAGNGEVR-----MLETFLRNHPPAFKGRYDPDGAQSWLKEVERIFRVMQCS 71
++ V+ PQ +G R +LE R F G DP A W ++R F+ +C
Sbjct: 149 SEVVEVNPQPNHGRKRGDYLSLLEHVSRLGTRHFMGSTDPIVADEWRSRLKRNFKSTRCP 208
Query: 72 EVQKVRFGTHMLAEEADDWWVSLLPVLEQNGAVVTWAVFRREFLNRYFPEDVRGRKEIEF 131
E + H L +A +WW+++ + V ++A F EF +YFP + R E +
Sbjct: 209 EDYQRDIAVHFLEGDAHNWWLTVEK--RRGDEVRSFADFEDEFNKKYFPPEAWDRLECAY 266
Query: 132 LELKQGDMSVVEYAAKFVELAKFYPHYAPETAEFSKCIKFENGLRADIKRAIGYQQIRIF 191
L+L QG+ +V EY +F L ++ E E ++ +F GLR +I+ +
Sbjct: 267 LDLVQGNRTVREYDEEFNRLRRYVGRELEE--EQAQLRRFIRGLRIEIRNHCLVRTFNSV 324
Query: 192 SDLVSRCRIYEEDTKAHYKVMSERRGKGHQNRPKPYSAPTDKGKQRLNDERRPSKRDAPA 251
S+LV R + EE + + E+ + KP K+R D+ +K DA
Sbjct: 325 SELVERAAMIEEGIEEERYLNREKAPIRNNQSTKP------ADKKRKFDKVDNTKSDAKT 378
Query: 252 EIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDV-----------VCYNCNE 300
C CG K H S C K C RCG K H + C + + C+ C +
Sbjct: 379 G-ECVTCG-KNH-SGTCWKAIGACGRCGSKDHAIQSCPKMEPGQSKVLGEETRTCFYCGK 435
Query: 301 EGHISTQC 308
GH+ +C
Sbjct: 436 TGHLKREC 443
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 86.3 bits (212), Expect = 3e-17
Identities = 81/298 (27%), Positives = 118/298 (39%), Gaps = 47/298 (15%)
Query: 31 VRMLETFLRNHPPAFKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDW 90
+RM+E R F P+ A SW V+R F +C +V H L +A W
Sbjct: 124 LRMMEQLQRIGTWYFSSGTSPEEADSWRSRVQRNFGSSRCPAEYRVDLAVHFLEGDAHLW 183
Query: 91 WVSLLPVLEQNGAVVTWAVFRREFLNRYFPEDVRGRKEIEFLELKQGDMSVVEYAAKFVE 150
W S+ Q A ++WA F EF +YFP++ R E FLEL QG+ SV EY +F
Sbjct: 184 WRSVTARRRQ--ADMSWADFVAEFNAKYFPQEALDRMEARFLELTQGEWSVREYDREFNR 241
Query: 151 LAKFYPHYAPETAEFSKCIKFENGLRADIKRAIGYQQIRIFSDLVSRCRIYEEDTKAHYK 210
L + + + ++ +F GLR D++ Q + LV EED +
Sbjct: 242 LLAYAGRGMED--DQAQMRRFLRGLRPDLRVQCRVSQYATKAALVETAAEVEEDLQRQVV 299
Query: 211 VMSERRGKGHQNRPKPYSAPTDKGKQRLNDERRPSKRDAPAEIVCFKCGEK---GHKSNV 267
+S + T +Q++ PSK PA+ G+K H S
Sbjct: 300 GVSP-------------AVQTKNTQQQVT----PSKGGKPAQ------GQKRKWDHPSRA 336
Query: 268 CTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKKVRVGGKVFALTG 325
F CG H +AD C E GH+ + GG+ A+ G
Sbjct: 337 GQGGRAGYFSCGSLDHTVAD----------CTERGHLHVKV-------AGGQFLAVLG 377
>At3g43830 putative protein
Length = 329
Score = 85.9 bits (211), Expect = 4e-17
Identities = 66/248 (26%), Positives = 97/248 (38%), Gaps = 46/248 (18%)
Query: 45 FKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPVLEQNGAV 104
F G DP A +W +E F+ M+C + L EA +WW + EQ G V
Sbjct: 116 FNGGTDPVKAYNWRNMLECKFKSMRCPVEFWRDLASCYLRGEAQEWW-ERVKQREQVGCV 174
Query: 105 VTWAVFRREFLNRYFPEDVRGRKEIEFLELKQGDMSVVEYAAKFVELAKFYPHYAPETAE 164
W+ F+ EF RY PE+ E++FL L+QG +V +Y +F L +F + E
Sbjct: 175 DQWSFFKEEFTRRYLPEETIDDLEMKFLRLQQGTKTVRKYEKEFHSLERFERR---KRGE 231
Query: 165 FSKCIKFENGLRADIKRAIGYQQIRIFSDLVSRCRIYE---EDTKAHYKVMSERRGKGHQ 221
KF +GLR DI+ + DLV + E E+ + ++ E+ G
Sbjct: 232 HELIHKFISGLRVDIRLCCHVRDFDNMIDLVEKAASLEIGLEEEARYKRIAQEKEAMG-- 289
Query: 222 NRPKPYSAPTDKGKQRLNDERRPSKRDAPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQK 281
G+ GH C E C+RCG
Sbjct: 290 -----------------------------------SYGQTGHSKRRC--QEVTCYRCGVA 312
Query: 282 GHMLADCK 289
GH+ DC+
Sbjct: 313 GHIARDCR 320
>At3g31970 hypothetical protein
Length = 1329
Score = 85.1 bits (209), Expect = 6e-17
Identities = 92/334 (27%), Positives = 130/334 (38%), Gaps = 47/334 (14%)
Query: 31 VRMLETFLRNHPPAFKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDW 90
+RM+E R F G P+ A SW VE F +C +V H L +A W
Sbjct: 144 LRMMEQLQRIGTRYFFGGTSPEEADSWKSRVEHNFGSSRCPAEYRVDLAVHFLEGDAHLW 203
Query: 91 WVSLLPVLEQNGAVVTWAVFRREFLNRYFPEDVRGRKEIEFLELKQGDMSVVEYAAKFVE 150
W S+ Q A ++WA F EF +YFP++ R E FLEL QG+ SV EY +F
Sbjct: 204 WRSVTARRRQ--AHMSWADFVAEFNAKYFPQEALDRMEARFLELTQGERSVREYEREFNR 261
Query: 151 LAKFYPHYAPETAEFSKCIKFENGLRADIKRAIGYQQIRIFSDLVSRCRIYEEDTKAHYK 210
L + + + ++ +F GLR D++ Q + LV EED
Sbjct: 262 LLVYAGRGMQD--DQAQMRRFLRGLRPDLRVRCRVLQYATKAALVETAAEVEEDL----- 314
Query: 211 VMSERRGKGHQNRPKPYSAPTDKGKQRLNDERRPSKRDAPAEIVCFKCGEKGHKSNVCTK 270
+R+ G KP ++ + PSK PA+ K E+GH C K
Sbjct: 315 ---QRQVVGVSPAVKP---------KKTQQQVAPSKGGKPAQ--GQKRKERGHLRPNCPK 360
Query: 271 DEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKKVRVGGKVFALTGTQTAN 330
++ Q + G V + HI+ A G T
Sbjct: 361 LQRMAVAVVQPA-----VQHGAQVQQRVQQLAHIAA---------------APQGYTT-- 398
Query: 331 EDRLIRGTCFFNSTPLIAIIDTGATHCFIALECA 364
R I T T + D+GA+HCFI E A
Sbjct: 399 --REIGSTSKRAITEAHVLFDSGASHCFITPESA 430
>At3g29480 hypothetical protein
Length = 718
Score = 84.3 bits (207), Expect = 1e-16
Identities = 71/294 (24%), Positives = 111/294 (37%), Gaps = 58/294 (19%)
Query: 2 AGRNDAAIAAALEAVAQAVQQQPQAGN---GEVRMLETFLRNHPPAFKGRYDPDGAQSWL 58
AG + A + VQQ+ +RM+E R F G P+ A SW
Sbjct: 5 AGGAQVPVQAPVAPRVAEVQQRAAVAEEVPSYLRMMEQLQRIGIGYFSGGTSPEEADSWR 64
Query: 59 KEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPVLEQNGAVVTWAVFRREFLNRY 118
VER F +C +V H L +A WW S+ Q A ++WA F EF +Y
Sbjct: 65 SRVERNFGSSRCPVEYRVDLAVHFLEGDAHLWWKSVTTRRRQ--ANMSWADFVAEFNAKY 122
Query: 119 FPEDVRGRKEIEFLELKQGDMSVVEYAAKFVELAKFYPHYAPETAEFSKCIKFENGLRAD 178
FP++ R E FLEL QG+ SV EY +F
Sbjct: 123 FPQEALDRMEAHFLELTQGERSVREYDREF------------------------------ 152
Query: 179 IKRAIGYQQIRIFSDLVSRCRIYEEDTKAHYKVMSERRGKGHQNRPKPYSAPTDKGKQRL 238
++ ++++ S + K H +V + GK Q + + +
Sbjct: 153 -------NRLLVYAECRSTRIPVVQPKKTHQQVAPSKGGKPAQGQKRSWD---------- 195
Query: 239 NDERRPSKRDAPAEIVCFKCGEKGHKSNVCTK--DEKKCFRCGQKGHMLADCKR 290
PS+ CF CG HK CT+ + ++ + C ++ H+ +C +
Sbjct: 196 ----HPSRAGQGGRAGCFFCGSLDHKVADCTQRAETREYYHCRKRRHLRPNCPK 245
>At4g03650 putative reverse transcriptase
Length = 839
Score = 79.0 bits (193), Expect = 4e-15
Identities = 56/174 (32%), Positives = 79/174 (45%), Gaps = 4/174 (2%)
Query: 31 VRMLETFLRNHPPAFKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDW 90
+RM++ R F G P+ A SW +VER F +C +V H L +A W
Sbjct: 142 LRMMKQLQRIGTEYFSGGTSPEEADSWRSQVERNFGSSRCPAEYRVDLTVHFLEGDAHLW 201
Query: 91 WVSLLPVLEQNGAVVTWAVFRREFLNRYFPEDVRGRKEIEFLELKQGDMSVVEYAAKFVE 150
W S+ Q A ++WA F EF +YFP + R E FLEL QG SV EY KF
Sbjct: 202 WRSVTARRRQ--ADMSWADFMAEFNAKYFPREALDRMEARFLELTQGVRSVREYDRKFNR 259
Query: 151 LAKFYPHYAPETAEFSKCIKFENGLRADIKRAIGYQQIRIFSDLVSRCRIYEED 204
L + + + ++ +F GLR +++ Q + LV EED
Sbjct: 260 LLVYAGRGMED--DQAQMRRFLRGLRPNLRVRCRVSQYATKAALVETAAEVEED 311
>At4g03840 putative transposon protein
Length = 973
Score = 68.2 bits (165), Expect = 8e-12
Identities = 42/142 (29%), Positives = 68/142 (47%), Gaps = 7/142 (4%)
Query: 18 QAVQQQPQAGNGEVR-----MLETFLRNHPPAFKGRYDPDGAQSWLKEVERIFRVMQCSE 72
+ V+ PQ +G R +LE R F G DP A W ++R F+ +C E
Sbjct: 115 EVVEVNPQPNHGRKRGDYLSLLEHVSRLGTRHFMGSTDPIVADEWRSRLKRNFKSTRCPE 174
Query: 73 VQKVRFGTHMLAEEADDWWVSLLPVLEQNGAVVTWAVFRREFLNRYFPEDVRGRKEIEFL 132
+ H L +A +WW+++ + V ++A F EF +YFP + R E +L
Sbjct: 175 DYQRDIAVHFLERDAHNWWLTV--EKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAYL 232
Query: 133 ELKQGDMSVVEYAAKFVELAKF 154
+L QG+ +V EY +F L ++
Sbjct: 233 DLVQGNRTVREYDEEFNRLRRY 254
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 68.2 bits (165), Expect = 8e-12
Identities = 38/107 (35%), Positives = 52/107 (48%), Gaps = 2/107 (1%)
Query: 31 VRMLETFLRNHPPAFKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDW 90
+RM+E R F G P+ A SW VER F +C ++ H L +A W
Sbjct: 142 LRMMEQLQRIGTGYFSGGTSPEVADSWRSRVERNFGSSRCPAEYRIDLAVHFLEGDAHLW 201
Query: 91 WVSLLPVLEQNGAVVTWAVFRREFLNRYFPEDVRGRKEIEFLELKQG 137
W S+ + A ++WA F EF +YFP++ R E FLEL QG
Sbjct: 202 WRSV--TARRRQADISWADFVAEFNAKYFPQEALDRMEAHFLELTQG 246
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 67.8 bits (164), Expect = 1e-11
Identities = 42/143 (29%), Positives = 69/143 (47%), Gaps = 7/143 (4%)
Query: 17 AQAVQQQPQAGNGEVR-----MLETFLRNHPPAFKGRYDPDGAQSWLKEVERIFRVMQCS 71
++ V+ PQ +G R +LE R F G DP A W ++R F+ +C
Sbjct: 114 SEVVEVNPQPNHGRKRGDYLSLLEHVSRLGTRHFMGSTDPIVADEWRSRLKRNFKSTRCP 173
Query: 72 EVQKVRFGTHMLAEEADDWWVSLLPVLEQNGAVVTWAVFRREFLNRYFPEDVRGRKEIEF 131
E + H L +A +WW+++ + V ++A F EF +YFP + R E +
Sbjct: 174 EDYQRDIAVHFLEGDAHNWWLTVEK--RRGDEVRSFADFEDEFNKKYFPPEAWDRLECAY 231
Query: 132 LELKQGDMSVVEYAAKFVELAKF 154
L+L QG+ +V EY +F L ++
Sbjct: 232 LDLVQGNRTVREYDEEFNRLRRY 254
>At3g42290 putative protein
Length = 462
Score = 66.2 bits (160), Expect = 3e-11
Identities = 68/253 (26%), Positives = 102/253 (39%), Gaps = 45/253 (17%)
Query: 54 AQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPVLEQNGAVVTWAVFRRE 113
A SW V+R F +C +V H L E+A WW S+ Q A ++WA F E
Sbjct: 163 ADSWRSRVKRNFSSSRCLMEYRVDLAVHFLEEDAHLWWKSVTARRRQ--ADMSWADFVVE 220
Query: 114 FLNRYFPEDVRGRKEIEFLELKQGDMSVVEYAAKFVELAKFYPHYAPETAEFSKCIKFEN 173
F +YF +D R E+ FLEL + SV EY +F + Y + + + ++ +F
Sbjct: 221 FNAKYFLQDELDRMEVRFLELTLVERSVWEYDREFNRVL-VYAGWGMKDGQ-AELRRFMR 278
Query: 174 GLRADIKRAIGYQQIRIFSDLVSRCRIYEEDTKAHYKVMSERRGKGHQNRPKPYSAPTDK 233
GLR D++ Q + + LV +T A +V + GK Q + + ++
Sbjct: 279 GLRPDLRVRCRMSQYALKAALV--------ETAA--EVAPRKGGKPMQGKKRSWN----- 323
Query: 234 GKQRLNDERRPSKRDAPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDV 293
S+ CF CG HK+ Q H+ A +
Sbjct: 324 ---------HLSRAGQGGRAGCFSCGNLAHKN------------VQQLAHIPAAPQ---- 358
Query: 294 VCYNCNEEGHIST 306
CYN E G ST
Sbjct: 359 -CYNTFEAGGTST 370
>At3g43090 putative protein
Length = 357
Score = 63.5 bits (153), Expect = 2e-10
Identities = 50/180 (27%), Positives = 71/180 (38%), Gaps = 9/180 (5%)
Query: 45 FKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLL---PVLEQN 101
F G DP A W K +E F +C + H L EEA WW +L PV
Sbjct: 131 FTGGIDPVKADDWRKLLENNFENARCPVEFQKDIIVHFLREEASHWWDGVLGNTPVQH-- 188
Query: 102 GAVVTWAVFRREFLNRYFPEDVRGRKEIEFLELKQGDMSVVEYAAKFVELAKFYPHYAPE 161
V+ W FR EF ++FP++ E +F EL+Q V E + L++F
Sbjct: 189 --VIFWEDFREEFNRKFFPQEAMDSLEDDFEELRQDTKKVRENERELSHLSRF--SVRAG 244
Query: 162 TAEFSKCIKFENGLRADIKRAIGYQQIRIFSDLVSRCRIYEEDTKAHYKVMSERRGKGHQ 221
E S + GLR DI ++ DLV + E K + + + Q
Sbjct: 245 RGEQSMIRRLMRGLRPDILTRCASREFFSMIDLVEKAARIESGLVLEAKYLKNTQARTTQ 304
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 59.7 bits (143), Expect = 3e-09
Identities = 57/237 (24%), Positives = 83/237 (34%), Gaps = 37/237 (15%)
Query: 31 VRMLETFLRNHPPAFKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDW 90
+RM+E R F G P+ A SW V R F +C +V H L +A W
Sbjct: 138 LRMMEQLQRIDTGYFSGGTSPEEADSWRSRVGRNFGSSRCPAEYRVDLAVHFLEGDAHLW 197
Query: 91 WVSLLPVLEQNGAVVTWAVFRREFLNRYFPEDVRGRKEIEFLELKQGDMSVVEYAAKFVE 150
W S+ Q ++WA F EF +YFP++ + +E Q M
Sbjct: 198 WRSVTARRRQTD--MSWADFVAEFKAKYFPQEALDPYAGQGMEDDQAQMR---------- 245
Query: 151 LAKFYPHYAPETAEFSKCIKFENGLRADIKRAIGYQQIRIFSDLVSRCRIYEEDTKAHYK 210
+F GLR D++ Q + LV EED +
Sbjct: 246 -------------------RFLRGLRPDLRVRCRVSQYATKAALVETAAEVEEDFQRQVV 286
Query: 211 VMSERRGKGHQNRPKPYSAPTDKGKQRLNDERR---PSKRDAPAEIVCFKCGEKGHK 264
+S + + P+ GK +R+ PS+ CF CG HK
Sbjct: 287 GVSP---VVQPKKTQQQVTPSKGGKPAQGQKRKWDHPSRAGQGGRARCFSCGSLDHK 340
>At2g07700 putative retroelement pol polyprotein
Length = 543
Score = 59.7 bits (143), Expect = 3e-09
Identities = 28/89 (31%), Positives = 49/89 (54%), Gaps = 2/89 (2%)
Query: 45 FKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPVLEQNGAV 104
FKG ++ A WL+++E F + +C + K + + L ++A W S+ G +
Sbjct: 53 FKGEHNTVVADKWLRDLEMNFEISRCPDNFKRQIAVNFLDKDARIWRESV--TARNQGQM 110
Query: 105 VTWAVFRREFLNRYFPEDVRGRKEIEFLE 133
++W FRREF +YFP + R R E +F++
Sbjct: 111 ISWEAFRREFEKKYFPPEARDRLEQQFMK 139
>At4g08100 putative polyprotein
Length = 1054
Score = 55.5 bits (132), Expect = 5e-08
Identities = 58/254 (22%), Positives = 102/254 (39%), Gaps = 43/254 (16%)
Query: 43 PAFKGRYDPDGAQSWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPVLE--Q 100
PAF G +PD W +++E +F +C + KV+ A WW L+ +
Sbjct: 97 PAFHGTDNPDTYLEWEQKIELVFLCQECLQSNKVKIAATKFYNYALSWWDQLVTSRRRTR 156
Query: 101 NGAVVTW----AVFRREFLNRYFPEDVRGRKEIEFLELKQGDMSVVEYAAKFVELAKFYP 156
+ + TW V R+ F+ Y+ ++ R L QG +V EY F+E+
Sbjct: 157 DYPIKTWNQLKFVMRKRFVPSYYHRELHQR----LRNLVQGSKTVEEY---FLEMETLML 209
Query: 157 HYAPETAEFSKCIKFENGLRADIKRAIGYQQIRIFSDLVSRCRIYEED----------TK 206
+ + +F GL +I+ + Q +++ + ++E+ TK
Sbjct: 210 RADLQEDGEAVMSRFMGGLNREIQDRLETQHYVELEEMLHKAVMFEQQIKRKNARSSHTK 269
Query: 207 AHY---KVMSERRGK-GHQNRPKPYSA-------PTDKGKQRLNDERRPSKRDAPAEIVC 255
+Y K ++ K G+QN KP+ P KGK+ + R +I C
Sbjct: 270 TNYSSGKPSYQKEEKFGYQNDSKPFVKPKPVDLDPKGKGKEVITRAR---------DIRC 320
Query: 256 FKCGEKGHKSNVCT 269
FK H ++ C+
Sbjct: 321 FKSQGLRHYASKCS 334
>At4g36020 glycine-rich protein
Length = 299
Score = 55.1 bits (131), Expect = 7e-08
Identities = 36/134 (26%), Positives = 54/134 (39%), Gaps = 25/134 (18%)
Query: 216 RGKGHQNRPKPYSAPTDKGKQRLNDERRPSKRDAPAEIVCFKCGEKGHKSNVC------- 268
+G + + +AP ++ N+ R R C+ CGE GH S C
Sbjct: 63 QGSDGKTKAVNVTAPGGGSLKKENNSRGNGARRGGGGSGCYNCGELGHISKDCGIGGGGG 122
Query: 269 -----TKDEKKCFRCGQKGHMLADC------------KRGDVVCYNCNEEGHISTQCTQP 311
++ + C+ CG GH DC K G+ CY C + GH++ CTQ
Sbjct: 123 GGERRSRGGEGCYNCGDTGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDCTQ- 181
Query: 312 KKVRVGGKVFALTG 325
K V G + A+ G
Sbjct: 182 KSVGNGDQRGAVKG 195
Score = 52.8 bits (125), Expect = 3e-07
Identities = 23/65 (35%), Positives = 35/65 (53%), Gaps = 10/65 (15%)
Query: 255 CFKCGEKGHKSNVCT---KDEKKCFRCGQKGHMLADC-KRG------DVVCYNCNEEGHI 304
C+ CG GH + C + + C++CG GH+ DC +RG D CY C +EGH
Sbjct: 232 CYSCGGVGHIARDCATKRQPSRGCYQCGGSGHLARDCDQRGSGGGGNDNACYKCGKEGHF 291
Query: 305 STQCT 309
+ +C+
Sbjct: 292 ARECS 296
Score = 49.3 bits (116), Expect = 4e-06
Identities = 29/93 (31%), Positives = 39/93 (41%), Gaps = 29/93 (31%)
Query: 255 CFKCGEKGHKSNVCTK----DEKK--------CFRCGQKGHMLADC-------------- 288
C+ CG+ GH + CT D++ C+ CG GH+ DC
Sbjct: 134 CYNCGDTGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDCTQKSVGNGDQRGAV 193
Query: 289 KRGDVVCYNCNEEGHISTQCTQ---PKKVRVGG 318
K G+ CY C + GH + CTQ VR GG
Sbjct: 194 KGGNDGCYTCGDVGHFARDCTQKVAAGNVRSGG 226
Score = 45.4 bits (106), Expect = 5e-05
Identities = 21/71 (29%), Positives = 31/71 (43%), Gaps = 15/71 (21%)
Query: 255 CFKCGEKGHKSNVCTKD------------EKKCFRCGQKGHMLADC---KRGDVVCYNCN 299
C+ CG+ GH + CT+ C+ CG GH+ DC ++ CY C
Sbjct: 200 CYTCGDVGHFARDCTQKVAAGNVRSGGGGSGTCYSCGGVGHIARDCATKRQPSRGCYQCG 259
Query: 300 EEGHISTQCTQ 310
GH++ C Q
Sbjct: 260 GSGHLARDCDQ 270
Score = 44.7 bits (104), Expect = 9e-05
Identities = 23/85 (27%), Positives = 35/85 (41%), Gaps = 26/85 (30%)
Query: 255 CFKCGEKGHKSNVCTK------DEKK--------CFRCGQKGHMLADCKR---------- 290
C+ CG+ GH + CT+ D++ C+ CG GH DC +
Sbjct: 166 CYTCGDVGHVARDCTQKSVGNGDQRGAVKGGNDGCYTCGDVGHFARDCTQKVAAGNVRSG 225
Query: 291 --GDVVCYNCNEEGHISTQCTQPKK 313
G CY+C GHI+ C ++
Sbjct: 226 GGGSGTCYSCGGVGHIARDCATKRQ 250
>At1g75560 DNA-binding protein
Length = 257
Score = 54.7 bits (130), Expect = 9e-08
Identities = 25/71 (35%), Positives = 37/71 (51%), Gaps = 7/71 (9%)
Query: 248 DAPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDV------VCYNCNEE 301
+ AE C+ C E GH ++ C+ +E C CG+ GH DC D +C NC ++
Sbjct: 88 ECTAESRCWNCREPGHVASNCS-NEGICHSCGKSGHRARDCSNSDSRAGDLRLCNNCFKQ 146
Query: 302 GHISTQCTQPK 312
GH++ CT K
Sbjct: 147 GHLAADCTNDK 157
Score = 53.1 bits (126), Expect = 3e-07
Identities = 22/63 (34%), Positives = 33/63 (51%), Gaps = 7/63 (11%)
Query: 252 EIVCFKCGEKGHKSNVCTKDEKK------CFRCGQKGHMLADCKRGDVVCYNCNEEGHIS 305
E +C CG+ GH++ C+ + + C C ++GH+ ADC D C NC GHI+
Sbjct: 111 EGICHSCGKSGHRARDCSNSDSRAGDLRLCNNCFKQGHLAADCTN-DKACKNCRTSGHIA 169
Query: 306 TQC 308
C
Sbjct: 170 RDC 172
Score = 48.1 bits (113), Expect = 8e-06
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Query: 254 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQC 308
+C C ++GH + CT D K C C GH+ DC R D VC C+ GH++ C
Sbjct: 139 LCNNCFKQGHLAADCTND-KACKNCRTSGHIARDC-RNDPVCNICSISGHVARHC 191
Score = 47.8 bits (112), Expect = 1e-05
Identities = 27/105 (25%), Positives = 49/105 (45%), Gaps = 10/105 (9%)
Query: 205 TKAHYKVMSERRGKGHQNRPKPYSAPTDKGKQRLNDERRPSKRDAPAEIVCFKCGEKGHK 264
++ ++ S R + R + AP+ + ER P + + + C C GH
Sbjct: 15 SRDRFRSRSPRDRRMRSERVSYHDAPSRR-------EREPRRAFSQGNL-CNNCKRPGHF 66
Query: 265 SNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCT 309
+ C+ + C CG GH+ A+C + C+NC E GH+++ C+
Sbjct: 67 ARDCS-NVSVCNNCGLPGHIAAECT-AESRCWNCREPGHVASNCS 109
Score = 32.3 bits (72), Expect = 0.47
Identities = 20/94 (21%), Positives = 29/94 (30%), Gaps = 34/94 (36%)
Query: 248 DAPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGD--------------- 292
D + C C GH + C D C C GH+ C +GD
Sbjct: 152 DCTNDKACKNCRTSGHIARDCRNDPV-CNICSISGHVARHCPKGDSNYSDRGSRVRDGGM 210
Query: 293 ------------------VVCYNCNEEGHISTQC 308
++C+NC GH + +C
Sbjct: 211 QRGGLSRMSRDREGVSAMIICHNCGGRGHRAYEC 244
Score = 30.4 bits (67), Expect = 1.8
Identities = 18/52 (34%), Positives = 23/52 (43%), Gaps = 1/52 (1%)
Query: 218 KGHQNRPKPYSAPTDKGKQRLNDERRPSKRDA-PAEIVCFKCGEKGHKSNVC 268
KG N S D G QR R R+ A I+C CG +GH++ C
Sbjct: 193 KGDSNYSDRGSRVRDGGMQRGGLSRMSRDREGVSAMIICHNCGGRGHRAYEC 244
>At3g43590 putative protein
Length = 551
Score = 53.9 bits (128), Expect = 1e-07
Identities = 28/70 (40%), Positives = 35/70 (50%), Gaps = 6/70 (8%)
Query: 253 IVCFKCGEKGHKSNVC---TKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCT 309
+ C+ CGE+GH S C TK K CF CG H C +G CY C + GH + C
Sbjct: 166 VSCYSCGEQGHTSFNCPTPTKRRKPCFICGSLEHGAKQCSKGH-DCYICKKTGHRAKDC- 223
Query: 310 QPKKVRVGGK 319
P K + G K
Sbjct: 224 -PDKYKNGSK 232
Score = 50.8 bits (120), Expect = 1e-06
Identities = 21/93 (22%), Positives = 38/93 (40%), Gaps = 31/93 (33%)
Query: 253 IVCFKCGEKGHKSNVC--------------------TKDEKKCFRCGQKGHMLADC---- 288
+ C++CG+ GH C +++ +C+RCG++GH +C
Sbjct: 285 VSCYRCGQLGHSGLACGRHYEESNENDSATPERLFNSREASECYRCGEEGHFARECPNSS 344
Query: 289 -------KRGDVVCYNCNEEGHISTQCTQPKKV 314
+ +CY CN GH + +C +V
Sbjct: 345 SISTSHGRESQTLCYRCNGSGHFARECPNSSQV 377
Score = 37.4 bits (85), Expect = 0.015
Identities = 25/112 (22%), Positives = 33/112 (29%), Gaps = 51/112 (45%)
Query: 254 VCFKCGEKGHKSNVCTKDEKK-------------------------------CFRCGQKG 282
VC +CG+ GH +C + K C+RCGQ G
Sbjct: 235 VCLRCGDFGHDMILCKYEYSKEDLKDVQCYICKSFGHLCCVEPGNSLSWAVSCYRCGQLG 294
Query: 283 HMLADC--------------------KRGDVVCYNCNEEGHISTQCTQPKKV 314
H C R CY C EEGH + +C +
Sbjct: 295 HSGLACGRHYEESNENDSATPERLFNSREASECYRCGEEGHFARECPNSSSI 346
Score = 35.4 bits (80), Expect = 0.055
Identities = 17/70 (24%), Positives = 27/70 (38%), Gaps = 11/70 (15%)
Query: 255 CFKCGEKGHKSNVCT-----------KDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGH 303
C++CGE+GH + C + + C+RC GH +C V E
Sbjct: 327 CYRCGEEGHFARECPNSSSISTSHGRESQTLCYRCNGSGHFARECPNSSQVSKRDRETST 386
Query: 304 ISTQCTQPKK 313
S + + K
Sbjct: 387 TSHKSRKKNK 396
Score = 32.3 bits (72), Expect = 0.47
Identities = 13/25 (52%), Positives = 15/25 (60%)
Query: 291 GDVVCYNCNEEGHISTQCTQPKKVR 315
G V CY+C E+GH S C P K R
Sbjct: 164 GWVSCYSCGEQGHTSFNCPTPTKRR 188
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 53.1 bits (126), Expect = 3e-07
Identities = 69/278 (24%), Positives = 110/278 (38%), Gaps = 58/278 (20%)
Query: 93 SLLPVLEQNGAVVTWAVFRREFLNRYFPEDVRGRKEIEFLELKQGDMSVVEYAAKFVELA 152
S L ++EQ + T F EF +YFP++ R E FLEL +G++SV EY +F L
Sbjct: 142 SYLRMMEQLQRIGT--DFVAEFNAKYFPQEALDRMEARFLELTKGELSVREYGREFKRLL 199
Query: 153 KFYPHYAPETAEFSKCIKFENGLRADIKRAIGYQQIRIFSDLVSRCRIYEEDTKAHYKVM 212
+ + + ++ +F GLR D++ RCR+ + TKA +
Sbjct: 200 VYAGRGMED--DQAQMRRFLRGLRPDLR---------------VRCRVSQYATKA---AL 239
Query: 213 SERRGKGHQNRPKPYSAPTDKGKQRLNDERRPSKRDAPAEIV------CFKCGEKGHKSN 266
+ + R +P+ + K ++ + PSK PA++ F+ G+ G +
Sbjct: 240 TAAEVEEDLQRQVVAVSPSVQPK-KIQQQVAPSKGGKPAQVQKRKWDHPFRAGQGGRAGS 298
Query: 267 VCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKKVRVGGKVFALTGT 326
D+ G A R C +ST C +GT
Sbjct: 299 AA--DDSSTSTAGGAAQS-AGAARSAAEC------AAVSTYC---------------SGT 334
Query: 327 QTANEDRLIRGTCFFNSTPLIAIIDTGATHCFIALECA 364
LI G C + + D+GA+HCFI E A
Sbjct: 335 TWLYYADLISGRCRGH-----VLFDSGASHCFITPESA 367
>At2g17870 putative glycine-rich, zinc-finger DNA-binding protein
Length = 301
Score = 51.6 bits (122), Expect = 7e-07
Identities = 24/73 (32%), Positives = 33/73 (44%), Gaps = 18/73 (24%)
Query: 255 CFKCGEKGHKSNVCTKD--------EKKCFRCGQKGHMLADCKR----------GDVVCY 296
C+ CG GH + VCT + C+ CG GH+ DC R G C+
Sbjct: 226 CYTCGGVGHIAKVCTSKIPSGGGGGGRACYECGGTGHLARDCDRRGSGSSGGGGGSNKCF 285
Query: 297 NCNEEGHISTQCT 309
C +EGH + +CT
Sbjct: 286 ICGKEGHFARECT 298
Score = 48.5 bits (114), Expect = 6e-06
Identities = 24/81 (29%), Positives = 33/81 (40%), Gaps = 27/81 (33%)
Query: 255 CFKCGEKGHKSNVC----------------TKDEKKCFRCGQKGHMLADCKR-------- 290
CF CGE GH + C + E +C+ CG GH DC++
Sbjct: 96 CFNCGEVGHMAKDCDGGSGGKSFGGGGGRRSGGEGECYMCGDVGHFARDCRQSGGGNSGG 155
Query: 291 ---GDVVCYNCNEEGHISTQC 308
G CY+C E GH++ C
Sbjct: 156 GGGGGRPCYSCGEVGHLAKDC 176
Score = 45.4 bits (106), Expect = 5e-05
Identities = 26/87 (29%), Positives = 33/87 (37%), Gaps = 25/87 (28%)
Query: 255 CFKCGEKGHKSNVCTKDE---------------KKCFRCGQKGHMLADCKR--------G 291
C+ CGE GH + C C+ CG GH DC++ G
Sbjct: 163 CYSCGEVGHLAKDCRGGSGGNRYGGGGGRGSGGDGCYMCGGVGHFARDCRQNGGGNVGGG 222
Query: 292 DVVCYNCNEEGHISTQCTQPKKVRVGG 318
CY C GHI+ CT K+ GG
Sbjct: 223 GSTCYTCGGVGHIAKVCT--SKIPSGG 247
Score = 44.7 bits (104), Expect = 9e-05
Identities = 26/98 (26%), Positives = 37/98 (37%), Gaps = 26/98 (26%)
Query: 247 RDAPAEIVCFKCGEKGHKSNVCTKDE-----------KKCFRCGQKGHMLADCK------ 289
R + E C+ CG+ GH + C + + C+ CG+ GH+ DC+
Sbjct: 124 RRSGGEGECYMCGDVGHFARDCRQSGGGNSGGGGGGGRPCYSCGEVGHLAKDCRGGSGGN 183
Query: 290 ---------RGDVVCYNCNEEGHISTQCTQPKKVRVGG 318
G CY C GH + C Q VGG
Sbjct: 184 RYGGGGGRGSGGDGCYMCGGVGHFARDCRQNGGGNVGG 221
Score = 40.0 bits (92), Expect = 0.002
Identities = 22/76 (28%), Positives = 26/76 (33%), Gaps = 16/76 (21%)
Query: 259 GEKGHKSNVCTKDEKKCFRCGQKGHMLADC----------------KRGDVVCYNCNEEG 302
G K N CF CG+ GHM DC G+ CY C + G
Sbjct: 80 GSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKSFGGGGGRRSGGEGECYMCGDVG 139
Query: 303 HISTQCTQPKKVRVGG 318
H + C Q GG
Sbjct: 140 HFARDCRQSGGGNSGG 155
Score = 39.3 bits (90), Expect = 0.004
Identities = 21/80 (26%), Positives = 30/80 (37%), Gaps = 16/80 (20%)
Query: 255 CFKCGEKGHKSNVCTKDE--------KKCFRCGQKGHMLADCKR--------GDVVCYNC 298
C+ CG GH + C ++ C+ CG GH+ C G CY C
Sbjct: 198 CYMCGGVGHFARDCRQNGGGNVGGGGSTCYTCGGVGHIAKVCTSKIPSGGGGGGRACYEC 257
Query: 299 NEEGHISTQCTQPKKVRVGG 318
GH++ C + GG
Sbjct: 258 GGTGHLARDCDRRGSGSSGG 277
>At4g07850 putative polyprotein
Length = 1138
Score = 50.8 bits (120), Expect = 1e-06
Identities = 64/314 (20%), Positives = 110/314 (34%), Gaps = 35/314 (11%)
Query: 71 SEVQKVRFGTHMLAEEADDWWVSLLPVLEQNG--AVVTWAVFRREFLNRYFPEDVRGRKE 128
+E KV+ + A WW ++ + G ++ TW + R+ P
Sbjct: 91 TEANKVKVAATEFYDYALSWWDQVVTTKRRLGDDSIETWNQLKNIMKRRFVPSHYHRELH 150
Query: 129 IEFLELKQGDMSVVEYAAKFVELAKFYPHYAPETAEFSKCIKFENGLRADIKRAIGYQQI 188
L QG+ +V EY F E+ + + +F GL DI
Sbjct: 151 QRLRNLVQGNRTVEEY---FKEMETLMLRADVQEECEATMSRFMGGLNRDILDRFEVIHY 207
Query: 189 RIFSDLVSRCRIYEEDTKAHYKVMSERRGKGHQNRPKP-YSAPTDKGKQRLNDERRPSKR 247
+L + ++E+ K R K N KP Y G Q+ E +P +
Sbjct: 208 ENLEELFHKAVMFEKQIK-------RRSAKPSYNSSKPSYQREEKSGFQK---EYKPFVK 257
Query: 248 DAPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQ 307
EI KG + V + KCF+C GH ++C ++ + G + ++
Sbjct: 258 PKVEEI-----SSKGKEKEVTRTRDLKCFKCHGLGHYASECSNKRIMI--IRDSGEVESE 310
Query: 308 CTQPK----------KVRVGGKVFAL--TGTQTANEDRLIRGTCFFNSTPLIAIIDTGAT 355
+P+ ++ V +V ++ + A + L C IID G+
Sbjct: 311 DEKPEESDVEEAPKGELLVTMRVLSVLNKAEEQAQRENLFHTRCLIKGKVCSLIIDGGSC 370
Query: 356 HCFIALECAYKLGL 369
+ KLGL
Sbjct: 371 TNVASETMVQKLGL 384
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,120,712
Number of Sequences: 26719
Number of extensions: 403268
Number of successful extensions: 1551
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 1198
Number of HSP's gapped (non-prelim): 253
length of query: 396
length of database: 11,318,596
effective HSP length: 101
effective length of query: 295
effective length of database: 8,619,977
effective search space: 2542893215
effective search space used: 2542893215
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136954.7


