

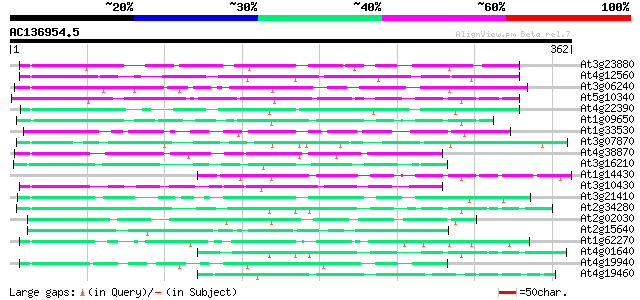
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.5 - phase: 0
(362 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g23880 unknown protein 82 6e-16
At4g12560 putative protein 80 2e-15
At3g06240 unknown protein 79 4e-15
At5g10340 putative protein 70 1e-12
At4g22390 unknown protein 67 2e-11
At1g09650 hypothetical protein 64 1e-10
At1g33530 hypothetical protein 63 2e-10
At3g07870 unknown protein 62 6e-10
At4g38870 putative protein 60 1e-09
At3g16210 hypothetical protein 59 3e-09
At1g14430 hypothetical protein 57 1e-08
At3g10430 hypothetical protein 57 2e-08
At3g21410 hypothetical protein 56 3e-08
At2g34280 hypothetical protein 53 2e-07
At2g02030 hypothetical protein 51 1e-06
At2g15640 hypothetical protein 50 2e-06
At1g62270 hypothetical protein 50 3e-06
At4g01640 hypothetical protein 48 7e-06
At4g19940 putative protein 47 2e-05
At4g19460 unknown protein 45 8e-05
>At3g23880 unknown protein
Length = 364
Score = 81.6 bits (200), Expect = 6e-16
Identities = 86/339 (25%), Positives = 139/339 (40%), Gaps = 67/339 (19%)
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMH---LKLSKSKGNLRLALF 63
LP+E++ EILLR V SL K V W +LIS+ +F H L+ SK+ + +
Sbjct: 14 LPLEMME-EILLRLPVKSLTRFKCVCSSWRSLISETLFALKHALILETSKATTSTKSPYG 72
Query: 64 SNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSC 123
R +++ + L ++T + D + D ++G+C
Sbjct: 73 VITTSRYHLKS-------------CCIHSLYNASTVYVSEHDG---ELLGRDYYQVVGTC 116
Query: 124 NGLICLHGCFHGSGYKKHSFCFWNPATRSK---SKTLLYVPSYLNRVRLGFGYDNSTDTY 180
+GL+C H + S Y WNP + + S + L V GFGYD S D Y
Sbjct: 117 HGLVCFHVDYDKSLY------LWNPTIKLQQRLSSSDLETSDDECVVTYGFGYDESEDDY 170
Query: 181 KTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFP--VELALRSRWDDIKYDGV 238
K V L K+++ +WR +SFP V +A +SR G+
Sbjct: 171 KVVAL-------LQQRHQVKIETKIYSTRQKLWRS-NTSFPSGVVVADKSR------SGI 216
Query: 239 YLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLP--------KLPFDNPN 290
Y++ +++W ++ + IIS D+ + + +L P + +
Sbjct: 217 YINGTLNWAATS---------SSSSWTIISYDMSRDEFKELPGPVCCGRGCFTMTLGDLR 267
Query: 291 ICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKI 329
C M C C K + +W MKEFG SW++ L I
Sbjct: 268 GCLSMVCYC-----KGANADVWVMKEFGEVYSWSKLLSI 301
>At4g12560 putative protein
Length = 408
Score = 79.7 bits (195), Expect = 2e-15
Identities = 88/341 (25%), Positives = 143/341 (41%), Gaps = 59/341 (17%)
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
+P++++ +I LR +L+ + +SKP LI+DP F++ HL G+ L +
Sbjct: 4 IPMDIVN-DIFLRLPAKTLVRCRALSKPCYHLINDPDFIESHLHRVLQTGD-HLMILLRG 61
Query: 67 NLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNGL 126
LRL Y+V + S+ D+++ + GS NGL
Sbjct: 62 ALRL----------YSVDL-------------DSLDSVSDVEHPMKRGGPTEVFGSSNGL 98
Query: 127 ICLHGCFHGSGYKKHSFCFWNPATRS------KSKTLLYVPSYLNRVRLGFGYDNSTDTY 180
I G +NP+TR S L S V G GYD+ +D Y
Sbjct: 99 I-------GLSNSPTDLAVFNPSTRQIHRLPPSSIDLPDGSSTRGYVFYGLGYDSVSDDY 151
Query: 181 KTV---MFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYD- 236
K V F I ++ LG + VKVF+L + W+ I+S L + + Y
Sbjct: 152 KVVRMVQFKIDSEDELGCSFPYE--VKVFSLKKNSWKRIESVASSIQLLFYFYYHLLYRR 209
Query: 237 --GVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPFDNPNICAL 294
GV NS+ W++ R NL I+ DL E + ++ P+ N N+
Sbjct: 210 GYGVLAGNSLHWVLPRRPGLIAFNL------IVRFDLALEEFEIVRFPEA-VANGNVDIQ 262
Query: 295 MD------CICFSYDFKETHFVIWQMKEFGVEESWTQFLKI 329
MD C+C ++ +++ +W MKE+ V +SWT+ +
Sbjct: 263 MDIGVLDGCLCLMCNYDQSYVDVWMMKEYNVRDSWTKVFTV 303
>At3g06240 unknown protein
Length = 427
Score = 79.0 bits (193), Expect = 4e-15
Identities = 96/367 (26%), Positives = 160/367 (43%), Gaps = 71/367 (19%)
Query: 4 SEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLR---- 59
S +LP E+IT EILLR S+ + VSK + TL SDP F K+HL L ++R
Sbjct: 33 SLVLPPEIIT-EILLRLPAKSIGRFRCVSKLFCTLSSDPGFAKIHLDLILRNESVRSLHR 91
Query: 60 -LALFSNKNLRLQIRAGGRGC--------SYTVTVAPTSVSLLLESTTSSIPIADDLQ-- 108
L + S+ L + G G +Y + P+ S ++ + + DD +
Sbjct: 92 KLIVSSHNLYSLDFNSIGDGIRDLAAVEHNYPLKDDPSIFSEMIRNYVGD-HLYDDRRVM 150
Query: 109 -------YQFSCVDCCGIIGSCNGLICLHGCFHGSGYKKHSFCFWNPAT-RSKSKTLLYV 160
Y+ + V+ I+GS NGL+C+ G G + +NP T SK +
Sbjct: 151 LKLNAKSYRRNWVE---IVGSSNGLVCIS---PGEG----AVFLYNPTTGDSKRLPENFR 200
Query: 161 PSYLNRVR-----LGFGYDNSTDTYKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRD 215
P + R GFG+D TD YK V T ++ L + V++L WR
Sbjct: 201 PKSVEYERDNFQTYGFGFDGLTDDYKLVKLVATSEDILDAS--------VYSLKADSWRR 252
Query: 216 IQSSFPVELALRSRWDDIKY-DGVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETE 274
I + L +D Y GV+ + +I W+ Q+ V+++ D++TE
Sbjct: 253 ICN-------LNYEHNDGSYTSGVHFNGAIHWVFTESRHNQR--------VVVAFDIQTE 297
Query: 275 TYTQLQLP------KLPFDNPNICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQF-L 327
+ ++ +P F N + +L +C + H IW M E+G +SW++ +
Sbjct: 298 EFREMPVPDEAEDCSHRFSNFVVGSLNGRLCVVNSCYDVHDDIWVMSEYGEAKSWSRIRI 357
Query: 328 KISYQNL 334
+ Y+++
Sbjct: 358 NLLYRSM 364
>At5g10340 putative protein
Length = 445
Score = 70.5 bits (171), Expect = 1e-12
Identities = 87/344 (25%), Positives = 146/344 (42%), Gaps = 49/344 (14%)
Query: 2 NISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHL--KLSKSKGNLR 59
++ E+LP ++I I++R DV +L+ K VSK W + I P F + L LS+S G+
Sbjct: 64 SMEELLPHDVIEYHIMVRLDVKTLLKFKSVSKQWMSTIQSPSFQERQLIHHLSQSSGDPH 123
Query: 60 LALFSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGI 119
+ L S + R +++ + L+ES+ +S+ I + + V C
Sbjct: 124 VLLVSLYD------PCARQQDPSISSFEALRTFLVESSAASVQIPTPWEDKLYFV--CNT 175
Query: 120 IGSCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNRVRL--------GF 171
SC+GLICL + + W+ T K L R GF
Sbjct: 176 --SCDGLICLFSFYELPSIVVNPTTRWH-RTFPKCNYQLVAADKGERHECFKVACPTPGF 232
Query: 172 GYDNSTDTYKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWD 231
G D + TYK V + + L + +VF + WR + + P L L ++
Sbjct: 233 GKDKISGTYKPVWLYNSAELDLND---KPTTCEVFDFATNAWRYVFPASP-HLILHTQ-- 286
Query: 232 DIKYDGVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPFDNPN- 290
D VY+ S+ W ++ + +++SLDL +ET+ + K PF N +
Sbjct: 287 ----DPVYVDGSLHWFTALSHE--------GETMVLSLDLHSETFQVIS--KAPFLNVSD 332
Query: 291 -----ICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKI 329
+C L D +C S + K + VIW + + ++W Q I
Sbjct: 333 EYYIVMCNLGDRLCVS-EQKWPNQVIWSLDD-SDHKTWKQIYSI 374
>At4g22390 unknown protein
Length = 401
Score = 66.6 bits (161), Expect = 2e-11
Identities = 78/334 (23%), Positives = 132/334 (39%), Gaps = 51/334 (15%)
Query: 8 PVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNKN 67
P +LI E+ LR +L+ + +SKP +LI P FV HL+ G + L
Sbjct: 5 PTDLIN-EMFLRLRATTLVKCRVLSKPCFSLIDSPEFVSSHLRRRLETGEHLMILLRGPR 63
Query: 68 LRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNGLI 127
L + +P +VS D+ + + GS NG+I
Sbjct: 64 LLRTVELD----------SPENVS--------------DIPHPLQAGGFTEVFGSFNGVI 99
Query: 128 CLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNR------VRLGFGYDNSTDTYK 181
G +NP+TR + + + R V G GYD+ D +K
Sbjct: 100 -------GLCNSPVDLAIFNPSTRKIHRLPIEPIDFPERDITREYVFYGLGYDSVGDDFK 152
Query: 182 TVMFGITMDEGLGGNRMRTAV-VKVFTLGDSIWRDIQSSFPVELALRSRWDDI---KYDG 237
V + G + V VKVF+L + W+ + F ++ S + + + G
Sbjct: 153 VVRI-VQCKLKEGKKKFPCPVEVKVFSLKKNSWKRVCLMFEFQILWISYYYHLLPRRGYG 211
Query: 238 VYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPF--DNPNICALM 295
V ++N + W++ R N II DL ++ L P+ + DN +I L
Sbjct: 212 VVVNNHLHWILPRRQGVIAFN------AIIKYDLASDDIGVLSFPQELYIEDNMDIGVLD 265
Query: 296 DCICFSYDFKETHFVIWQMKEFGVEESWTQFLKI 329
C+C + +H +W +KE+ +SWT+ ++
Sbjct: 266 GCVCLMCYDEYSHVDVWVLKEYEDYKSWTKLYRV 299
>At1g09650 hypothetical protein
Length = 382
Score = 64.3 bits (155), Expect = 1e-10
Identities = 89/327 (27%), Positives = 131/327 (39%), Gaps = 66/327 (20%)
Query: 5 EILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFS 64
E LP E++ IL R D + L+ K VSK W + I P F + + GN + + S
Sbjct: 11 ESLPHEVVEC-ILERLDADPLLRFKAVSKQWKSTIESPFFQRRQFTQRQQSGNPDVLMVS 69
Query: 65 N-KNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSC 123
++ +I A T V +S S+ + T +D +Y S SC
Sbjct: 70 RCADINSEIEA------LTTLVLGSSSSVKI-PTPWEEEEEEDKEYSVS-------QDSC 115
Query: 124 NGLICLHGCFHGSGYKKHSFCFWNPATR-SKSKTLLYVPSYLNRV------------RLG 170
+GL+CL F K F F NPATR L + L + RLG
Sbjct: 116 DGLVCLFDKF------KSGFVF-NPATRWYHPLPLCQLQQLLIAIGDGFYDLGYRVSRLG 168
Query: 171 FGYDNSTDTYKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRW 230
FG D T TYK V +++ GL +VF + WR + + P +
Sbjct: 169 FGKDKLTGTYKPVWLYNSIEIGL----ENATTCEVFDFSTNAWRYVSPAAPYRIVGCPA- 223
Query: 231 DDIKYDGVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPFDNPN 290
V + S+ W +C E+ I+S DL TET+ + K PF N +
Sbjct: 224 ------PVCVDGSLHWFT----EC-------EETKILSFDLHTETFQVVS--KAPFANVD 264
Query: 291 -----ICALMDCICFSYDFKETHFVIW 312
+C L + +C S + K + VIW
Sbjct: 265 GFDIVMCNLDNRLCVS-EMKLPNQVIW 290
>At1g33530 hypothetical protein
Length = 441
Score = 63.2 bits (152), Expect = 2e-10
Identities = 80/324 (24%), Positives = 137/324 (41%), Gaps = 68/324 (20%)
Query: 10 ELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNKNLR 69
+++ EIL R V L+ LK +SK W +LI + HL+L + K L K ++
Sbjct: 99 DVLVEEILQRLPVKYLVRLKSISKGWKSLIESDHLAEKHLRLLEKKYGL-------KEIK 151
Query: 70 LQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNGLICL 129
+ + R S ++ + S + + S +DDL + GSCNGL+C+
Sbjct: 152 ITVE---RSTSKSICIKFFSRRSGMNAINSD---SDDL---------LRVPGSCNGLVCV 196
Query: 130 HGCFHGSGYKKHSFCFW--NPATRSKSKTLLYVPSYLNRVRLGFGYDNSTDTYKTVMFGI 187
Y+ S + NP T T P ++ +GFG D T TYK ++
Sbjct: 197 --------YELDSVYIYLLNPMT---GVTRTLTPPRGTKLSVGFGIDVVTGTYKVMVL-- 243
Query: 188 TMDEGLGGNRMRTAVVKVFTLGDSIWRD-IQSSFPVELALRSRWDDIKYDGVYLSNSISW 246
G +R+ T VF L + WR +++ P+ L S + + V+++ S+ W
Sbjct: 244 -----YGFDRVGTV---VFDLDTNKWRQRYKTAGPMPL---SCIPTPERNPVFVNGSLFW 292
Query: 247 LVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPFDNPNIC-------ALMDCIC 299
L L ++ I+ +DL TE + L P D+ ++ +L D +C
Sbjct: 293 L-----------LASDFSEILVMDLHTEKFRTLSQPN-DMDDVDVSSGYIYMWSLEDRLC 340
Query: 300 FSYDFKETHFVIWQMKEFGVEESW 323
S + H +W + + + E W
Sbjct: 341 VSNVRQGLHSYVWVLVQDELSEKW 364
>At3g07870 unknown protein
Length = 417
Score = 61.6 bits (148), Expect = 6e-10
Identities = 93/393 (23%), Positives = 155/393 (38%), Gaps = 86/393 (21%)
Query: 5 EILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFS 64
E LP ++I +I R ++S+ L FV + W ++++ H +LS S + +
Sbjct: 26 ESLPEDIIA-DIFSRLPISSIARLMFVCRSWRSVLTQ------HGRLSSSSSSP-----T 73
Query: 65 NKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTT----SSIPIADDLQYQFSCVDCCGII 120
L L + R + + ++ + + T SS+P D ++
Sbjct: 74 KPCLLLHCDSPIRNGLHFLDLSEEEKRIKTKKFTLRFASSMPEFD-------------VV 120
Query: 121 GSCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNRVR-----LGFGYDN 175
GSCNGL+CL + S +NP T + L +P N+ GFG+
Sbjct: 121 GSCNGLLCLSDSLYND-----SLYLYNPFTTNS----LELPECSNKYHDQELVFGFGFHE 171
Query: 176 STDTYKTVMF----GITMD-----EGLGGNRMRTAVVKVFTLGDSI------WRDIQSSF 220
T YK + G + + G G + + + V++ TL WR + +
Sbjct: 172 MTKEYKVLKIVYFRGSSSNNNGIYRGRGRIQYKQSEVQILTLSSKTTDQSLSWRSLGKA- 230
Query: 221 PVELALRSRWDDIKYDGVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQ 280
P + RS S ++ H ++++ +FV S DLE E + ++
Sbjct: 231 PYKFVKRS------------SEALVNGRLHFVTRPRRHVPDRKFV--SFDLEDEEFKEIP 276
Query: 281 LPK---LPFDNPNICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKI-SYQNLGI 336
P L N + L C+C IW MK +GV+ESW + I +Y G+
Sbjct: 277 KPDCGGLNRTNHRLVNLKGCLCAVVYGNYGKLDIWVMKTYGVKESWGKEYSIGTYLPKGL 336
Query: 337 NYILGR---------NGFLVLPVCLSENGATLI 360
L R NG +V +CL ENG L+
Sbjct: 337 KQNLDRPMWIWKNAENGKVVRVLCLLENGEILL 369
>At4g38870 putative protein
Length = 426
Score = 60.5 bits (145), Expect = 1e-09
Identities = 74/282 (26%), Positives = 119/282 (41%), Gaps = 34/282 (12%)
Query: 4 SEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALF 63
SE+LPV+LI EIL + + L+ VSK W ++I DP F+K+ L S L
Sbjct: 50 SELLPVDLIM-EILKKLSLKPLIRFLCVSKLWASIIRDPYFMKLFLNES---------LK 99
Query: 64 SNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCV--DCCGIIG 121
K+L RA G ++ ++ + S++SS A + Y +C I
Sbjct: 100 RPKSLVFVFRAQSLGSIFSSVHLKSTREISSSSSSSS---ASSITYHVTCYTQQRMTISP 156
Query: 122 SCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNRVRLGFGYDNSTDTYK 181
S +GLIC S +NP TR +S TL + + + GYD YK
Sbjct: 157 SVHGLICYG--------PPSSLVIYNPCTR-RSITLPKIKAGRRAINQYIGYDPLDGNYK 207
Query: 182 TVMF--GITMDEGLGGNRMRTAVVKVFTLG--DSIWRDIQSSFPVELALRSRWDDIKYDG 237
V G+ M L R ++V TLG DS WR I P + +++ +G
Sbjct: 208 VVCITRGMPM---LRNRRGLAEEIQVLTLGTRDSSWRMIHDIIPPHSPVS---EELCING 261
Query: 238 VYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQL 279
V + + ++ +E+F +I + ++++L
Sbjct: 262 VLYYRAFIGTKLNESAIMSFDVRSEKFDLIKVPCNFRSFSKL 303
>At3g16210 hypothetical protein
Length = 360
Score = 59.3 bits (142), Expect = 3e-09
Identities = 71/284 (25%), Positives = 107/284 (37%), Gaps = 66/284 (23%)
Query: 3 ISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLAL 62
+S+ LP EL EIL+R + L + V K W LI+DP F + + +S +K
Sbjct: 1 MSKFLPEEL-AIEILVRLSMKDLARFRCVCKTWRDLINDPGFTETYRDMSPAK----FVS 55
Query: 63 FSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGS 122
F +KN + G P S++ EST +
Sbjct: 56 FYDKNFYMLDVEGKHPVITNKLDFPLDQSMIDEST---------------------CVLH 94
Query: 123 CNGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPS---YLNRVRLGFGYDNSTDT 179
C+G +C+ K H+ WNP SK VP+ Y + LGFGYD D
Sbjct: 95 CDGTLCV-------TLKNHTLMVWNPF----SKQFKIVPNPGIYQDSNILGFGYDPVHDD 143
Query: 180 YKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIW-RDIQSSFPVELALRSRWDDIKYDGV 238
YK V F +R+ + VF W ++ S+P W G
Sbjct: 144 YKVVTF---------IDRLDVSTAHVFEFRTGSWGESLRISYP-------DWHYRDRRGT 187
Query: 239 YLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLP 282
+L + W+ + ++F I+ +L T Y +L LP
Sbjct: 188 FLDQYLYWIAYRS--------SADRF-ILCFNLSTHEYRKLPLP 222
>At1g14430 hypothetical protein
Length = 849
Score = 57.4 bits (137), Expect = 1e-08
Identities = 66/257 (25%), Positives = 105/257 (40%), Gaps = 46/257 (17%)
Query: 122 SCNGLICLHGCFHGSGYKKHSFCFWN---PATRSKSKTLLYVPSYLNRV----RLGFGYD 174
SC+GLIC + + SG + W+ P R K L S + V LGFG D
Sbjct: 582 SCDGLICFYNLYK-SGIVVNPATRWHRSFPPPRYKQVILDKSSSEYSSVFPLPLLGFGKD 640
Query: 175 NSTDTYKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIK 234
TYK V +++ GL +VF ++W+ I S P +
Sbjct: 641 IINGTYKPVCLYNSLERGL----RNATTCEVFDFRTNVWKYITPSSPYRII-------AF 689
Query: 235 YDGVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPFDNPN---- 290
+D VY+ S+ W +C ++ ++SLDL TET+ + K+PFD N
Sbjct: 690 HDPVYVDGSLHWFT----EC-------KEIKVLSLDLHTETFQVIS--KVPFDIANCDNI 736
Query: 291 -ICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKISYQNLGINYILGRNGFLVLP 349
+C L + +C S + K V+W + W + + + G++ V P
Sbjct: 737 AMCNLDNRLCIS-ENKWPDQVVWSFD--SCNKKWHKMFTLDLNIFSHGF--GKHFLAVSP 791
Query: 350 VCLSE----NGATLIGN 362
V + E NG L+ +
Sbjct: 792 VAVLEKKKKNGKLLLSS 808
>At3g10430 hypothetical protein
Length = 370
Score = 57.0 bits (136), Expect = 2e-08
Identities = 72/279 (25%), Positives = 116/279 (40%), Gaps = 60/279 (21%)
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLIS-DPIFVKMHLKLSKSKGNLRLALFSN 65
LP +LI EIL R SL+ K K W LIS D F+ HL S +
Sbjct: 5 LPFDLIL-EILQRTPAESLLRFKSTCKKWYELISNDKRFMYKHLDKS-----------TK 52
Query: 66 KNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQ-FSCVDCCGIIGSCN 124
+ LR++ R V + +L ST I ++L+++ F+ + C G++
Sbjct: 53 RFLRIENRE-------RVQILDPVTEILAVST-----IPNELRHKYFTLIHCDGLM---- 96
Query: 125 GLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVP---SYLNRVRLGFGYDNS-TDTY 180
L C+ G + WNP R K K + P Y LGFGYD + D Y
Sbjct: 97 ----LGMCYEELG-SDPNLAVWNPVMR-KIKWIKPSPPLVCYWGSDYLGFGYDKTFRDNY 150
Query: 181 KTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDGVYL 240
K + F D+ + + +++ WR I++ F E+ D++ DGV +
Sbjct: 151 KILRFTYLGDDDDDESYPK---CQIYEFNSGSWRSIEAKFDGEI-------DVEVDGVSV 200
Query: 241 SNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQL 279
+ S+ W+ L ++ I+S D ET+ ++
Sbjct: 201 NGSMYWI----------ELQEKKNFILSFDFSKETFNRI 229
>At3g21410 hypothetical protein
Length = 374
Score = 55.8 bits (133), Expect = 3e-08
Identities = 75/339 (22%), Positives = 127/339 (37%), Gaps = 67/339 (19%)
Query: 6 ILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSN 65
+LP EL EIL R SL+ LK K W L +D F+ HL L + + +N
Sbjct: 10 LLPFELFE-EILCRVPTKSLLRLKLTCKRWLALFNDKRFIYKHLALVREH-----IIRTN 63
Query: 66 KNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNG 125
+ +++ G S+ S + + ++ ++ V C G+
Sbjct: 64 QMVKIINPVVGACSSF--------------SLPNKFQVKGEI---YTMVPCDGL------ 100
Query: 126 LICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNRVRLGFGYDN-STDTYKTVM 184
L+C+ ++ S WNP L PS+ G GYD S D+YK +
Sbjct: 101 LLCI--------FETGSMAVWNPCLNQVRWIFLLNPSFRGCSCYGIGYDGLSRDSYKILR 152
Query: 185 FGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDGVYLSNSI 244
F V ++ L + W+ + S + LR + G+ L ++
Sbjct: 153 F-------YANTGSYKPEVDIYELKSNSWKTFKVSLDWHVVLRCK-------GLSLKGNM 198
Query: 245 SWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPFDNPNICALM----DCICF 300
W+ K I S + TET+ L + +D N+ AL D +
Sbjct: 199 YWIAKWNRK--------PDIFIQSFNFSTETFEPLCSLPVRYDVHNVVALSAFKGDNLSL 250
Query: 301 SYDFKETHFV-IWQMKEF--GVEESWTQFLKISYQNLGI 336
+ KET + +W + GV WT+ ++ +L +
Sbjct: 251 LHQSKETSKIDVWVTNKVKNGVSILWTKLFSVTRPDLPV 289
>At2g34280 hypothetical protein
Length = 391
Score = 53.1 bits (126), Expect = 2e-07
Identities = 91/386 (23%), Positives = 144/386 (36%), Gaps = 90/386 (23%)
Query: 5 EILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFS 64
++LP +++ IL R DV SL+ K VSK W + I F + L + N + L S
Sbjct: 2 DLLPYDVVE-HILERLDVKSLLNCKSVSKQWRSTIRCRAFQERQLMHRRQSCNPDVLLVS 60
Query: 65 NKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCN 124
+ ++ + V + SV +L + + SC+
Sbjct: 61 VADEFYLLKRVHQVMRTLVLGSSVSVRILTPWEDTLYKVCQ---------------SSCD 105
Query: 125 GLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNR----------VRLGFGYD 174
GLICL+ + S NP TR + L L+R +LGFG D
Sbjct: 106 GLICLYNTY-------ASNIVVNPTTRWHREFPLSTFQRLDRYEHQVGSVSWAKLGFGKD 158
Query: 175 NSTDTYKTV-------MFGITMDEG-------------LGGNRMRTAVVKVFTLGDSIWR 214
TYK V + G+ D+ GN + +VF WR
Sbjct: 159 KINGTYKPVWLYNSAELGGLNDDDEDYDDEDKNNISMICQGNNNTSTFCEVFDFTTKAWR 218
Query: 215 DIQSSFPVELALRSRWDDIKY-DGVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLET 273
+ + P + + Y D VY+ S+ WL + TT ++S DL
Sbjct: 219 FVVPASPYRI--------LPYQDPVYVDGSLHWLT--------EGETTN---VLSFDLHA 259
Query: 274 ETYTQLQLPKLPFDNPN--------ICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQ 325
ET+ + K PF + + +C L D +C S VIW + ++W +
Sbjct: 260 ETFQVVS--KAPFLHHHDYSSRELIMCNLDDRLCVSEKMWPNQ-VIWSLD--SDHKTWKE 314
Query: 326 FLKISYQNLGI-NYILGRNGFLVLPV 350
I +L I + + G NGF + P+
Sbjct: 315 IYSI---DLNITSSLFGSNGFALRPL 337
>At2g02030 hypothetical protein
Length = 334
Score = 50.8 bits (120), Expect = 1e-06
Identities = 77/316 (24%), Positives = 117/316 (36%), Gaps = 60/316 (18%)
Query: 12 ITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNKNLRLQ 71
I EIL+R V SL + VSK W TLI+ F K H+ L KSKG L + +
Sbjct: 43 IVEEILVRLPVKSLTRFQTVSKHWRTLITSKYFGKRHMALEKSKGCKLLFVCDD----FV 98
Query: 72 IRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQF-SCVDCCGIIGSCNGLICLH 130
RA TV + TSVS D+ ++F I SC+GL+C +
Sbjct: 99 DRAEDTLFLKTVALEKTSVS-----------EGDEQAFEFEGYKGFLDISESCDGLVCFY 147
Query: 131 GCFHGSGY--------------KKHSFCFWNPATRSKSKTLLYVPSYLNRV----RLGFG 172
+ C + P + + + L+ V ++G G
Sbjct: 148 DTTRAVEVMNPATTMFIELPLSRIQQLCIYKPNPEVELEPVQDPNPVLDPVMTCSQIGVG 207
Query: 173 YDNSTDTYKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDD 232
D+ + +YK V T + +V L WR + ++ +L
Sbjct: 208 KDSVSGSYKLVWMYNT-------SPATPPTCEVLDLDGKKWRFVNTT-----SLDHHQIL 255
Query: 233 IKYDGVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPF------ 286
V+ + S+ WL ++ T Q +I LDL TE + +Q P PF
Sbjct: 256 CDQRPVFANGSLYWLT-----GDEEGYATTQTKLIVLDLHTEMFQVIQTP--PFITRDAS 308
Query: 287 -DNPNICALMDCICFS 301
D +C L +C S
Sbjct: 309 GDKIGLCNLDGRLCIS 324
>At2g15640 hypothetical protein
Length = 426
Score = 50.1 bits (118), Expect = 2e-06
Identities = 67/281 (23%), Positives = 106/281 (36%), Gaps = 37/281 (13%)
Query: 12 ITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNKNLRLQ 71
+T EIL R S+ VSK W ++ P F ++ L S +K L A+ N
Sbjct: 10 LTVEILSRLPAKSVARFHCVSKQWGSIFGSPYFKELFLTRSSTKPRLLFAMAEKVNEE-- 67
Query: 72 IRAGGRGCSYTVTVAP-------TSVSLLLESTTSSIPIADDLQYQFSCVDCCGI-IGSC 123
+ C + P S S L+ + + + D Y C D IG
Sbjct: 68 -----KNCVWRFFSTPQLENPYEKSSSTLVAAAEFHVKFSPDKLYICHCYDLKYFSIGYA 122
Query: 124 NGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNRVRLG-FGYDNSTDTYKT 182
+GLI L+G G C NP T + + Y R FG+D YK
Sbjct: 123 SGLIYLYG---DRGEATPLIC--NPTT---GRYAILPNRYTYRKAYSFFGFDPIDKQYKA 174
Query: 183 VMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDGVYLSN 242
+ I G G +++ T F G WR I LR DIK +G+ ++
Sbjct: 175 L--SIFYPSGPGHSKILT-----FGAGHMKWRKI------NCPLRYDRHDIKSEGICING 221
Query: 243 SISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPK 283
+ +L + + +VI+ D+ +E +T + + +
Sbjct: 222 VLYYLGSTSDCVKDGHGIVSDYVIVCFDIRSEKFTFIDVER 262
>At1g62270 hypothetical protein
Length = 383
Score = 49.7 bits (117), Expect = 3e-06
Identities = 83/353 (23%), Positives = 140/353 (39%), Gaps = 70/353 (19%)
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
LP +L+ +IL R SL L+ K WN L +D IF KMH ++ + F
Sbjct: 12 LPWDLVE-DILARVPATSLKRLRSTCKQWNFLFNDQIFTKMHFDKAEKQ-------FLVL 63
Query: 67 NLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVD--------CCG 118
LRL YTV S+SL L +I + +++ + S +D
Sbjct: 64 ILRL----------YTV----CSMSLDLRGLHDNIDPSIEVKGELSLIDPHCSSRKTFVS 109
Query: 119 IIGSCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLN-RVRLGFGYDNST 177
+ CNGL+ C +G WNP T VP N + LG+G S
Sbjct: 110 KVFHCNGLLL---CTTMTG-----LVVWNPCTDQTRWIKTEVPHNRNDKYALGYGNYKSC 161
Query: 178 DTYKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDG 237
YK + F +D + + ++++ + + WR + + P + + + +G
Sbjct: 162 YNYKIMKF---LD-------LESFDLEIYEVNSNSWRVLGTVTP-DFTI-----PLDAEG 205
Query: 238 VYLSNSISWLVCHRYK--CQQKNLTTEQFV---IISLDLETETY-TQLQLP---KLPFDN 288
V L + W+ H+ + +++ E F+ +IS D TE + ++ LP + +D
Sbjct: 206 VSLRGNSYWIASHKREEIEEEEEEENEYFINDFLISFDFTTERFGPRVSLPFKCESSWDT 265
Query: 289 PNICALMD--CICFSYDFKETHFVIWQMKEFGVEE----SWTQFLKISYQNLG 335
++ + + F D IW + SW+ FLKI G
Sbjct: 266 ISLSCVREERLSLFFQDDGTLKMEIWMTNNITETKTTTMSWSPFLKIDLYTYG 318
>At4g01640 hypothetical protein
Length = 300
Score = 48.1 bits (113), Expect = 7e-06
Identities = 66/273 (24%), Positives = 104/273 (37%), Gaps = 73/273 (26%)
Query: 122 SCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNR----------VRLGF 171
SC+GLICL+ + + NP TR + L L+R +LGF
Sbjct: 21 SCDGLICLYNTYAWN-------IVVNPTTRWHREFPLSTFQRLDRYEHQVGSVSWAKLGF 73
Query: 172 GYDNSTDTYKTV-------MFGITMDEG--------LGGNRMRTAVVKVFTLGDSIWRDI 216
G D TYK V + G+ D+ GN + +VF WR +
Sbjct: 74 GKDKINGTYKPVWLYNSAELGGLNDDDEDYDDISIICQGNNNTSTFCQVFDFTTKAWRFV 133
Query: 217 QSSFPVELALRSRWDDIKY-DGVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETET 275
+ P + + Y D VY+ S+ WL + ++S DL TET
Sbjct: 134 VPASPYRI--------LPYQDPVYVDGSLHWLTTN---------------VLSFDLHTET 170
Query: 276 YTQLQLPKLPFDNPN--------ICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFL 327
+ + K PF + + +C L D +C S VIW + ++W +
Sbjct: 171 FQVVS--KAPFLHHHDYSSRELIMCNLDDRLCVSEKMWPNQ-VIWSLD--SDHKTWKEIY 225
Query: 328 KISYQNLGI-NYILGRNGFLVLPVCLSENGATL 359
I +L I + + G NGF + P+ + + L
Sbjct: 226 SI---DLNITSSLFGSNGFALRPLAVFDKDKLL 255
>At4g19940 putative protein
Length = 411
Score = 47.0 bits (110), Expect = 2e-05
Identities = 75/291 (25%), Positives = 112/291 (37%), Gaps = 60/291 (20%)
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
+P +L+ EIL R SLM K VSK W++LI F LKLS S L + L S+
Sbjct: 35 IPFDLVI-EILTRLPAKSLMRFKSVSKLWSSLICSRNFTNRLLKLS-SPPRLFMCLSSSD 92
Query: 67 NLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQ------YQFSCVDCCGII 120
N L + +++ P S + T SS I DL YQ S V
Sbjct: 93 NSHL------KTVLLSLSSPPDS-----DITMSSSVIDQDLTMPGMKGYQISHV------ 135
Query: 121 GSCNGLICLHGCFHGSGYKKHSFCFWNPATRS-------KSKTLLYVPSYLNRVRLGFGY 173
GL+CL KK S +N TR + T+L ++ G+
Sbjct: 136 --FRGLMCL--------VKKSSAQIYNTTTRQLVVLPDIEESTILAEEHKSKKIMYHIGH 185
Query: 174 DNSTDTYKTVMFGITMDEGLGGNRMRTA--VVKVFTLGDSIWRDIQSSFPVELALRSRWD 231
D D YK V + + + V+ + G WR I +P + L
Sbjct: 186 DPVYDQYKVVCIVSRASDEVEEYTFLSEHWVLLLEGEGSRRWRKISCKYPPHVPLG---- 241
Query: 232 DIKYDGVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLP 282
G+ LS + +L R ++ V++ D +E ++ LQ+P
Sbjct: 242 ----QGLTLSGRMHYLAWVR--------VSDNRVLVIFDTHSEEFSMLQVP 280
>At4g19460 unknown protein
Length = 796
Score = 44.7 bits (104), Expect = 8e-05
Identities = 61/239 (25%), Positives = 91/239 (37%), Gaps = 37/239 (15%)
Query: 122 SCNGLICLHGCFHGSGYKKHSFCFW-NPATRSKSKTLL------YVPSYLNRVRLGFGYD 174
SC+GL+CL+ SGY + W P + L+ Y LGFG D
Sbjct: 566 SCDGLVCLYN-HDKSGYVVNPTTRWYRPLPLCDYQKLMIDLRDGYYKLKRKFFMLGFGKD 624
Query: 175 NSTDTYKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIK 234
TYK V + + GL +VF + WR + + P +A
Sbjct: 625 KFRGTYKPVWLYNSAEIGL----KNATTCEVFDFSTNAWRYVTLAAPYRIAAYPA----- 675
Query: 235 YDGVYLSNSISWLV-CHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPFDNPNICA 293
VYL S+ W C K NL TE F ++ + + K+ F N
Sbjct: 676 --PVYLDGSLYWFTECKETKVLPFNLHTETFQVVC----KAPFANVDPYKIVFYN----- 724
Query: 294 LMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKISYQNLGINYILGRNGFLVLPVCL 352
L + +C S + K + VIW ++W + I L +N++ G VLP+ L
Sbjct: 725 LDNRLCVS-EMKWPNQVIWSFN--SGNKTWDKLCSIV---LNLNFVTGI--CAVLPLAL 775
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.140 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,383,492
Number of Sequences: 26719
Number of extensions: 360050
Number of successful extensions: 1051
Number of sequences better than 10.0: 171
Number of HSP's better than 10.0 without gapping: 106
Number of HSP's successfully gapped in prelim test: 66
Number of HSP's that attempted gapping in prelim test: 883
Number of HSP's gapped (non-prelim): 204
length of query: 362
length of database: 11,318,596
effective HSP length: 101
effective length of query: 261
effective length of database: 8,619,977
effective search space: 2249813997
effective search space used: 2249813997
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136954.5


