

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.12 + phase: 0
(369 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
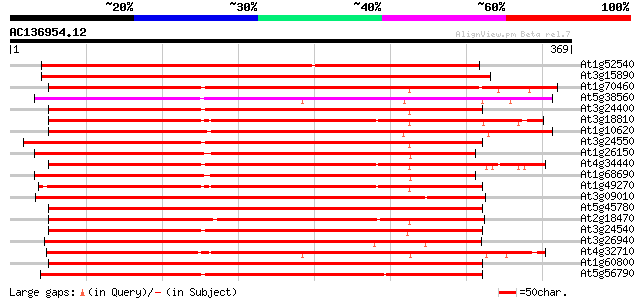
Score E
Sequences producing significant alignments: (bits) Value
At1g52540 unknown protein 335 2e-92
At3g15890 protein kinase, putative 322 3e-88
At1g70460 putative protein kinase 293 8e-80
At5g38560 putative protein 283 8e-77
At3g24400 protein kinase, putative 281 5e-76
At3g18810 protein kinase, putative 280 9e-76
At1g10620 putative serine/threonine protein kinase emb|CAA18823.1 279 2e-75
At3g24550 protein kinase, putative 277 6e-75
At1g26150 Pto kinase interactor, putative 273 8e-74
At4g34440 serine/threonine protein kinase - like 273 1e-73
At1g68690 protein kinase, putative 272 2e-73
At1g49270 hypothetical protein 271 3e-73
At3g09010 putative receptor ser/thr protein kinase 270 1e-72
At5g45780 receptor-like protein kinase 269 2e-72
At2g18470 putative protein kinase 269 2e-72
At3g24540 protein kinase, putative 268 5e-72
At3g26940 protein kinase, putative 267 6e-72
At4g32710 putative protein kinase 266 1e-71
At1g60800 Putative Serine/Threonine protein kinase 266 2e-71
At5g56790 putative protein kinase 264 7e-71
>At1g52540 unknown protein
Length = 350
Score = 335 bits (859), Expect = 2e-92
Identities = 163/288 (56%), Positives = 215/288 (74%), Gaps = 1/288 (0%)
Query: 22 SWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAV 81
SWRIF+ KELH ATN F+ D KLGEG FGSVYWG+ DG QIAVK+LKA +S+ E++FAV
Sbjct: 24 SWRIFSLKELHAATNSFNYDNKLGEGRFGSVYWGQLWDGSQIAVKRLKAWSSREEIDFAV 83
Query: 82 EVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSI 141
EVE+L R+RHKNLL +RGYC +RLIVYDYMPNLSL+SHLHGQ++ E L+W +RM+I
Sbjct: 84 EVEILARIRHKNLLSVRGYCAEGQERLIVYDYMPNLSLVSHLHGQHSSESLLDWTRRMNI 143
Query: 142 AIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVK 201
A+ SA+ I YLHH TP I+H D++ASNVLLDS+F V DFG+ KL+P+ ++ +T+
Sbjct: 144 AVSSAQAIAYLHHFATPRIVHGDVRASNVLLDSEFEARVTDFGYDKLMPDDGANKSTK-G 202
Query: 202 GTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK 261
+GYL+PE GK S+ DVYSFG+LLLELVTG++P E++ KR ITEW PL+ +
Sbjct: 203 NNIGYLSPECIESGKESDMGDVYSFGVLLLELVTGKRPTERVNLTTKRGITEWVLPLVYE 262
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLL 309
+F ++VD +L G + E ++K+ V V +C Q E EKRP M +VV +L
Sbjct: 263 RKFGEIVDQRLNGKYVEEELKRIVLVGLMCAQRESEKRPTMSEVVEML 310
>At3g15890 protein kinase, putative
Length = 361
Score = 322 bits (824), Expect = 3e-88
Identities = 155/296 (52%), Positives = 210/296 (70%), Gaps = 1/296 (0%)
Query: 22 SWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAV 81
SWR+F+ KELH ATN F+ D KLGEG FGSVYWG+ DG QIAVK+LK +++ E++FAV
Sbjct: 23 SWRVFSLKELHAATNSFNYDNKLGEGRFGSVYWGQLWDGSQIAVKRLKEWSNREEIDFAV 82
Query: 82 EVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSI 141
EVE+L R+RHKNLL +RGYC +RL+VY+YM NLSL+SHLHGQ++ E L+W KRM I
Sbjct: 83 EVEILARIRHKNLLSVRGYCAEGQERLLVYEYMQNLSLVSHLHGQHSAECLLDWTKRMKI 142
Query: 142 AIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIP-EGVSHMTTRV 200
AI SA+ I YLH TPHI+H D++ASNVLLDS+F V DFG+ KL+P + T+
Sbjct: 143 AISSAQAIAYLHDHATPHIVHGDVRASNVLLDSEFEARVTDFGYGKLMPDDDTGDGATKA 202
Query: 201 KGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLIT 260
K GY++PE GK SE+ DVYSFGILL+ LV+G++P+E+L R ITEW PL+
Sbjct: 203 KSNNGYISPECDASGKESETSDVYSFGILLMVLVSGKRPLERLNPTTTRCITEWVLPLVY 262
Query: 261 KGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQEPDQ 316
+ F ++VD +L ++K+ V V +C Q++P+KRP M +VV +L + ++
Sbjct: 263 ERNFGEIVDKRLSEEHVAEKLKKVVLVGLMCAQTDPDKRPTMSEVVEMLVNESKEK 318
>At1g70460 putative protein kinase
Length = 710
Score = 293 bits (751), Expect = 8e-80
Identities = 156/349 (44%), Positives = 222/349 (62%), Gaps = 17/349 (4%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
FTY+EL T GFS LGEGGFG VY G+ +DG +AVK+LK + + + EF EVE+
Sbjct: 341 FTYEELTDITEGFSKHNILGEGGFGCVYKGKLNDGKLVAVKQLKVGSGQGDREFKAEVEI 400
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+ RV H++L+ L GYC+ D +RL++Y+Y+PN +L HLHG+ G L W +R+ IAIGS
Sbjct: 401 ISRVHHRHLVSLVGYCIADSERLLIYEYVPNQTLEHHLHGK--GRPVLEWARRVRIAIGS 458
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
A+G+ YLH + P IIHRDIK++N+LLD +F VADFG AKL +H++TRV GT G
Sbjct: 459 AKGLAYLHEDCHPKIIHRDIKSANILLDDEFEAQVADFGLAKLNDSTQTHVSTRVMGTFG 518
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK---- 261
YLAPEYA GK+++ DV+SFG++LLEL+TGRKP+++ + ++ EWA PL+ K
Sbjct: 519 YLAPEYAQSGKLTDRSDVFSFGVVLLELITGRKPVDQYQPLGEESLVEWARPLLHKAIET 578
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQEPDQGKVT- 320
G F ++VD +L ++ EN+V + + AA CV+ KRP M QVV L E D G ++
Sbjct: 579 GDFSELVDRRLEKHYVENEVFRMIETAAACVRHSGPKRPRMVQVVRAL-DSEGDMGDISN 637
Query: 321 ------KMRIDSVKYNDELLALDQPS---DDDYDGNSSYGVFSAIDVQK 360
DS +YN++ + + + DD D G +S D +K
Sbjct: 638 GNKVGQSSAYDSGQYNNDTMKFRKMAFGFDDSSDSGMYSGDYSVQDSRK 686
>At5g38560 putative protein
Length = 681
Score = 283 bits (725), Expect = 8e-77
Identities = 157/360 (43%), Positives = 214/360 (58%), Gaps = 21/360 (5%)
Query: 17 GVANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAE 76
G+ +N F+Y EL T+GFS+ LGEGGFG VY G SDG ++AVK+LK S+ E
Sbjct: 318 GMVSNQRSWFSYDELSQVTSGFSEKNLLGEGGFGCVYKGVLSDGREVAVKQLKIGGSQGE 377
Query: 77 MEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQ 136
EF EVE++ RV H++L+ L GYC+ + RL+VYDY+PN +L HLH G + W+
Sbjct: 378 REFKAEVEIISRVHHRHLVTLVGYCISEQHRLLVYDYVPNNTLHYHLHA--PGRPVMTWE 435
Query: 137 KRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPE--GVS 194
R+ +A G+A GI YLH + P IIHRDIK+SN+LLD+ F LVADFG AK+ E +
Sbjct: 436 TRVRVAAGAARGIAYLHEDCHPRIIHRDIKSSNILLDNSFEALVADFGLAKIAQELDLNT 495
Query: 195 HMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEW 254
H++TRV GT GY+APEYA GK+SE DVYS+G++LLEL+TGRKP++ ++ EW
Sbjct: 496 HVSTRVMGTFGYMAPEYATSGKLSEKADVYSYGVILLELITGRKPVDTSQPLGDESLVEW 555
Query: 255 AEPL----ITKGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLL- 309
A PL I F ++VDP+L NF ++ + V AA CV+ KRP M QVV L
Sbjct: 556 ARPLLGQAIENEEFDELVDPRLGKNFIPGEMFRMVEAAAACVRHSAAKRPKMSQVVRALD 615
Query: 310 ---------KGQEPDQGKVTKMRIDSVK---YNDELLALDQPSDDDYDGNSSYGVFSAID 357
G P Q +V R S + + S D +D + S+ + + D
Sbjct: 616 TLEEATDITNGMRPGQSQVFDSRQQSAQIRMFQRMAFGSQDYSSDFFDRSQSHSSWGSRD 675
>At3g24400 protein kinase, putative
Length = 694
Score = 281 bits (718), Expect = 5e-76
Identities = 139/290 (47%), Positives = 196/290 (66%), Gaps = 6/290 (2%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
F Y+EL ATNGFS+ LG+GGFG V+ G +G ++AVK+LK +S+ E EF EV +
Sbjct: 319 FNYEELSRATNGFSEANLLGQGGFGYVFKGMLRNGKEVAVKQLKEGSSQGEREFQAEVGI 378
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+ RV H++L+ L GYC+ D QRL+VY+++PN +L HLHG+ G + W R+ IA+GS
Sbjct: 379 ISRVHHRHLVALVGYCIADAQRLLVYEFVPNNTLEFHLHGK--GRPTMEWSSRLKIAVGS 436
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
A+G+ YLH P IIHRDIKASN+L+D F VADFG AK+ + +H++TRV GT G
Sbjct: 437 AKGLSYLHENCNPKIIHRDIKASNILIDFKFEAKVADFGLAKIASDTNTHVSTRVMGTFG 496
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK---- 261
YLAPEYA GK++E DV+SFG++LLEL+TGR+PI+ ++ +WA PL+ +
Sbjct: 497 YLAPEYASSGKLTEKSDVFSFGVVLLELITGRRPIDVNNVHADNSLVDWARPLLNQVSEL 556
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKG 311
G F +VD KL +D+ ++ + V AA CV+S +RP M QV +L+G
Sbjct: 557 GNFEVVVDKKLNNEYDKEEMARMVACAAACVRSTAPRRPRMDQVARVLEG 606
>At3g18810 protein kinase, putative
Length = 700
Score = 280 bits (716), Expect = 9e-76
Identities = 151/349 (43%), Positives = 220/349 (62%), Gaps = 29/349 (8%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
FTY EL AT GFS LG+GGFG V+ G +G +IAVK LKA + + E EF EV++
Sbjct: 325 FTYDELAAATQGFSQSRLLGQGGFGYVHKGILPNGKEIAVKSLKAGSGQGEREFQAEVDI 384
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+ RV H+ L+ L GYC+ QR++VY+++PN +L HLHG+ +G+V L+W R+ IA+GS
Sbjct: 385 ISRVHHRFLVSLVGYCIAGGQRMLVYEFLPNDTLEFHLHGK-SGKV-LDWPTRLKIALGS 442
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
A+G+ YLH + P IIHRDIKASN+LLD F VADFG AKL + V+H++TR+ GT G
Sbjct: 443 AKGLAYLHEDCHPRIIHRDIKASNILLDESFEAKVADFGLAKLSQDNVTHVSTRIMGTFG 502
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK---- 261
YLAPEYA GK+++ DV+SFG++LLELVTGR+P++ L G ++ ++ +WA P+
Sbjct: 503 YLAPEYASSGKLTDRSDVFSFGVMLLELVTGRRPVD-LTGEMEDSLVDWARPICLNAAQD 561
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLK----------- 310
G + ++VDP+L ++ +++ Q V AA V+ +RP M Q+V L+
Sbjct: 562 GDYSELVDPRLENQYEPHEMAQMVACAAAAVRHSARRRPKMSQIVRALEGDATLDDLSEG 621
Query: 311 ---GQEPDQGKVTKMRIDSVKYNDEL-----LALDQPSDDDYDGNSSYG 351
GQ G+ + DS Y+ ++ +ALD +Y +S YG
Sbjct: 622 GKAGQSSFLGRGSSSDYDSSTYSADMKKFRKVALD---SHEYGASSEYG 667
>At1g10620 putative serine/threonine protein kinase emb|CAA18823.1
Length = 718
Score = 279 bits (713), Expect = 2e-75
Identities = 145/342 (42%), Positives = 220/342 (63%), Gaps = 12/342 (3%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
FTY+EL T GF + +GEGGFG VY G +G +A+K+LK+++++ EF EVE+
Sbjct: 358 FTYEELSQITEGFCKSFVVGEGGFGCVYKGILFEGKPVAIKQLKSVSAEGYREFKAEVEI 417
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+ RV H++L+ L GYC+ + R ++Y+++PN +L HLHG+ L W +R+ IAIG+
Sbjct: 418 ISRVHHRHLVSLVGYCISEQHRFLIYEFVPNNTLDYHLHGKNLPV--LEWSRRVRIAIGA 475
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
A+G+ YLH + P IIHRDIK+SN+LLD +F VADFG A+L SH++TRV GT G
Sbjct: 476 AKGLAYLHEDCHPKIIHRDIKSSNILLDDEFEAQVADFGLARLNDTAQSHISTRVMGTFG 535
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEP----LITK 261
YLAPEYA GK+++ DV+SFG++LLEL+TGRKP++ + ++ EWA P I K
Sbjct: 536 YLAPEYASSGKLTDRSDVFSFGVVLLELITGRKPVDTSQPLGEESLVEWARPRLIEAIEK 595
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQE-----PDQ 316
G ++VDP+L ++ E++V + + AA CV+ KRP M QVV L ++ +
Sbjct: 596 GDISEVVDPRLENDYVESEVYKMIETAASCVRHSALKRPRMVQVVRALDTRDDLSDLTNG 655
Query: 317 GKVTKMRI-DSVKYNDELLALDQPSDDDYDGNSSYGVFSAID 357
KV + R+ DS +Y++E+ + S+D D ++ G + + D
Sbjct: 656 VKVGQSRVYDSGQYSNEIRIFRRASEDSSDLGTNTGYYPSQD 697
>At3g24550 protein kinase, putative
Length = 652
Score = 277 bits (709), Expect = 6e-75
Identities = 142/307 (46%), Positives = 201/307 (65%), Gaps = 7/307 (2%)
Query: 10 VCRPTSYG-VANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKL 68
V P S G V S FTY+EL ATNGFS+ LG+GGFG V+ G G ++AVK+L
Sbjct: 251 VLPPPSPGLVLGFSKSTFTYEELSRATNGFSEANLLGQGGFGYVHKGILPSGKEVAVKQL 310
Query: 69 KAMNSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYA 128
KA + + E EF EVE++ RV H++L+ L GYC+ QRL+VY+++PN +L HLHG+
Sbjct: 311 KAGSGQGEREFQAEVEIISRVHHRHLVSLIGYCMAGVQRLLVYEFVPNNNLEFHLHGK-- 368
Query: 129 GEVQLNWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKL 188
G + W R+ IA+GSA+G+ YLH + P IIHRDIKASN+L+D F VADFG AK+
Sbjct: 369 GRPTMEWSTRLKIALGSAKGLSYLHEDCNPKIIHRDIKASNILIDFKFEAKVADFGLAKI 428
Query: 189 IPEGVSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLK 248
+ +H++TRV GT GYLAPEYA GK++E DV+SFG++LLEL+TGR+P++ +
Sbjct: 429 ASDTNTHVSTRVMGTFGYLAPEYAASGKLTEKSDVFSFGVVLLELITGRRPVDANNVYVD 488
Query: 249 RTITEWAEPLITK----GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQ 304
++ +WA PL+ + G F + D K+ +D ++ + V AA CV+ +RP M Q
Sbjct: 489 DSLVDWARPLLNRASEEGDFEGLADSKMGNEYDREEMARMVACAAACVRHSARRRPRMSQ 548
Query: 305 VVSLLKG 311
+V L+G
Sbjct: 549 IVRALEG 555
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 273 bits (699), Expect = 8e-74
Identities = 137/294 (46%), Positives = 193/294 (65%), Gaps = 8/294 (2%)
Query: 17 GVANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAE 76
G S +F+Y+EL ATNGFSD+ LGEGGFG VY G D +AVK+LK + +
Sbjct: 409 GGFGQSRELFSYEELVIATNGFSDENLLGEGGFGRVYKGVLPDERVVAVKQLKIGGGQGD 468
Query: 77 MEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQ 136
EF EV+ + RV H+NLL + GYC+ +++RL++YDY+PN +L HLHG L+W
Sbjct: 469 REFKAEVDTISRVHHRNLLSMVGYCISENRRLLIYDYVPNNNLYFHLHGTPG----LDWA 524
Query: 137 KRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHM 196
R+ IA G+A G+ YLH + P IIHRDIK+SN+LL+++F LV+DFG AKL + +H+
Sbjct: 525 TRVKIAAGAARGLAYLHEDCHPRIIHRDIKSSNILLENNFHALVSDFGLAKLALDCNTHI 584
Query: 197 TTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAE 256
TTRV GT GY+APEYA GK++E DV+SFG++LLEL+TGRKP++ ++ EWA
Sbjct: 585 TTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVVLLELITGRKPVDASQPLGDESLVEWAR 644
Query: 257 PLITKG----RFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVV 306
PL++ F + DPKL N+ ++ + + AA C++ KRP M Q+V
Sbjct: 645 PLLSNATETEEFTALADPKLGRNYVGVEMFRMIEAAAACIRHSATKRPRMSQIV 698
>At4g34440 serine/threonine protein kinase - like
Length = 670
Score = 273 bits (698), Expect = 1e-73
Identities = 149/354 (42%), Positives = 213/354 (60%), Gaps = 31/354 (8%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
FTY EL AT GF+ LG+GGFG V+ G G ++AVK LK + + E EF EV++
Sbjct: 300 FTYDELSIATEGFAQSNLLGQGGFGYVHKGVLPSGKEVAVKSLKLGSGQGEREFQAEVDI 359
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+ RV H++L+ L GYC+ QRL+VY+++PN +L HLHG+ G L+W R+ IA+GS
Sbjct: 360 ISRVHHRHLVSLVGYCISGGQRLLVYEFIPNNTLEFHLHGK--GRPVLDWPTRVKIALGS 417
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
A G+ YLH + P IIHRDIKA+N+LLD F VADFG AKL + +H++TRV GT G
Sbjct: 418 ARGLAYLHEDCHPRIIHRDIKAANILLDFSFETKVADFGLAKLSQDNYTHVSTRVMGTFG 477
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK---- 261
YLAPEYA GK+S+ DV+SFG++LLEL+TGR P++ L G ++ ++ +WA PL K
Sbjct: 478 YLAPEYASSGKLSDKSDVFSFGVMLLELITGRPPLD-LTGEMEDSLVDWARPLCLKAAQD 536
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQ--------- 312
G + + DP+L N+ ++ Q + AA ++ +RP M Q+V L+G
Sbjct: 537 GDYNQLADPRLELNYSHQEMVQMASCAAAAIRHSARRRPKMSQIVRALEGDMSMDDLSEG 596
Query: 313 -EPDQ------GKVTKMRIDSVKYNDEL-----LALD--QPSDDDYDGNSSYGV 352
P Q G V+ D+ Y ++ LAL+ + +Y G S YG+
Sbjct: 597 TRPGQSTYLSPGSVSS-EYDASSYTADMKKFKKLALENKEYQSSEYGGTSEYGL 649
>At1g68690 protein kinase, putative
Length = 708
Score = 272 bits (696), Expect = 2e-73
Identities = 137/294 (46%), Positives = 193/294 (65%), Gaps = 7/294 (2%)
Query: 17 GVANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAE 76
G NS +F+Y+EL ATNGFS + LGEGGFG VY G DG +AVK+LK + +
Sbjct: 356 GGLGNSKALFSYEELVKATNGFSQENLLGEGGFGCVYKGILPDGRVVAVKQLKIGGGQGD 415
Query: 77 MEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQ 136
EF EVE L R+ H++L+ + G+C+ D+RL++YDY+ N L HLHG+ + L+W
Sbjct: 416 REFKAEVETLSRIHHRHLVSIVGHCISGDRRLLIYDYVSNNDLYFHLHGEKS---VLDWA 472
Query: 137 KRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHM 196
R+ IA G+A G+ YLH + P IIHRDIK+SN+LL+ +F V+DFG A+L + +H+
Sbjct: 473 TRVKIAAGAARGLAYLHEDCHPRIIHRDIKSSNILLEDNFDARVSDFGLARLALDCNTHI 532
Query: 197 TTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAE 256
TTRV GT GY+APEYA GK++E DV+SFG++LLEL+TGRKP++ ++ EWA
Sbjct: 533 TTRVIGTFGYMAPEYASSGKLTEKSDVFSFGVVLLELITGRKPVDTSQPLGDESLVEWAR 592
Query: 257 PLITKG----RFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVV 306
PLI+ F + DPKL GN+ E+++ + + A CV+ KRP M Q+V
Sbjct: 593 PLISHAIETEEFDSLADPKLGGNYVESEMFRMIEAAGACVRHLATKRPRMGQIV 646
>At1g49270 hypothetical protein
Length = 699
Score = 271 bits (694), Expect = 3e-73
Identities = 144/297 (48%), Positives = 203/297 (67%), Gaps = 10/297 (3%)
Query: 20 NNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEF 79
NNS FTY+EL +AT GFS D LG+GGFG V+ G +G +IAVK LKA + + E EF
Sbjct: 320 NNS--TFTYEELASATQGFSKDRLLGQGGFGYVHKGILPNGKEIAVKSLKAGSGQGEREF 377
Query: 80 AVEVEVLGRVRHKNLLGLRGYCVG-DDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKR 138
EVE++ RV H++L+ L GYC QRL+VY+++PN +L HLHG+ +G V ++W R
Sbjct: 378 QAEVEIISRVHHRHLVSLVGYCSNAGGQRLLVYEFLPNDTLEFHLHGK-SGTV-MDWPTR 435
Query: 139 MSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTT 198
+ IA+GSA+G+ YLH + P IIHRDIKASN+LLD +F VADFG AKL + +H++T
Sbjct: 436 LKIALGSAKGLAYLHEDCHPKIIHRDIKASNILLDHNFEAKVADFGLAKLSQDNNTHVST 495
Query: 199 RVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPL 258
RV GT GYLAPEYA GK++E DV+SFG++LLEL+TGR P++ L G ++ ++ +WA PL
Sbjct: 496 RVMGTFGYLAPEYASSGKLTEKSDVFSFGVMLLELITGRGPVD-LSGDMEDSLVDWARPL 554
Query: 259 ITK----GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKG 311
+ G + ++VDP L ++ ++ + V AA V+ +RP M Q+V L+G
Sbjct: 555 CMRVAQDGEYGELVDPFLEHQYEPYEMARMVACAAAAVRHSGRRRPKMSQIVRTLEG 611
>At3g09010 putative receptor ser/thr protein kinase
Length = 393
Score = 270 bits (689), Expect = 1e-72
Identities = 129/296 (43%), Positives = 193/296 (64%), Gaps = 1/296 (0%)
Query: 18 VANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEM 77
+ N+ R+F+Y L +AT+ F ++G GG+G V+ G DG Q+AVK L A + +
Sbjct: 26 ICTNNVRVFSYNSLRSATDSFHPTNRIGGGGYGVVFKGVLRDGTQVAVKSLSAESKQGTR 85
Query: 78 EFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQK 137
EF E+ ++ + H NL+ L G C+ + R++VY+Y+ N SL S L G + V L+W K
Sbjct: 86 EFLTEINLISNIHHPNLVKLIGCCIEGNNRILVYEYLENNSLASVLLGSRSRYVPLDWSK 145
Query: 138 RMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMT 197
R +I +G+A G+ +LH EV PH++HRDIKASN+LLDS+F P + DFG AKL P+ V+H++
Sbjct: 146 RAAICVGTASGLAFLHEEVEPHVVHRDIKASNILLDSNFSPKIGDFGLAKLFPDNVTHVS 205
Query: 198 TRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEP 257
TRV GT+GYLAPEYA+ G++++ DVYSFGIL+LE+++G G + EW
Sbjct: 206 TRVAGTVGYLAPEYALLGQLTKKADVYSFGILVLEVISGNSSTRAAFGDEYMVLVEWVWK 265
Query: 258 LITKGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQE 313
L + R + VDP+L F ++V + + VA C Q+ +KRPNMKQV+ +L+ +E
Sbjct: 266 LREERRLLECVDPELT-KFPADEVTRFIKVALFCTQAAAQKRPNMKQVMEMLRRKE 320
>At5g45780 receptor-like protein kinase
Length = 570
Score = 269 bits (688), Expect = 2e-72
Identities = 130/287 (45%), Positives = 192/287 (66%), Gaps = 1/287 (0%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
F+++E+ TAT+ FS LG+GGFG VY G +G +AVK+LK E++F EVE+
Sbjct: 244 FSFREIQTATSNFSPKNILGQGGFGMVYKGYLPNGTVVAVKRLKDPIYTGEVQFQTEVEM 303
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+G H+NLL L G+C+ ++R++VY YMPN S+ L Y + L+W +R+SIA+G+
Sbjct: 304 IGLAVHRNLLRLFGFCMTPEERMLVYPYMPNGSVADRLRDNYGEKPSLDWNRRISIALGA 363
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
A G++YLH + P IIHRD+KA+N+LLD F +V DFG AKL+ + SH+TT V+GT+G
Sbjct: 364 ARGLVYLHEQCNPKIIHRDVKAANILLDESFEAIVGDFGLAKLLDQRDSHVTTAVRGTIG 423
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKR-TITEWAEPLITKGRF 264
++APEY G+ SE DV+ FG+L+LEL+TG K I++ G +++ I W L + RF
Sbjct: 424 HIAPEYLSTGQSSEKTDVFGFGVLILELITGHKMIDQGNGQVRKGMILSWVRTLKAEKRF 483
Query: 265 RDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKG 311
+MVD L+G FD+ +++ V +A LC Q P RP M QV+ +L+G
Sbjct: 484 AEMVDRDLKGEFDDLVLEEVVELALLCTQPHPNLRPRMSQVLKVLEG 530
>At2g18470 putative protein kinase
Length = 633
Score = 269 bits (687), Expect = 2e-72
Identities = 130/291 (44%), Positives = 198/291 (67%), Gaps = 7/291 (2%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
FTY+EL AT GF+D LG+GGFG V+ G G ++AVK LKA + + E EF EV++
Sbjct: 272 FTYQELAAATGGFTDANLLGQGGFGYVHKGVLPSGKEVAVKSLKAGSGQGEREFQAEVDI 331
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+ RV H+ L+ L GYC+ D QR++VY+++PN +L HLHG+ ++ + R+ IA+G+
Sbjct: 332 ISRVHHRYLVSLVGYCIADGQRMLVYEFVPNKTLEYHLHGKNLPVMEFS--TRLRIALGA 389
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
A+G+ YLH + P IIHRDIK++N+LLD +F +VADFG AKL + +H++TRV GT G
Sbjct: 390 AKGLAYLHEDCHPRIIHRDIKSANILLDFNFDAMVADFGLAKLTSDNNTHVSTRVMGTFG 449
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK---- 261
YLAPEYA GK++E DV+S+G++LLEL+TG++P++ + T+ +WA PL+ +
Sbjct: 450 YLAPEYASSGKLTEKSDVFSYGVMLLELITGKRPVDN-SITMDDTLVDWARPLMARALED 508
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQ 312
G F ++ D +L GN++ ++ + V AA ++ KRP M Q+V L+G+
Sbjct: 509 GNFNELADARLEGNYNPQEMARMVTCAAASIRHSGRKRPKMSQIVRALEGE 559
>At3g24540 protein kinase, putative
Length = 509
Score = 268 bits (684), Expect = 5e-72
Identities = 133/290 (45%), Positives = 191/290 (65%), Gaps = 6/290 (2%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 85
FTY EL ATN FS+ LGEGGFG VY G ++G ++AVK+LK +++ E EF EV +
Sbjct: 167 FTYGELARATNKFSEANLLGEGGFGFVYKGILNNGNEVAVKQLKVGSAQGEKEFQAEVNI 226
Query: 86 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGS 145
+ ++ H+NL+ L GYC+ QRL+VY+++PN +L HLHG+ G + W R+ IA+ S
Sbjct: 227 ISQIHHRNLVSLVGYCIAGAQRLLVYEFVPNNTLEFHLHGK--GRPTMEWSLRLKIAVSS 284
Query: 146 AEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLG 205
++G+ YLH P IIHRDIKA+N+L+D F VADFG AK+ + +H++TRV GT G
Sbjct: 285 SKGLSYLHENCNPKIIHRDIKAANILIDFKFEAKVADFGLAKIALDTNTHVSTRVMGTFG 344
Query: 206 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLIT----K 261
YLAPEYA GK++E DVYSFG++LLEL+TGR+P++ ++ +WA PL+ +
Sbjct: 345 YLAPEYAASGKLTEKSDVYSFGVVLLELITGRRPVDANNVYADDSLVDWARPLLVQALEE 404
Query: 262 GRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKG 311
F + D KL +D ++ + V AA CV+ +RP M QVV +L+G
Sbjct: 405 SNFEGLADIKLNNEYDREEMARMVACAAACVRYTARRRPRMDQVVRVLEG 454
>At3g26940 protein kinase, putative
Length = 432
Score = 267 bits (683), Expect = 6e-72
Identities = 137/293 (46%), Positives = 192/293 (64%), Gaps = 6/293 (2%)
Query: 24 RIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEV 83
+IF+Y+EL ATN F ++ +G GGFG+VY GR S G IAVK L + + EF VEV
Sbjct: 60 QIFSYRELAIATNSFRNESLIGRGGFGTVYKGRLSTGQNIAVKMLDQSGIQGDKEFLVEV 119
Query: 84 EVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAI 143
+L + H+NL+ L GYC DQRL+VY+YMP S+ HL+ G+ L+W+ RM IA+
Sbjct: 120 LMLSLLHHRNLVHLFGYCAEGDQRLVVYEYMPLGSVEDHLYDLSEGQEALDWKTRMKIAL 179
Query: 144 GSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIP-EGVSHMTTRVKG 202
G+A+G+ +LH+E P +I+RD+K SN+LLD D+ P ++DFG AK P + +SH++TRV G
Sbjct: 180 GAAKGLAFLHNEAQPPVIYRDLKTSNILLDHDYKPKLSDFGLAKFGPSDDMSHVSTRVMG 239
Query: 203 TLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRK---PIEKLPGGLKRTITEWAEPLI 259
T GY APEYA GK++ D+YSFG++LLEL++GRK P + G R + WA PL
Sbjct: 240 THGYCAPEYANTGKLTLKSDIYSFGVVLLELISGRKALMPSSECVGNQSRYLVHWARPLF 299
Query: 260 TKGRFRDMVDPKL--RGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLK 310
GR R +VDP+L +G F + + + VA LC+ E RP++ QVV LK
Sbjct: 300 LNGRIRQIVDPRLARKGGFSNILLYRGIEVAFLCLAEEANARPSISQVVECLK 352
>At4g32710 putative protein kinase
Length = 731
Score = 266 bits (680), Expect = 1e-71
Identities = 149/346 (43%), Positives = 209/346 (60%), Gaps = 22/346 (6%)
Query: 25 IFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVE 84
+F+Y+EL AT GFS++ LGEGGFG V+ G +G ++AVK+LK + + E EF EV+
Sbjct: 376 MFSYEELSKATGGFSEENLLGEGGFGYVHKGVLKNGTEVAVKQLKIGSYQGEREFQAEVD 435
Query: 85 VLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIG 144
+ RV HK+L+ L GYCV D+RL+VY+++P +L HLH + G V L W+ R+ IA+G
Sbjct: 436 TISRVHHKHLVSLVGYCVNGDKRLLVYEFVPKDTLEFHLH-ENRGSV-LEWEMRLRIAVG 493
Query: 145 SAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPE---GVSHMTTRVK 201
+A+G+ YLH + +P IIHRDIKA+N+LLDS F V+DFG AK + +H++TRV
Sbjct: 494 AAKGLAYLHEDCSPTIIHRDIKAANILLDSKFEAKVSDFGLAKFFSDTNSSFTHISTRVV 553
Query: 202 GTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK 261
GT GY+APEYA GKV++ DVYSFG++LLEL+TGR I +++ +WA PL+TK
Sbjct: 554 GTFGYMAPEYASSGKVTDKSDVYSFGVVLLELITGRPSIFAKDSSTNQSLVDWARPLLTK 613
Query: 262 G----RFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQ----- 312
F +VD +L N+D Q+ AA C++ RP M QVV L+G+
Sbjct: 614 AISGESFDFLVDSRLEKNYDTTQMANMAACAAACIRQSAWLRPRMSQVVRALEGEVALRK 673
Query: 313 -EPDQGKVTKMRID-----SVKYNDELLALDQPSDDDYDGNSSYGV 352
E VT + + +Y D S D Y S YGV
Sbjct: 674 VEETGNSVTYSSSENPNDITPRYGTNKRRFDTGSSDGY--TSEYGV 717
>At1g60800 Putative Serine/Threonine protein kinase
Length = 632
Score = 266 bits (679), Expect = 2e-71
Identities = 130/288 (45%), Positives = 188/288 (65%), Gaps = 2/288 (0%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMN-SKAEMEFAVEVE 84
+T+KEL +ATN F+ LG GG+G VY G +DG +AVK+LK N + E++F EVE
Sbjct: 289 YTFKELRSATNHFNSKNILGRGGYGIVYKGHLNDGTLVAVKRLKDCNIAGGEVQFQTEVE 348
Query: 85 VLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIG 144
+ H+NLL LRG+C + +R++VY YMPN S+ S L GE L+W +R IA+G
Sbjct: 349 TISLALHRNLLRLRGFCSSNQERILVYPYMPNGSVASRLKDNIRGEPALDWSRRKKIAVG 408
Query: 145 SAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTL 204
+A G++YLH + P IIHRD+KA+N+LLD DF +V DFG AKL+ SH+TT V+GT+
Sbjct: 409 TARGLVYLHEQCDPKIIHRDVKAANILLDEDFEAVVGDFGLAKLLDHRDSHVTTAVRGTV 468
Query: 205 GYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIE-KLPGGLKRTITEWAEPLITKGR 263
G++APEY G+ SE DV+ FGILLLEL+TG+K ++ K + +W + L +G+
Sbjct: 469 GHIAPEYLSTGQSSEKTDVFGFGILLLELITGQKALDFGRSAHQKGVMLDWVKKLHQEGK 528
Query: 264 FRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKG 311
+ ++D L FD ++++ V VA LC Q P RP M +V+ +L+G
Sbjct: 529 LKQLIDKDLNDKFDRVELEEIVQVALLCTQFNPSHRPKMSEVMKMLEG 576
>At5g56790 putative protein kinase
Length = 669
Score = 264 bits (674), Expect = 7e-71
Identities = 141/293 (48%), Positives = 191/293 (65%), Gaps = 5/293 (1%)
Query: 21 NSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFA 80
N R FTY EL TAT GFS L EGGFGSV+ G DG IAVK+ K +++ + EF
Sbjct: 373 NPPRWFTYSELETATKGFSKGSFLAEGGFGSVHLGTLPDGQIIAVKQYKIASTQGDREFC 432
Query: 81 VEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMS 140
EVEVL +H+N++ L G CV D +RL+VY+Y+ N SL SHL+G G L W R
Sbjct: 433 SEVEVLSCAQHRNVVMLIGLCVEDGKRLLVYEYICNGSLHSHLYGM--GREPLGWSARQK 490
Query: 141 IAIGSAEGILYLHHEVTPH-IIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTR 199
IA+G+A G+ YLH E I+HRD++ +N+LL DF PLV DFG A+ PEG + TR
Sbjct: 491 IAVGAARGLRYLHEECRVGCIVHRDMRPNNILLTHDFEPLVGDFGLARWQPEGDKGVETR 550
Query: 200 VKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIE-KLPGGLKRTITEWAEPL 258
V GT GYLAPEYA G+++E DVYSFG++L+EL+TGRK ++ K P G ++ +TEWA PL
Sbjct: 551 VIGTFGYLAPEYAQSGQITEKADVYSFGVVLVELITGRKAMDIKRPKG-QQCLTEWARPL 609
Query: 259 ITKGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKG 311
+ K +++DP+L + E +V A LC++ +P RP M QV+ +L+G
Sbjct: 610 LQKQAINELLDPRLMNCYCEQEVYCMALCAYLCIRRDPNSRPRMSQVLRMLEG 662
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,678,465
Number of Sequences: 26719
Number of extensions: 379977
Number of successful extensions: 4164
Number of sequences better than 10.0: 998
Number of HSP's better than 10.0 without gapping: 876
Number of HSP's successfully gapped in prelim test: 122
Number of HSP's that attempted gapping in prelim test: 932
Number of HSP's gapped (non-prelim): 1116
length of query: 369
length of database: 11,318,596
effective HSP length: 101
effective length of query: 268
effective length of database: 8,619,977
effective search space: 2310153836
effective search space used: 2310153836
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136954.12


