

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136953.9 - phase: 0
(567 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
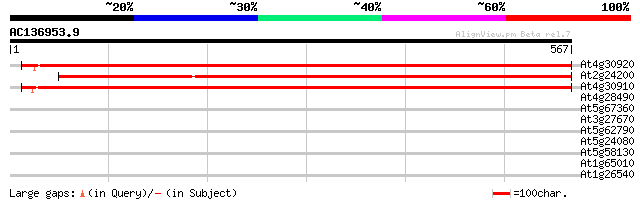
Score E
Sequences producing significant alignments: (bits) Value
At4g30920 leucyl aminopeptidase like protein 821 0.0
At2g24200 putative leucine aminopeptidase 816 0.0
At4g30910 leucyl aminopeptidase like protein 780 0.0
At4g28490 receptor-like protein kinase 5 precursor (RLK5) 38 0.013
At5g67360 cucumisin-like serine protease (gb|AAC18851.1) 34 0.25
At3g27670 unknown protein 32 1.2
At5g62790 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) 30 4.7
At5g24080 receptor-like protein kinase 30 4.7
At5g58130 unknown protein 29 6.1
At1g65010 hypothetical protein 29 8.0
At1g26540 unknown protein 29 8.0
>At4g30920 leucyl aminopeptidase like protein
Length = 583
Score = 821 bits (2121), Expect = 0.0
Identities = 415/560 (74%), Positives = 483/560 (86%), Gaps = 6/560 (1%)
Query: 13 SKSSSSSLFLT----SRIRFASFPKRSFHSTTKLMSQS-RYTTLGLTHPTNIEAPKISFS 67
S SS SS F+ SR+R A F +S+++ M+ + + TLGLT +++ PKISFS
Sbjct: 24 SPSSLSSCFVRFQFPSRLRLA-FAVTPLYSSSRAMAHTISHATLGLTQANSVDHPKISFS 82
Query: 68 ATDVDVTEWKGDILAVGVTEKDLTRDAKSRFENSILNKIDLKLDGLLSEASSEEDFSGKV 127
++DVTEWKGDILAVGVTEKD+ +D S+FEN IL K+D L GLL++ SSEEDFSGK
Sbjct: 83 GKEIDVTEWKGDILAVGVTEKDMAKDVNSKFENPILKKLDAHLGGLLADVSSEEDFSGKP 142
Query: 128 GQSTVVRIKGLGSKRVGLIGLGQLPSTTALYKGLGEAVVAVAKSAQASNVAIVLASSEGL 187
GQSTV+R+ GLGSKRVGLIGLG+ ST + ++ LGEAV A AK++QAS+VA+VLASSE +
Sbjct: 143 GQSTVLRLPGLGSKRVGLIGLGKSASTPSAFQSLGEAVAAAAKASQASSVAVVLASSESV 202
Query: 188 SSESKLSTAYAIASGAVLGLFEDQRYKSESKKPAVRSIDIIGLGTGPDLEKKLKYAGDVS 247
S+ESKL +A AIASG VLGLFED RYKSESKKP+++S+DIIG G+GP+LEKKLKYA VS
Sbjct: 203 SNESKLCSASAIASGTVLGLFEDSRYKSESKKPSLKSVDIIGFGSGPELEKKLKYAEHVS 262
Query: 248 FGIIFGRELVNSPANVLTPGVLAEEASKVASTYSDVFTAKILDADQCKELKMGSYLGVAA 307
+G+IFG+ELVNSPANVLTP VLAEEA +AS YSDV TA IL+ +QCKELKMGSYL VAA
Sbjct: 263 YGVIFGKELVNSPANVLTPAVLAEEALNLASMYSDVMTANILNEEQCKELKMGSYLAVAA 322
Query: 308 ASANPPRFIHLTYKPPSGSVKVKLALVGKGLTFDSGGYNIKTGPGCSIELMKFDMGGSAA 367
ASANPP FIHL YKP SG VK KLALVGKGLTFDSGGYNIKTGPGC IELMKFDMGGSAA
Sbjct: 323 ASANPPHFIHLIYKPSSGPVKTKLALVGKGLTFDSGGYNIKTGPGCLIELMKFDMGGSAA 382
Query: 368 VLGAAKALGQIKPLGVEVHFIVAACENMISGTGMRPGDVVTASNGKTIEVNNTDAEGRLT 427
VLGAAKA+GQIKP GVEVHFIVAACENMISGTGMRPGDV+TASNGKTIEVNNTDAEGRLT
Sbjct: 383 VLGAAKAIGQIKPPGVEVHFIVAACENMISGTGMRPGDVLTASNGKTIEVNNTDAEGRLT 442
Query: 428 LADALVYACNQGVEKIIDLATLTGACVVALGPSIAGVFSPNDELVKEVLEASEVSGEKLW 487
LADALVYACNQGV+K++DLATLTGAC++ALG S+AG+++P+D+L KEV+ ASE SGEKLW
Sbjct: 443 LADALVYACNQGVDKVVDLATLTGACIIALGTSMAGIYTPSDKLAKEVIAASERSGEKLW 502
Query: 488 RLPIEESYWESMKSGVADMVNTGGRPGGSITAALFLKQFVDEKVQWLHIDMAGPVWNDKK 547
R+P+EESYWE MKSGVADMVNTGGR GGSITAALFLKQFV E V+W+HIDMAGPVWN+KK
Sbjct: 503 RMPMEESYWEMMKSGVADMVNTGGRAGGSITAALFLKQFVSEDVEWMHIDMAGPVWNEKK 562
Query: 548 RSATGFGVSTLVEWVLKNSS 567
++ATGFGV+TLVEWV +SS
Sbjct: 563 KAATGFGVATLVEWVQNHSS 582
>At2g24200 putative leucine aminopeptidase
Length = 520
Score = 816 bits (2109), Expect = 0.0
Identities = 405/518 (78%), Positives = 462/518 (89%), Gaps = 2/518 (0%)
Query: 50 TLGLTHPTNIEAPKISFSATDVDVTEWKGDILAVGVTEKDLTRDAKSRFENSILNKIDLK 109
TLGLT P + E KISF+A ++DV EWKGDIL VGVTEKDL +D S+FEN IL+K+D
Sbjct: 4 TLGLTQPNSTEPHKISFTAKEIDVIEWKGDILVVGVTEKDLAKDGNSKFENPILSKVDAH 63
Query: 110 LDGLLSEASSEEDFSGKVGQSTVVRIKGLGSKRVGLIGLGQLPSTTALYKGLGEAVVAVA 169
L GLL++ SSEEDF+GK GQSTV+R+ GLGSKR+ LIGLGQ S+ + LGEAV V+
Sbjct: 64 LSGLLAQVSSEEDFTGKPGQSTVLRLPGLGSKRIALIGLGQSVSSPVAFHSLGEAVATVS 123
Query: 170 KSAQASNVAIVLASSEGLSSESKLSTAYAIASGAVLGLFEDQRYKSESKKPAVRSIDIIG 229
K++Q+++ AIVLASS +S ESKLS+ A+ASG VLGLFED RYKSESKKP+++++DIIG
Sbjct: 124 KASQSTSAAIVLASS--VSDESKLSSVSALASGIVLGLFEDGRYKSESKKPSLKAVDIIG 181
Query: 230 LGTGPDLEKKLKYAGDVSFGIIFGRELVNSPANVLTPGVLAEEASKVASTYSDVFTAKIL 289
GTG ++EKKLKYA DVS+G+IFGREL+NSPANVLTP VLAEEA+KVASTYSDVFTA IL
Sbjct: 182 FGTGAEVEKKLKYAEDVSYGVIFGRELINSPANVLTPAVLAEEAAKVASTYSDVFTANIL 241
Query: 290 DADQCKELKMGSYLGVAAASANPPRFIHLTYKPPSGSVKVKLALVGKGLTFDSGGYNIKT 349
+ +QCKELKMGSYL VAAASANPP FIHL YKPP+GSVK KLALVGKGLTFDSGGYNIKT
Sbjct: 242 NEEQCKELKMGSYLAVAAASANPPHFIHLVYKPPNGSVKTKLALVGKGLTFDSGGYNIKT 301
Query: 350 GPGCSIELMKFDMGGSAAVLGAAKALGQIKPLGVEVHFIVAACENMISGTGMRPGDVVTA 409
GPGCSIELMKFDMGGSAAVLGAAKA+G+IKP GVEVHFIVAACENMISGTGMRPGDV+TA
Sbjct: 302 GPGCSIELMKFDMGGSAAVLGAAKAIGEIKPPGVEVHFIVAACENMISGTGMRPGDVITA 361
Query: 410 SNGKTIEVNNTDAEGRLTLADALVYACNQGVEKIIDLATLTGACVVALGPSIAGVFSPND 469
SNGKTIEVNNTDAEGRLTLADALVYACNQGV+KI+DLATLTGACV+ALG S+AG+++P+D
Sbjct: 362 SNGKTIEVNNTDAEGRLTLADALVYACNQGVDKIVDLATLTGACVIALGTSMAGIYTPSD 421
Query: 470 ELVKEVLEASEVSGEKLWRLPIEESYWESMKSGVADMVNTGGRPGGSITAALFLKQFVDE 529
EL KEV+ ASE SGEKLWR+P+EESYWE MKSGVADMVNTGGR GGSITAALFLKQFV E
Sbjct: 422 ELAKEVIAASERSGEKLWRMPLEESYWEMMKSGVADMVNTGGRAGGSITAALFLKQFVSE 481
Query: 530 KVQWLHIDMAGPVWNDKKRSATGFGVSTLVEWVLKNSS 567
KVQW+HIDMAGPVWN+KK+S TGFGV+TLVEWV KNSS
Sbjct: 482 KVQWMHIDMAGPVWNEKKKSGTGFGVATLVEWVQKNSS 519
>At4g30910 leucyl aminopeptidase like protein
Length = 581
Score = 780 bits (2015), Expect = 0.0
Identities = 401/560 (71%), Positives = 468/560 (82%), Gaps = 6/560 (1%)
Query: 13 SKSSSSSLF----LTSRIRFASFPKRSFHSTTKLMSQS-RYTTLGLTHPTNIEAPKISFS 67
S SS SS F L SR+R SF + ++K + + + TLGLT +++ PKISFS
Sbjct: 23 SPSSLSSCFVRFQLLSRLR-VSFAITPLYCSSKATAHTIAHATLGLTQANSVDHPKISFS 81
Query: 68 ATDVDVTEWKGDILAVGVTEKDLTRDAKSRFENSILNKIDLKLDGLLSEASSEEDFSGKV 127
++DVTEWKGDILAVGVTEKD+ +DA S+FEN IL K+D L GLL++ S+EEDFSGK
Sbjct: 82 GKEIDVTEWKGDILAVGVTEKDMAKDANSKFENPILKKLDAHLGGLLADVSAEEDFSGKP 141
Query: 128 GQSTVVRIKGLGSKRVGLIGLGQLPSTTALYKGLGEAVVAVAKSAQASNVAIVLASSEGL 187
GQSTV+R+ GLGSKRVGLIGLG+ ST + ++ LGEAV A AK++QAS+VA+VLASSE
Sbjct: 142 GQSTVLRLPGLGSKRVGLIGLGKSASTPSAFQSLGEAVAAAAKASQASSVAVVLASSESF 201
Query: 188 SSESKLSTAYAIASGAVLGLFEDQRYKSESKKPAVRSIDIIGLGTGPDLEKKLKYAGDVS 247
S ESKLS+A IASG VLGLFED RYKSESKKP+++S+ IG GTGP+LE KLKYA VS
Sbjct: 202 SDESKLSSASDIASGTVLGLFEDSRYKSESKKPSLKSVVFIGFGTGPELENKLKYAEHVS 261
Query: 248 FGIIFGRELVNSPANVLTPGVLAEEASKVASTYSDVFTAKILDADQCKELKMGSYLGVAA 307
+G+IF +ELVNSPANVL+P VLAEEAS +AS YS+V TA IL +QCKELKMGSYL VAA
Sbjct: 262 YGVIFTKELVNSPANVLSPAVLAEEASNLASMYSNVMTANILKEEQCKELKMGSYLAVAA 321
Query: 308 ASANPPRFIHLTYKPPSGSVKVKLALVGKGLTFDSGGYNIKTGPGCSIELMKFDMGGSAA 367
ASANPP FIHL YKP SG VK KLALVGKGLTFDSGGYNIK GP IELMK D+GGSAA
Sbjct: 322 ASANPPHFIHLIYKPSSGPVKTKLALVGKGLTFDSGGYNIKIGPELIIELMKIDVGGSAA 381
Query: 368 VLGAAKALGQIKPLGVEVHFIVAACENMISGTGMRPGDVVTASNGKTIEVNNTDAEGRLT 427
VLGAAKA+G+IKP GVEVHFIVAACENMISGTGMRPGDV+TASNGKTIEVN+TD+EGRLT
Sbjct: 382 VLGAAKAIGEIKPPGVEVHFIVAACENMISGTGMRPGDVITASNGKTIEVNDTDSEGRLT 441
Query: 428 LADALVYACNQGVEKIIDLATLTGACVVALGPSIAGVFSPNDELVKEVLEASEVSGEKLW 487
LADALVYACNQGV+KI+D+ATLTG +VALGPS+AG+++ +DEL KEV+ AS+ SGEKLW
Sbjct: 442 LADALVYACNQGVDKIVDIATLTGEIIVALGPSMAGMYTASDELAKEVIAASQRSGEKLW 501
Query: 488 RLPIEESYWESMKSGVADMVNTGGRPGGSITAALFLKQFVDEKVQWLHIDMAGPVWNDKK 547
R+P+EESYWE MKSGVADMVN GGR GGSITAALFLK+FV E V+WLHIDMAG VWN+KK
Sbjct: 502 RMPMEESYWEMMKSGVADMVNFGGRAGGSITAALFLKRFVSENVEWLHIDMAGRVWNEKK 561
Query: 548 RSATGFGVSTLVEWVLKNSS 567
++ATGFGV+TLVEWV NSS
Sbjct: 562 KAATGFGVATLVEWVQNNSS 581
>At4g28490 receptor-like protein kinase 5 precursor (RLK5)
Length = 999
Score = 38.1 bits (87), Expect = 0.013
Identities = 39/133 (29%), Positives = 65/133 (48%), Gaps = 15/133 (11%)
Query: 38 STTKLMSQSRYTTLGLTHPTNIEAPKISFSATDVDVTEWKGDILAVGVTEKDLT--RDAK 95
S KL Q + GL + +E SF+ G I + K+L+ R +K
Sbjct: 411 SNNKLSGQIPHGFWGLPRLSLLELSDNSFT----------GSIPKTIIGAKNLSNLRISK 460
Query: 96 SRFENSILNKIDLKLDGLLSEASSEEDFSGKVGQSTVVRIKGLGSKRVGLIGL-GQLPST 154
+RF SI N+I L+G++ + +E DFSG++ +S +V++K L + L G++P
Sbjct: 461 NRFSGSIPNEIG-SLNGIIEISGAENDFSGEIPES-LVKLKQLSRLDLSKNQLSGEIPRE 518
Query: 155 TALYKGLGEAVVA 167
+K L E +A
Sbjct: 519 LRGWKNLNELNLA 531
>At5g67360 cucumisin-like serine protease (gb|AAC18851.1)
Length = 757
Score = 33.9 bits (76), Expect = 0.25
Identities = 34/118 (28%), Positives = 55/118 (45%), Gaps = 7/118 (5%)
Query: 331 LALVGKGLTFDSGGYNIKTGPGCSIELMKFDMGGSA--AVLGAAKALGQIKPLGVEVHFI 388
LA++G G F G ++ G +L+ F G+A A G G + P V+ +
Sbjct: 344 LAILGNGKNFT--GVSLFKGEALPDKLLPFIYAGNASNATNGNLCMTGTLIPEKVKGKIV 401
Query: 389 VAACENMISGTGMRPGDVVTASNGKTIEVNNTDAEGRLTLADALVYACNQGVEKIIDL 446
+ C+ I+ ++ GDVV A+ G + + NT A G +ADA + EK D+
Sbjct: 402 M--CDRGINAR-VQKGDVVKAAGGVGMILANTAANGEELVADAHLLPATTVGEKAGDI 456
>At3g27670 unknown protein
Length = 1815
Score = 31.6 bits (70), Expect = 1.2
Identities = 24/95 (25%), Positives = 42/95 (43%), Gaps = 4/95 (4%)
Query: 26 IRFASFPKRSFHSTTKLMSQS-RYTTLGLTHPTNIEAPKISFSATDVDVTEW-KGDILAV 83
++F S RSFH ++ R L NI IS + + W + +IL++
Sbjct: 852 VKFGS--SRSFHDVHFQYEEAFRVVVKSLQLSRNISLALISLQSLKAFMRRWMRANILSI 909
Query: 84 GVTEKDLTRDAKSRFENSILNKIDLKLDGLLSEAS 118
T K+L+ D S+ N+I+ + + + AS
Sbjct: 910 DATTKELSSDKTSKATNNIMKALPAASHNIKASAS 944
>At5g62790 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR)
Length = 477
Score = 29.6 bits (65), Expect = 4.7
Identities = 17/59 (28%), Positives = 30/59 (50%), Gaps = 6/59 (10%)
Query: 497 ESMKSGVADMVNTGGRPGGSITAALF------LKQFVDEKVQWLHIDMAGPVWNDKKRS 549
+++K D+ GR GG++T L ++ F+DEK+ +L I + DK R+
Sbjct: 383 DNVKYPSMDLAYAAGRAGGTMTGVLSAANEKAVEMFIDEKISYLDIFKVVELTCDKHRN 441
>At5g24080 receptor-like protein kinase
Length = 872
Score = 29.6 bits (65), Expect = 4.7
Identities = 17/53 (32%), Positives = 30/53 (56%), Gaps = 2/53 (3%)
Query: 32 PKRSFHSTTKLMSQSRYTTLGLTHPTNIEAPKISFSA-TDVDVTEWKGDILAV 83
P R H + K++ Q +LGLT+ N++ P ++S + D++ GD+ AV
Sbjct: 176 PSRHGHYSLKMLQQHTSLSLGLTYNINLD-PHANYSYWSGPDISNVTGDVTAV 227
>At5g58130 unknown protein
Length = 748
Score = 29.3 bits (64), Expect = 6.1
Identities = 43/161 (26%), Positives = 67/161 (40%), Gaps = 19/161 (11%)
Query: 87 EKDLTRDAKSRFENSILNKIDLKLDGLLSEASSEEDFS-GKVGQSTVVRIKGLGSKRVGL 145
+ D D SR SILNK + SEE +S G+ G T + + L
Sbjct: 231 DMDSALDMLSRKRKSILNK----------KTPSEEGYSEGRKGNLTH---PSKNRQTISL 277
Query: 146 IGLGQLPSTTALY-KGLGEAVVAVAKSAQASNVAIVLASSEGLSSESKLSTAYAIASGAV 204
G+ S+ A+ K VV S + S + S + +S K S +A+G
Sbjct: 278 EETGRQESSQAIRGKKKPSEVVPDKSSDEPSRTKDLEQSIDNISWSQKSSWKSLMANGNS 337
Query: 205 ----LGLFEDQRYKSESKKPAVRSIDIIGLGTGPDLEKKLK 241
+ F S++ +PA R+ D+ GL + +L+KK K
Sbjct: 338 NDFSVSSFLPGVGSSKAVQPAPRNTDLAGLPSRENLKKKTK 378
>At1g65010 hypothetical protein
Length = 1318
Score = 28.9 bits (63), Expect = 8.0
Identities = 18/65 (27%), Positives = 36/65 (54%), Gaps = 3/65 (4%)
Query: 58 NIEAPKISFSATDVDVTEWKGDI--LAVGVTEKDLTRDAKSRFENSILNKIDLKLDGLLS 115
++EA K++ S T+ V EWK + L V E + ++ + S S++ ++ +L+ +L
Sbjct: 267 DLEAAKMAESCTNSSVEEWKNKVHELEKEVEESNRSKSSASESMESVMKQL-AELNHVLH 325
Query: 116 EASSE 120
E S+
Sbjct: 326 ETKSD 330
>At1g26540 unknown protein
Length = 695
Score = 28.9 bits (63), Expect = 8.0
Identities = 56/253 (22%), Positives = 94/253 (37%), Gaps = 23/253 (9%)
Query: 103 LNKIDLKLDGLLSEASSEEDFSGKVGQSTVVRIKGLGSKR---VGLIGLGQLPSTTALYK 159
L + + D ++ +S EE F G ++ + G S+R V L + ++ L +
Sbjct: 3 LQTMKITKDCVVEVSSEEEGFEGAWFRAVLEENPGNSSRRKLRVRYSTLLDMDGSSPLIE 62
Query: 160 GLGEAVVAVAKSAQASNVAIVLASSEGLSSESKLSTAYAIASGAVLGLFEDQRYKSESKK 219
+ + + + +VL EGL ++ + +G V+ ED Y
Sbjct: 63 HIEQRFIRPVPPEENQQKDVVL--EEGLLVDADHKDGWW--TGVVVKKMEDDNYLVYFDL 118
Query: 220 PAVRSIDIIGLGTGPDLEKKLKYAGDVSFGIIFGRELVNSPANVLTPGVLAEEASKVAST 279
P DII L L + G G E+ S ++ +PG + E S +
Sbjct: 119 PP----DIIQFER-KQLRTHLIWTG----GTWIQPEIEESNKSMFSPGTMVEVFSAKEAV 169
Query: 280 YSDVFTAKILDADQCKEL---KMGSYLGVAAASANPPRFIH---LTYKPPSGSVKVKLAL 333
+S K D D K+ YL A P ++ + PP SV K AL
Sbjct: 170 WSPAMVVKETDVDDKKKFIVKDCNRYLSCNGDEARPTNIVNSRRVRPIPPPSSVD-KYAL 228
Query: 334 VGKGLTFDSGGYN 346
+ TF G++
Sbjct: 229 LESVETFSGLGWH 241
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.132 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,973,266
Number of Sequences: 26719
Number of extensions: 517191
Number of successful extensions: 1032
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1028
Number of HSP's gapped (non-prelim): 11
length of query: 567
length of database: 11,318,596
effective HSP length: 105
effective length of query: 462
effective length of database: 8,513,101
effective search space: 3933052662
effective search space used: 3933052662
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC136953.9


