

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136953.3 + phase: 0 /pseudo
(630 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
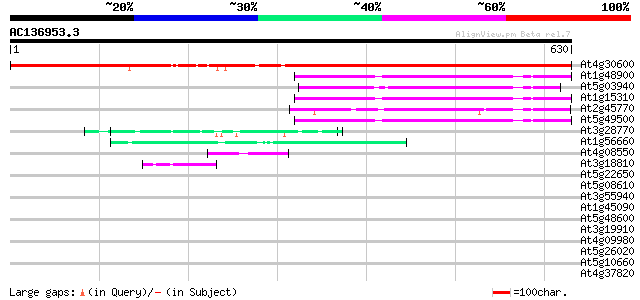
Score E
Sequences producing significant alignments: (bits) Value
At4g30600 signal recognition particle receptor-like protein 898 0.0
At1g48900 unknown protein (At1g48900) 133 3e-31
At5g03940 signal recognition particle 54CP protein precursor 132 4e-31
At1g15310 putative signal recognition particle 54 kDa subunit 131 1e-30
At2g45770 putative signal recognition particle receptor (alpha s... 117 2e-26
At5g49500 SRP54 (signal recognition particle 54 KDa) protein 110 2e-24
At3g28770 hypothetical protein 45 9e-05
At1g56660 hypothetical protein 44 2e-04
At4g08550 hypothetical protein 44 3e-04
At3g18810 protein kinase, putative 42 8e-04
At5g22650 putative histone deacetylase (HD2B) 41 0.002
At5g08610 RNA helicase-like protein 41 0.002
At3g55940 phosphoinositide-specific phospholipase C - like protein 40 0.004
At1g45090 hypothetical protein 40 0.005
At5g48600 chromosome condensation protein 39 0.007
At3g19910 unknown protein 39 0.007
At4g09980 unknown protein 39 0.009
At5g26020 putative protein 39 0.011
At5g10660 vacuolar calcium binding protein - like 38 0.015
At4g37820 unknown protein 38 0.015
>At4g30600 signal recognition particle receptor-like protein
Length = 634
Score = 898 bits (2321), Expect = 0.0
Identities = 489/644 (75%), Positives = 539/644 (82%), Gaps = 25/644 (3%)
Query: 1 MLEQLLIFTRGGLILWSCNEIGNALKGSPIDTLIRSCLLEERSGASSYNYDAPGAAYSLK 60
MLEQLLIFTRGGLILW+C EIGNALKGSPIDTLIRSCLLEERSGA S+NYDAPGAAY+LK
Sbjct: 1 MLEQLLIFTRGGLILWTCKEIGNALKGSPIDTLIRSCLLEERSGAVSFNYDAPGAAYTLK 60
Query: 61 WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFSQVYDPTRTVYRDFDEIFKQLKIEA 120
WTFHNDLGLVFVAVYQRILHLLYVDDLL+ VK+ FS+VYDP R Y DFDE F+QL+IEA
Sbjct: 61 WTFHNDLGLVFVAVYQRILHLLYVDDLLSMVKQSFSEVYDPKRMAYDDFDETFRQLRIEA 120
Query: 121 EARAEDLKKSNPV---IVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSEN 177
EARAE+L+K+ V + ++ QV+ G K+ S GG K+DGDG N K S
Sbjct: 121 EARAEELRKTKQVGKPVTSVKKQGQVSKPGLEGGNKRVSSEGGSKKDDGDGGNKAKVSTL 180
Query: 178 DRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNKGGNGK----K 233
+NGN + + + N NGKEN S +N A D+++LQK +R+KG G+ K
Sbjct: 181 TNGH--SNGN-HQMEDDSQETNDLANGKENTS-SNVAVDLSKLQK-LRSKGVRGRGGVRK 235
Query: 234 TDAVVTK----AEP-KKVVKKNRVWDEK-PVETKLDFTDHVDIDGDADKDRKVDYLAKEQ 287
TD++ K AEP KK KKNRVWD+ P ++KLDFTD +D +G+ D VD +A +Q
Sbjct: 236 TDSIGNKSSKVAEPAKKATKKNRVWDDAAPKQSKLDFTDSIDENGNNDH---VDIVAADQ 292
Query: 288 GESMMDKDEIFSSDSEDEEDDDDDNAGKKSKP-DAKKKGWFSSMFQSIAGKANLEKSDLE 346
GESMMDK+E+FSSDSE E DDD G KP AKKKGWFSS+FQSI GKANLE++DL
Sbjct: 293 GESMMDKEEVFSSDSESE---DDDEPGSDEKPAQAKKKGWFSSVFQSITGKANLERTDLG 349
Query: 347 PALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTVQAAMEDALVRILTP 406
PALKALK+RLMTKNVAEEIAEKLCESV ASLEGKKL+SFTRISSTVQAAMEDALVRILTP
Sbjct: 350 PALKALKERLMTKNVAEEIAEKLCESVEASLEGKKLSSFTRISSTVQAAMEDALVRILTP 409
Query: 407 RRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHNVNVMMAACDTFRS 466
RRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQH V+VMMAACDTFRS
Sbjct: 410 RRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHKVSVMMAACDTFRS 469
Query: 467 GAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDVVLVDTAGRMQDNEPLM 526
GAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEA+RNGSDVVLVDTAGRMQDNEPLM
Sbjct: 470 GAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEATRNGSDVVLVDTAGRMQDNEPLM 529
Query: 527 RALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGILLTKFDT 586
RALSKLI LN PDLVLFVGEALVGNDAVDQLSKFNQKL+DLSTS PRLIDGILLTKFDT
Sbjct: 530 RALSKLINLNQPDLVLFVGEALVGNDAVDQLSKFNQKLSDLSTSGNPRLIDGILLTKFDT 589
Query: 587 IDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
IDDKVGAALSMVYISG+PVMFVGCGQSYTDLKKLNVK+IVKTLL
Sbjct: 590 IDDKVGAALSMVYISGSPVMFVGCGQSYTDLKKLNVKAIVKTLL 633
>At1g48900 unknown protein (At1g48900)
Length = 495
Score = 133 bits (334), Expect = 3e-31
Identities = 83/310 (26%), Positives = 152/310 (48%), Gaps = 18/310 (5%)
Query: 321 AKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGK 380
A+ G + Q ++ +++ L L + L+ +V+ + +++ ++ + +
Sbjct: 4 AELGGRITRAIQQMSNVTIIDEKALNECLNEITRALLQSDVSFPLVKEMQSNIKKIVNLE 63
Query: 381 KLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKST 440
LA+ ++ A+ L ++L P + A ++ K VV+FVG+ G GK+T
Sbjct: 64 DLAAGHNKRRIIEQAIFSELCKMLDPGKPA-------FAPKKAKASVVMFVGLQGAGKTT 116
Query: 441 NLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAI 500
K AY+ Q+ + DTFR+GA +QL+ +A + +IP + E DP +A E +
Sbjct: 117 TCTKYAYYHQKKGYKPALVCADTFRAGAFDQLKQNATKAKIPFYGSYTESDPVKIAVEGV 176
Query: 501 QEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKF 560
+ D+++VDT+GR + L + ++ PDLV+FV ++ +G A DQ F
Sbjct: 177 DTFKKENCDLIIVDTSGRHKQEASLFEEMRQVAEATKPDLVIFVMDSSIGQAAFDQAQAF 236
Query: 561 NQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKL 620
Q +A + +++TK D K G ALS V + +PV+F+G G+ + +
Sbjct: 237 KQSVA----------VGAVIITKMDG-HAKGGGALSAVAATKSPVIFIGTGEHMDEFEVF 285
Query: 621 NVKSIVKTLL 630
+VK V LL
Sbjct: 286 DVKPFVSRLL 295
>At5g03940 signal recognition particle 54CP protein precursor
Length = 564
Score = 132 bits (333), Expect = 4e-31
Identities = 84/294 (28%), Positives = 155/294 (52%), Gaps = 18/294 (6%)
Query: 325 GWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLAS 384
G + + + G+ L K ++ ++ ++ L+ +V+ + + +SV+ G +
Sbjct: 83 GGLEAAWSKLKGEEVLTKDNIAEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGMGVIR 142
Query: 385 FTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAK 444
+ + + D LV+++ S ++ AK P V++ G+ GVGK+T AK
Sbjct: 143 GVKPDQQLVKIVHDELVKLMGGEVS-----ELQFAKSG--PTVILLAGLQGVGKTTVCAK 195
Query: 445 VAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEAS 504
+A +L++ + M+ A D +R A++QL ++ +P++ G + PA +AK+ ++EA
Sbjct: 196 LACYLKKQGKSCMLIAGDVYRPAAIDQLVILGEQVGVPVYTAGTDVKPADIAKQGLKEAK 255
Query: 505 RNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKL 564
+N DVV++DTAGR+Q ++ +M L + NP VL V +A+ G +A ++ FN ++
Sbjct: 256 KNNVDVVIMDTAGRLQIDKGMMDELKDVKKFLNPTEVLLVVDAMTGQEAAALVTTFNVEI 315
Query: 565 ADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLK 618
I G +LTK D D + GAALS+ +SG P+ VG G+ DL+
Sbjct: 316 G----------ITGAILTKLDG-DSRGGAALSVKEVSGKPIKLVGRGERMEDLE 358
>At1g15310 putative signal recognition particle 54 kDa subunit
Length = 479
Score = 131 bits (329), Expect = 1e-30
Identities = 84/310 (27%), Positives = 151/310 (48%), Gaps = 18/310 (5%)
Query: 321 AKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGK 380
A+ G + Q + +++ L L + L+ +V+ + EK+ ++ +
Sbjct: 4 AELGGRITRAIQQMNNVTIIDEKVLNDFLNEITRALLQSDVSFGLVEKMQTNIKKIVNLD 63
Query: 381 KLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKST 440
LA+ ++ A+ L R+L P + A ++ KP VV+FVG+ G GK+T
Sbjct: 64 DLAAGHNKRLIIEQAIFKELCRMLDPGKPA-------FAPKKAKPSVVMFVGLQGAGKTT 116
Query: 441 NLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAI 500
K AY+ Q+ + DTFR+GA +QL+ +A + +IP + E DP +A E +
Sbjct: 117 TCTKYAYYHQKKGYKAALVCADTFRAGAFDQLKQNATKAKIPFYGSYTESDPVKIAVEGV 176
Query: 501 QEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKF 560
+ D+++VDT+GR + L + ++ PDLV+FV ++ +G A +Q F
Sbjct: 177 DRFKKEKCDLIIVDTSGRHKQAASLFEEMRQVAEATEPDLVIFVMDSSIGQAAFEQAEAF 236
Query: 561 NQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKL 620
+ ++ + +++TK D K G ALS V + +PV+F+G G+ + +
Sbjct: 237 KETVS----------VGAVIITKMDG-HAKGGGALSAVAATKSPVIFIGTGEHMDEFEVF 285
Query: 621 NVKSIVKTLL 630
+VK V LL
Sbjct: 286 DVKPFVSRLL 295
>At2g45770 putative signal recognition particle receptor (alpha
subunit)
Length = 366
Score = 117 bits (293), Expect = 2e-26
Identities = 94/328 (28%), Positives = 165/328 (49%), Gaps = 34/328 (10%)
Query: 315 KKSKPDAKKK-GWFSSMFQSIAGKANL----EKSDLEPALKALKDRLMTKNVAEEIAEKL 369
+K+K D +K FS +++A L ++ + L L++ L+ + +I ++
Sbjct: 58 EKAKSDVEKVFSGFSKTRENLAVIDELLLFWNLAETDRVLDELEEALLVSDFGPKITVRI 117
Query: 370 CESVAASLEGKKLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVV 429
E + + KL S S ++ A++++++ +L + S L+ RKP V++
Sbjct: 118 VERLREDIMSGKLKS----GSEIKDALKESVLEMLAKKNSKTELQ-----LGFRKPAVIM 168
Query: 430 FVGVNGVGKSTNLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPI-FEKGY 488
VGVNG GK+T+L K+A+ L+ V+MAA DTFR+ A +QL A R I +G
Sbjct: 169 IVGVNGGGKTTSLGKLAHRLKNEGTKVLMAAGDTFRAAASDQLEIWAERTGCEIVVAEGD 228
Query: 489 EKDPAVVAKEAIQEASRNGSDVVLVDTAGRMQDNEPLM-------RALSKLIYLNNPDLV 541
+ A V +A++ G DVVL DT+GR+ N LM +A+ K++ P+ +
Sbjct: 229 KAKAATVLSKAVKRGKEEGYDVVLCDTSGRLHTNYSLMEELIACKKAVGKIV-SGAPNEI 287
Query: 542 LFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYIS 601
L V + G + + Q +FN+ + I G++LTK D + G +S+V
Sbjct: 288 LLVLDGNTGLNMLPQAREFNEVVG----------ITGLILTKLDG-SARGGCVVSVVEEL 336
Query: 602 GAPVMFVGCGQSYTDLKKLNVKSIVKTL 629
G PV F+G G++ DL+ + ++ V +
Sbjct: 337 GIPVKFIGVGEAVEDLQPFDPEAFVNAI 364
>At5g49500 SRP54 (signal recognition particle 54 KDa) protein
Length = 497
Score = 110 bits (275), Expect = 2e-24
Identities = 76/312 (24%), Positives = 147/312 (46%), Gaps = 20/312 (6%)
Query: 321 AKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGK 380
A+ G S Q + +++ L L + L+ +V+ + +++ ++ + +
Sbjct: 4 AELGGRIMSAIQKMNNVTIIDEKALNDCLNEITRALLQSDVSFPLVKEMQTNIKKIVNLE 63
Query: 381 KLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNG--VGK 438
LA+ ++ A+ L ++L P +S A ++ KP VV+FVG+ G + K
Sbjct: 64 DLAAGHNKRRIIEQAIFSELCKMLDPGKSA-------FAPKKAKPSVVMFVGLQGEVLEK 116
Query: 439 STNLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKE 498
+ + +++ + DTFR+GA +QL+ +A + +IP + DP +A E
Sbjct: 117 PQLVPSMLIIIRRKGYKPALVCADTFRAGAFDQLKQNATKSKIPYYGSYTGSDPVKIAVE 176
Query: 499 AIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLS 558
+ + D+++VDT+GR + L + ++ PDLV+FV ++ +G A +Q
Sbjct: 177 GVDRFKKENCDLIIVDTSGRHKQQASLFEEMRQISEATKPDLVIFVMDSSIGQTAFEQAR 236
Query: 559 KFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLK 618
F Q +A + +++TK D K G LS V + +PV+F+G G+ + +
Sbjct: 237 AFKQTVA----------VGAVIITKMDG-HAKGGGTLSAVAATKSPVIFIGTGEHMDEFE 285
Query: 619 KLNVKSIVKTLL 630
+ K V LL
Sbjct: 286 VFDAKPFVSRLL 297
>At3g28770 hypothetical protein
Length = 2081
Score = 45.4 bits (106), Expect = 9e-05
Identities = 59/262 (22%), Positives = 105/262 (39%), Gaps = 31/262 (11%)
Query: 114 KQLKIEAEARAEDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKK 173
+ +K + + + E K+ N + + KQ KG KK S +K + K
Sbjct: 912 ESVKYKKDEKKEGNKEENKDTINTSSKQ----KGKDKKKKKKESKNSNMKKKEEDKKEYV 967
Query: 174 NSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNKGGNGKK 233
N+E + +N N + + + KE + A NR +K+ K
Sbjct: 968 NNELKKQE--DNKKETTKSENSKLKEENKDNKEKKESEDSA-SKNREKKEYEEKKSK--- 1021
Query: 234 TDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMD 293
TK E KK KK++ + K + D + +K+ D AK++ E +
Sbjct: 1022 -----TKEEAKKEKKKSQ-------DKKREEKDSEERKSKKEKEESRDLKAKKKEEETKE 1069
Query: 294 KDEIFSSDSEDEED--DDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKA 351
K E + S+ +ED + +DN K + D K+K +S + K +K D+E
Sbjct: 1070 KKESENHKSKKKEDKKEHEDNKSMKKEEDKKEK---KKHEESKSRKKEEDKKDME----K 1122
Query: 352 LKDRLMTKNVAEEIAEKLCESV 373
L+D+ K ++ +K + V
Sbjct: 1123 LEDQNSNKKKEDKNEKKKSQHV 1144
Score = 43.5 bits (101), Expect = 4e-04
Identities = 65/308 (21%), Positives = 117/308 (37%), Gaps = 45/308 (14%)
Query: 85 DDLLAAVKREFSQVYDPTRTVYRDFDEIFKQLKIEAEARAEDLKKSNPVIVGGNRKQQVT 144
DD K+E +Q+Y E +EA+ + ++ K++ NR +
Sbjct: 722 DDKSVDDKQEEAQIYG---------GESKDDKSVEAKGKKKESKENKKTKTNENRVRNKE 772
Query: 145 WKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNG 204
G+ + G K D K+ + +N + + N + RS G
Sbjct: 773 ENVQGNKKESEKVEKGEKKESKDAKS-VETKDNKKLSSTENRDEAKERS----------G 821
Query: 205 KENDSVNNGAFDVNRLQKKVRNKGGN-----GKKTDA--------VVTKAEPKKVVKKNR 251
++N + D ++ K +N+ G G K D+ V KA ++ +KK R
Sbjct: 822 EDNKEDKEESKDYQSVEAKEKNENGGVDTNVGNKEDSKDLKDDRSVEVKANKEESMKKKR 881
Query: 252 VW----DEKPVETKLDFTDHVDIDGDADKDRKVDYL--AKEQGESMMDKDEIFSSDSEDE 305
D+ + DF +++DID V Y K++G +KD I +S +
Sbjct: 882 EEVQRNDKSSTKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKEENKDTINTSSKQKG 941
Query: 306 ED-----DDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKN 360
+D + N+ K K + KK+ + + + K KS+ + KD K
Sbjct: 942 KDKKKKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDN-KEKK 1000
Query: 361 VAEEIAEK 368
+E+ A K
Sbjct: 1001 ESEDSASK 1008
Score = 38.1 bits (87), Expect = 0.015
Identities = 41/232 (17%), Positives = 93/232 (39%), Gaps = 14/232 (6%)
Query: 92 KREFSQVYDPTRTVYRDFDEIFKQLKIEAEARAEDLKKSNPVIVGGNRKQQVTWKGDGSD 151
K+E + ++ +++ ++ D+ K+ E+++R ++ K + + Q K + +
Sbjct: 1080 KKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKL---EDQNSNKKKEDKN 1136
Query: 152 GKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVN 211
KK +K + D K K+N E + + + + N V + K+
Sbjct: 1137 EKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKS---QKNEVDKKEKKSSKDQQKKK 1193
Query: 212 NGAFDVNRLQKKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDID 271
+ +K +N+ K+T K + + +KN+ D+K TK ++
Sbjct: 1194 EKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDKKNTTKQSGGKKESME 1253
Query: 272 GDADKDRKVDYLAKEQGESMMDKDE-----IFSSDSEDEEDDDDDNAGKKSK 318
++ ++ + K Q + D DE + +DS+ + D +SK
Sbjct: 1254 SES---KEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESK 1302
Score = 36.2 bits (82), Expect = 0.057
Identities = 48/248 (19%), Positives = 88/248 (35%), Gaps = 42/248 (16%)
Query: 108 DFDEIFKQLKIEAEARA-------EDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGG 160
D DE ++ ++A+++A ED KK V +K+ K D KKN +
Sbjct: 1297 DSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTKQS 1356
Query: 161 GLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRL 220
G K + K+ +S + ++ ++ S +DS + N +
Sbjct: 1357 GGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEI 1416
Query: 221 QKKV--------RNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKP--------------- 257
+ N+ K+T K + + +KN+ D+K
Sbjct: 1417 LMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTEQSGGKKESMES 1476
Query: 258 --VETKLDFTDHVDIDGDADK---------DRKVDYLAKEQGESMMDKDEI-FSSDSEDE 305
E + G++D+ D + D A QG+S K+EI +DS+ +
Sbjct: 1477 ESKEAENQQKSQATTQGESDESKNEILMQADSQADTHANSQGDSDESKNEILMQADSQAD 1536
Query: 306 EDDDDDNA 313
D D +
Sbjct: 1537 SQTDSDES 1544
Score = 36.2 bits (82), Expect = 0.057
Identities = 55/242 (22%), Positives = 95/242 (38%), Gaps = 10/242 (4%)
Query: 138 NRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVV 197
N++ + K D S G K + + + G NS N + GN + + V
Sbjct: 604 NKEDKKELKDDESVGAKTNNETSLEEKREQTQKGHDNSINSKIVDNKGGNADSNKEKEVH 663
Query: 198 GNVSVNGKENDSVNNGAFDVN-RLQKKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEK 256
S N +S + +V + KG GK+ + E KK+ K D K
Sbjct: 664 VGDSTNDNNMESKEDTKSEVEVKKNDGSSEKGEEGKENNK--DSMEDKKLENKESQTDSK 721
Query: 257 PVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNA-GK 315
++ D + I G KD D + +G+ K+ + +E+ + ++N G
Sbjct: 722 DDKSVDDKQEEAQIYGGESKD---DKSVEAKGKKKESKENKKTKTNENRVRNKEENVQGN 778
Query: 316 KSKPDAKKKG--WFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESV 373
K + + +KG S +S+ K N + S E +A K+R N ++ K +SV
Sbjct: 779 KKESEKVEKGEKKESKDAKSVETKDNKKLSSTENRDEA-KERSGEDNKEDKEESKDYQSV 837
Query: 374 AA 375
A
Sbjct: 838 EA 839
Score = 35.0 bits (79), Expect = 0.13
Identities = 43/211 (20%), Positives = 69/211 (32%), Gaps = 26/211 (12%)
Query: 114 KQLKIEAEARAEDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKK 173
K K + E+R DLK +K+ K + KK +K + D K KK
Sbjct: 1047 KSKKEKEESR--DLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKK 1104
Query: 174 NSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNKGGNGKK 233
+ E+ KE D + + KK +K K
Sbjct: 1105 HEESKSRK-----------------------KEEDKKDMEKLEDQNSNKKKEDKNEKKKS 1141
Query: 234 TDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMD 293
+ K E K KK + E + + ++D K K D K++ E
Sbjct: 1142 QHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSK-DQQKKKEKEMKES 1200
Query: 294 KDEIFSSDSEDEEDDDDDNAGKKSKPDAKKK 324
+++ + ED + KK K K+K
Sbjct: 1201 EEKKLKKNEEDRKKQTSVEENKKQKETKKEK 1231
Score = 31.2 bits (69), Expect = 1.8
Identities = 56/276 (20%), Positives = 104/276 (37%), Gaps = 16/276 (5%)
Query: 119 EAEARAEDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSEND 178
E E + E ++ S+ + N + + K + K GS+ G + NG +E
Sbjct: 334 EVEGQGESIEDSD---IEKNLESKEDVKSEVEAAKNAGSSMTGKLEEAQRNNGVSTNETM 390
Query: 179 RSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNKGGN-----GKK 233
S +G N + N + KEN + + + + NK GN G+
Sbjct: 391 NSENKGSGESTNDKMVNATTNDEDHKKENKEETHENNGESVKGENLENKAGNEESMKGEN 450
Query: 234 TDAVVTKAEPK----KVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGE 289
+ V E K K N ++ + ++ V ++ + K V+ + G+
Sbjct: 451 LENKVGNEELKGNASVEAKTNNESSKEEKREESQRSNEVYMNKETTKGENVNIQGESIGD 510
Query: 290 SMMDKDEIFSSDSEDEED--DDDDNAGKKSKPDAKKKGWFSSMFQSIAG-KANLEKSDLE 346
S D D + + D + D N+ K+ +A+ S+ +++ A+ +K + +
Sbjct: 511 STKDNSLENKEDVKPKVDANESDGNSTKERHQEAQVNNGVSTEDKNLDNIGADEQKKNDK 570
Query: 347 PALKALKDRLMTKNVAEEIAEKLCESVA-ASLEGKK 381
D TK EE ESV +LE K+
Sbjct: 571 SVEVTTNDGDHTKEKREETQGNNGESVKNENLENKE 606
Score = 30.4 bits (67), Expect = 3.1
Identities = 51/274 (18%), Positives = 98/274 (35%), Gaps = 21/274 (7%)
Query: 107 RDFDEIFKQLKIEAEARAEDLKKSNPV-IVGGNRKQQVTWKGDGSDGKKNGSAGGGLKND 165
+D +++ Q + + + KKS V +V ++ + + K + KN+
Sbjct: 1118 KDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNE 1177
Query: 166 GDGKNGK--KNSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQ-- 221
D K K K+ + + + L+ N + +EN + N+ +
Sbjct: 1178 VDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDD 1237
Query: 222 KKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVD 281
KK K GKK E + K E+K + D D+ D + D
Sbjct: 1238 KKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQAD 1297
Query: 282 YLAKEQGESMMDKDEIFSSDSEDEED-----------DDDDNAGKKSKPDAKKKGWFSSM 330
+ + E +M D ++ +EED + +K+KP KK +
Sbjct: 1298 -SDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKK----NT 1352
Query: 331 FQSIAGKANLEKSDLEPALKALKDRLMTKNVAEE 364
+ GK +S+ + A K + T+ ++E
Sbjct: 1353 TKQSGGKKESMESESKEAENQQKSQATTQADSDE 1386
Score = 30.0 bits (66), Expect = 4.1
Identities = 44/240 (18%), Positives = 82/240 (33%), Gaps = 19/240 (7%)
Query: 139 RKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVN-NGNGYNLRSNGVV 197
+K Q T + D + K + D + + E+ ++ + R+N
Sbjct: 1263 QKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRNNEED 1322
Query: 198 GNVSVNGKENDSVNNGAFDVNRLQ--KKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDE 255
+ EN + N+ + KK K GKK E + K
Sbjct: 1323 RKKQTSVAENKKQKETKEEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQA 1382
Query: 256 KPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEED-------- 307
E+K + D D+ D + D + + E +M D ++ +EED
Sbjct: 1383 DSDESKNEILMQADSQADSHSDSQAD-SDESKNEILMQADSQATTQRNNEEDRKKQTSVA 1441
Query: 308 ---DDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEE 364
+ +K+KP KK + + GK +S+ + A K + T+ ++E
Sbjct: 1442 ENKKQKETKEEKNKPKDDKK----NTTEQSGGKKESMESESKEAENQQKSQATTQGESDE 1497
Score = 29.3 bits (64), Expect = 6.9
Identities = 24/109 (22%), Positives = 40/109 (36%), Gaps = 9/109 (8%)
Query: 125 EDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKND---------GDGKNGKKNS 175
ED K N + GN +G K + +KN+ GD NG+
Sbjct: 1779 EDSVKDNVTEIQGNDNSLTNSTSSEPNGDKLDTNKDSMKNNTMEAQGGSNGDSTNGETEE 1838
Query: 176 ENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKV 224
+ + +NN N ++ SN N G +D ++ + K+V
Sbjct: 1839 TKESNVSMNNQNMQDVGSNENSMNNQTTGTGDDIISTTTDTESNTSKEV 1887
>At1g56660 hypothetical protein
Length = 522
Score = 44.3 bits (103), Expect = 2e-04
Identities = 76/341 (22%), Positives = 135/341 (39%), Gaps = 28/341 (8%)
Query: 114 KQLKIEAEARAEDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKK 173
K K E + + KK P K+Q D K G G K D + ++ +K
Sbjct: 176 KNKKKEKDESGTEEKKKKPK----KEKKQKEESKSNEDKKVKGKKEKGEKGDLEKEDEEK 231
Query: 174 NSENDRS--AIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFD-VNRLQKKVRNKGGN 230
E+D + + + N + K+ D + + KK++ K G
Sbjct: 232 KKEHDETDQEMKEKDSKKNKKKEKDESCAEEKKKKPDKEKKEKDESTEKEDKKLKGKKGK 291
Query: 231 GKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGES 290
G+K E + KK + D E + DH + +KD+ AK++ E+
Sbjct: 292 GEKP-------EKEDEGKKTKEHDATEQEMDDEAADHKEGKKKKNKDK-----AKKK-ET 338
Query: 291 MMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALK 350
++D E+ +++D++DD+ + KK+K KK + K N ++++
Sbjct: 339 VID--EVCEKETKDKDDDEGETKQKKNKKKEKKSEKGEKDVKEDKKKENPLETEVMSRDI 396
Query: 351 ALKDRLMTKNVAEEIAEKLCESV--AASLEGKKLASFTRISSTVQAAMEDALVRILTPRR 408
L++ K ++ EK V S EGKK + + + E + ++
Sbjct: 397 KLEEPEAEKKEEDDTEEKKKSKVEGGESEEGKKKKKKDKKKNKKKDTKEPKMTEDEEEKK 456
Query: 409 --SIDILRDVHAAKEQRKPY-VVVFVGVNGVGK-STNLAKV 445
S D+ + AKE++K V G N +GK T LAK+
Sbjct: 457 DDSKDVKIEGSKAKEEKKDKDVKKKKGGNDIGKLKTKLAKI 497
Score = 41.6 bits (96), Expect = 0.001
Identities = 60/243 (24%), Positives = 101/243 (40%), Gaps = 36/243 (14%)
Query: 114 KQLKIEAEARAEDLKKSNPVIVGGNRKQQVTWKGDGSD---GKKNGSAGGGLKNDGDGKN 170
K+ K+ + + ++L +P G N + ++ K + KK+ + G K D + K
Sbjct: 9 KEEKLHVKIKTQEL---DPKEKGENVEVEMEVKAKSIEKVKAKKDEESSGKSKKDKEKKK 65
Query: 171 GKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNKGGN 230
GK N S + + + + +V G + V V +K+ +K G
Sbjct: 66 GK----NVDSEVKEDKDDDKKKDGKMVSKKHEEGHGDLEVKESDVKVEEHEKE--HKKGK 119
Query: 231 GKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGES 290
KK + + + E KK KKN+ EK D G +K++K D K + S
Sbjct: 120 EKKHEELEEEKEGKK--KKNK--KEK------------DESGPEEKNKKADKEKKHEDVS 163
Query: 291 MMDKDEIFSSDSEDEEDDDDDNAG---KKSKPDAKKKGWFSSMF---QSIAGKANL-EKS 343
+K+E+ D + + + D +G KK KP +KK S + + GK EK
Sbjct: 164 -QEKEELEEEDGKKNKKKEKDESGTEEKKKKPKKEKKQKEESKSNEDKKVKGKKEKGEKG 222
Query: 344 DLE 346
DLE
Sbjct: 223 DLE 225
Score = 38.9 bits (89), Expect = 0.009
Identities = 53/225 (23%), Positives = 96/225 (42%), Gaps = 29/225 (12%)
Query: 162 LKNDGDGKNGKKNSENDRSAIVN-NGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRL 220
L+ + +GK K E D S N + + V +E D N
Sbjct: 126 LEEEKEGKKKKNKKEKDESGPEEKNKKADKEKKHEDVSQEKEELEEEDGKKN-------- 177
Query: 221 QKKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKV 280
+KK +++ G +K + + K+ K N ++K V+ K + + D++ + D+++K
Sbjct: 178 KKKEKDESGTEEKKKKPKKEKKQKEESKSN---EDKKVKGKKEKGEKGDLEKE-DEEKKK 233
Query: 281 DYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKKGWFSSM---FQSIAGK 337
++ E + M +KD S ++ +E D+ KK KPD +KK S + + GK
Sbjct: 234 EH--DETDQEMKEKD---SKKNKKKEKDESCAEEKKKKPDKEKKEKDESTEKEDKKLKGK 288
Query: 338 ANL-EKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKK 381
EK + E K K+ T+ +++ + A EGKK
Sbjct: 289 KGKGEKPEKEDEGKKTKEHDATE-------QEMDDEAADHKEGKK 326
>At4g08550 hypothetical protein
Length = 587
Score = 43.9 bits (102), Expect = 3e-04
Identities = 26/92 (28%), Positives = 46/92 (49%), Gaps = 10/92 (10%)
Query: 223 KVRNKGGNGKKTDAVVTKAEPK-KVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVD 281
K +N K + + + PK V KK + KPV V+I+ + ++ +D
Sbjct: 27 KQQNVEEERKSAEIIAKEVSPKHNVEKKEEEFTRKPV---------VEIEEEEEEMESID 77
Query: 282 YLAKEQGESMMDKDEIFSSDSEDEEDDDDDNA 313
+E+G++ + DEI S DS D++DD + +A
Sbjct: 78 IHEEEEGDNNVSLDEIMSVDSSDDDDDSESSA 109
>At3g18810 protein kinase, putative
Length = 700
Score = 42.4 bits (98), Expect = 8e-04
Identities = 27/83 (32%), Positives = 38/83 (45%), Gaps = 5/83 (6%)
Query: 150 SDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDS 209
SD + S G N+ DG NG N++N+ + NNGN N +NG + + NG N+
Sbjct: 59 SDSQSPPSPQGN--NNNDGNNGNNNNDNNNN---NNGNNNNDNNNGNNKDNNNNGNNNNG 113
Query: 210 VNNGAFDVNRLQKKVRNKGGNGK 232
NN D N N N +
Sbjct: 114 NNNNGNDNNGNNNNGNNNDNNNQ 136
Score = 37.7 bits (86), Expect = 0.020
Identities = 25/76 (32%), Positives = 35/76 (45%), Gaps = 8/76 (10%)
Query: 148 DGSDGKKNG----SAGGGLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVN 203
DG++G N + G ND + N K N+ N + NNGN N N N + N
Sbjct: 74 DGNNGNNNNDNNNNNNGNNNNDNNNGNNKDNNNNGNN---NNGNNNNGNDNN-GNNNNGN 129
Query: 204 GKENDSVNNGAFDVNR 219
+N++ NNG NR
Sbjct: 130 NNDNNNQNNGGGSNNR 145
Score = 35.8 bits (81), Expect = 0.074
Identities = 24/79 (30%), Positives = 33/79 (41%), Gaps = 8/79 (10%)
Query: 152 GKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGN-VSVNGKENDSV 210
G N G N+ + N N+ ND NNGN + +NG N + NG +N+
Sbjct: 69 GNNNNDGNNGNNNNDNNNNNNGNNNNDN----NNGNNKDNNNNGNNNNGNNNNGNDNNGN 124
Query: 211 NNGAFDVNRLQKKVRNKGG 229
NN N +N GG
Sbjct: 125 NNNG---NNNDNNNQNNGG 140
>At5g22650 putative histone deacetylase (HD2B)
Length = 306
Score = 41.2 bits (95), Expect = 0.002
Identities = 35/104 (33%), Positives = 49/104 (46%), Gaps = 11/104 (10%)
Query: 236 AVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKD 295
A V KA+ K K V KP E K + +D D D D+ + D E MD D
Sbjct: 129 AAVVKADTKPKAKPAEV---KPAEEKPE-SDEEDESDDEDESEEDD-----DSEKGMDVD 179
Query: 296 EIFSSDSEDE--EDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGK 337
E S D E+E ED++++ KK +P KK+ S ++GK
Sbjct: 180 EDDSDDDEEEDSEDEEEEETPKKPEPINKKRPNESVSKTPVSGK 223
>At5g08610 RNA helicase-like protein
Length = 827
Score = 41.2 bits (95), Expect = 0.002
Identities = 75/365 (20%), Positives = 145/365 (39%), Gaps = 45/365 (12%)
Query: 139 RKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSN-GVV 197
+ + +++G +GS+ G + NDR+ V++G+ + RS+ V
Sbjct: 169 KSSESSFRGRSDRNVDSGSSFRGRSDKNVDSGSSFRGRNDRN--VDSGSSFRGRSDRNVD 226
Query: 198 GNVSVNGKENDSVNNGAFDVNRLQKKVRN----KGGNGKKTDAVVTKAEPKKVVKKNRVW 253
S G+ + +V++G+ R + V + +G N + ++ + EP + NR
Sbjct: 227 SGSSFRGRSDRNVDSGSSFRGRNDRNVDSGSSFRGRNDRNVESGFRR-EPGS--ENNRGL 283
Query: 254 DEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDD-- 311
++ L+ D D D E + + D++ S DS DE+D++D+
Sbjct: 284 GKQTRGLSLEEEDSSDDD--------------ENRVGLGNIDDLPSEDSSDEDDENDEPL 329
Query: 312 ---NAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEP-ALKALKDR-LMTKNVAEEIA 366
A K+ K G S K ++ L P +LKA+KD T V +E
Sbjct: 330 IKKAASAKAVQTDKPTGEHVKTSDSYLSKTRFDQFPLSPLSLKAIKDAGFETMTVVQE-- 387
Query: 367 EKLCESVAASLEGKKLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPY 426
++ L+GK + + + + V L P +++ A+++ R+P
Sbjct: 388 ----ATLPIILQGKDVLAKAKTGT-------GKTVAFLLPAIEA-VIKSPPASRDSRQPP 435
Query: 427 VVVFVGVNGVGKSTNLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEK 486
++V V ++ A A L +++ ++ + EQ R QI +
Sbjct: 436 IIVLVVCPTRELASQAAAEANTLLKYHPSIGVQVVIGGTKLPTEQRRMQTNPCQILVATP 495
Query: 487 GYEKD 491
G KD
Sbjct: 496 GRLKD 500
>At3g55940 phosphoinositide-specific phospholipase C - like protein
Length = 584
Score = 40.0 bits (92), Expect = 0.004
Identities = 29/96 (30%), Positives = 46/96 (47%), Gaps = 9/96 (9%)
Query: 292 MDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKA 351
+DK++ D +D++DDDDD+ G D KK IA +A K + LK
Sbjct: 288 VDKNDSNGDDDDDDDDDDDDDDG----DDKIKKNAPPEYKHLIAIEAGKPKGGITECLKV 343
Query: 352 LKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTR 387
D++ +++EE EK E A K++ FT+
Sbjct: 344 DPDKVRRLSLSEEQLEKASEKYA-----KQIVRFTQ 374
>At1g45090 hypothetical protein
Length = 1210
Score = 39.7 bits (91), Expect = 0.005
Identities = 40/137 (29%), Positives = 62/137 (45%), Gaps = 15/137 (10%)
Query: 84 VDDLLAAVKREFSQVYDPTRTVYRDFDEIFKQLKIEAEARAEDLKK----SNPVIVGGNR 139
VD++L +K E Q ++ V D D KQ++ A LK+ S+PV N+
Sbjct: 358 VDNILRMLKSE--QEWNEKMWVGGD-DAPKKQVRPSATGERVILKRQRRVSDPV---ENK 411
Query: 140 KQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGN 199
K++++ DG G G GG + GDG+ G + + +G G NG G
Sbjct: 412 KRKISHSDDGEGGPSGGDGEGG-PSGGDGEGGPSGGDGEGGPSGGDGEG---GPNGADGE 467
Query: 200 VSVNGKENDSVNNGAFD 216
NG + + V + AFD
Sbjct: 468 GGPNGADGE-VGDEAFD 483
>At5g48600 chromosome condensation protein
Length = 1241
Score = 39.3 bits (90), Expect = 0.007
Identities = 48/163 (29%), Positives = 80/163 (48%), Gaps = 34/163 (20%)
Query: 124 AEDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNG--SAGGGLKNDG--------DGKNGK- 172
A+DL ++ + GGNR+ + DG+ +K+G S GGG G G +G+
Sbjct: 661 AKDLDQATRIAYGGNREFRRVVALDGALFEKSGTMSGGGGKARGGRMGTSIRATGVSGEA 720
Query: 173 -KNSENDRSAIV---NN-----GNG---YNLRSNGVVG---NVSVNGKENDSVNNGAFDV 217
N+EN+ S IV NN GN Y N V G ++ + +E +S+N+ +
Sbjct: 721 VANAENELSKIVDMLNNIREKVGNAVRQYRAAENEVSGLEMELAKSQREIESLNS---EH 777
Query: 218 NRLQKKVRN-KGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPVE 259
N L+K++ + + + KTD + E KK++ K +EK +E
Sbjct: 778 NYLEKQLASLEAASQPKTDEIDRLKELKKIISK----EEKEIE 816
>At3g19910 unknown protein
Length = 340
Score = 39.3 bits (90), Expect = 0.007
Identities = 35/142 (24%), Positives = 56/142 (38%), Gaps = 27/142 (19%)
Query: 197 VGNVSVNGKENDSVN--NGAFDVNRLQKKVRNKG--------GNGKKTDAVVTKAEPKKV 246
VGNV V ++ +N +G NR V + G G G+ AV E
Sbjct: 11 VGNVVVTAEQATKINETDGRLPENRQTGVVSDTGSGSERGEQGVGESAVAVAVPVEESGS 70
Query: 247 VKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKV-----------------DYLAKEQGE 289
+ + + ++ FT+ ID D R + DY + E G
Sbjct: 71 ISVGELPAPRSSSARVPFTNLSQIDADLALARTLQEQERAYMMLTMNSEISDYGSWETGS 130
Query: 290 SMMDKDEIFSSDSEDEEDDDDD 311
+ D+DE ++EDE+DD+D+
Sbjct: 131 YVYDEDEFDDPENEDEDDDEDE 152
>At4g09980 unknown protein
Length = 963
Score = 38.9 bits (89), Expect = 0.009
Identities = 39/159 (24%), Positives = 66/159 (40%), Gaps = 12/159 (7%)
Query: 147 GDGSDG--KKNGSAGGGLKNDGDGKNGK----KNSENDRSAIVNNGNGYNLRSNGVVGNV 200
G SD +K GGL NDGD K+ K ++ V+N +G + R ++
Sbjct: 80 GSSSDSSKRKRWDEAGGLVNDGDHKSSKLSDSRHDSGGERVSVSNEHGESRRDLKSDRSL 139
Query: 201 SVNGKENDSVNNGAFDVNRLQKKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPVET 260
+ ++ S + G D +R + G +G + V ++ K + EK +
Sbjct: 140 KTSSRDEKSKSRGVKDDDRGSPLKKTSGKDGSEVVREVGRSNRSKTPDADY---EKEKYS 196
Query: 261 KLDFTDHVDIDGDADKDRKVDYLA---KEQGESMMDKDE 296
+ D DG +D+DR + L K + S DKD+
Sbjct: 197 RKDERSRGRDDGWSDRDRDQEGLKDNWKRRHSSSGDKDQ 235
>At5g26020 putative protein
Length = 241
Score = 38.5 bits (88), Expect = 0.011
Identities = 38/171 (22%), Positives = 66/171 (38%), Gaps = 43/171 (25%)
Query: 206 ENDSVNNGAFDVNRLQKKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFT 265
E N+ A L++KVR++ G V + E K ++ +
Sbjct: 87 EQQQANDEAGGSKELKRKVRDETGTPMTQRKKVCEPEASKTQRRGK-------------- 132
Query: 266 DHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEED---------------DDD 310
GD K + K + E+ ++F EDE+D DDD
Sbjct: 133 ------GDEASASKTERRKKSEAEASKKPKKVFEISDEDEDDDDEEDDYGDNGADDVDDD 186
Query: 311 DNAGKKSKPDAKK---KGWFSSMFQSIAGKANLEKSDLE----PALKALKD 354
++ GK SKP+ ++ K W + + ++ + EK D E P+ ++L D
Sbjct: 187 EDNGKSSKPNKRRQYSKVW-NGLSINMQSQLEYEKFDAEDETLPSFQSLVD 236
>At5g10660 vacuolar calcium binding protein - like
Length = 407
Score = 38.1 bits (87), Expect = 0.015
Identities = 47/227 (20%), Positives = 90/227 (38%), Gaps = 37/227 (16%)
Query: 166 GDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKVR 225
G + G + S +S V + ++ +G+ GN S K+ S N + KK
Sbjct: 137 GSSRGGLRGSSTVKSPPVASRGSSGVKKSGLSGNSSSKSKKEGSGN--------VPKKSS 188
Query: 226 NKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDR------- 278
K + + + +++VK VET + +DH + + DKD+
Sbjct: 189 GKEISPDSSPLASAHEDEEEIVK---------VETDVHISDHGEEPKEEDKDQFAQPDES 239
Query: 279 -------KVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMF 331
V +EQ ++D+D+ E +E ++ + + + + KKK
Sbjct: 240 GEEKETSPVAASTEEQKGELIDEDKSTEQIEEPKEPENIEENNSEEEEEVKKKSDDEENS 299
Query: 332 QSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLE 378
+++A +D+ A+ + + K A E+ E+ ES AA E
Sbjct: 300 ETVA-----TTTDMNEAVNVEESKEEEKEEA-EVKEEEGESSAAKEE 340
Score = 35.4 bits (80), Expect = 0.097
Identities = 44/227 (19%), Positives = 83/227 (36%), Gaps = 28/227 (12%)
Query: 123 RAEDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAI 182
R KS PV G+ + + S K G + GK +S SA
Sbjct: 144 RGSSTVKSPPVASRGSSGVKKSGLSGNSSSKSKKEGSGNVPKKSSGKEISPDSSPLASAH 203
Query: 183 VNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKV--------RNKG---GNG 231
+ + ++ + + KE D D + +K+ KG
Sbjct: 204 EDEEEIVKVETDVHISDHGEEPKEEDKDQFAQPDESGEEKETSPVAASTEEQKGELIDED 263
Query: 232 KKTDAVVTKAEPKKVVKKNRVWDEK------------PVETKLDFTDHVDIDGDADKDRK 279
K T+ + EP+ + + N +E+ V T D + V+++ +++++
Sbjct: 264 KSTEQIEEPKEPENIEENNSEEEEEVKKKSDDEENSETVATTTDMNEAVNVEESKEEEKE 323
Query: 280 VDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDN-----AGKKSKPDA 321
+ +E+GES K+E + ++ EE ++ GKK P A
Sbjct: 324 EAEVKEEEGESSAAKEETTETMAQVEELPEEGTKNEVVQGKKESPTA 370
>At4g37820 unknown protein
Length = 532
Score = 38.1 bits (87), Expect = 0.015
Identities = 57/298 (19%), Positives = 110/298 (36%), Gaps = 33/298 (11%)
Query: 135 VGGNRKQQVTWKGD--------GSDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNG 186
+ +++ +KGD G++ K N K + +G++ ++EN +G
Sbjct: 155 ISNEEAREINYKGDDASSEVMHGTEEKSNE------KVEVEGESKSNSTENVSVHEDESG 208
Query: 187 NGYNLRSNGVVGNVSVNGKENDSVNNGAFDV-NRLQKKVRNKGGNGKKTDAVVTKAEPKK 245
+ V+ VS+N EN S + + + L K KG + + T
Sbjct: 209 PKNEVLEGSVIKEVSLNTTENGSDDGEQQETKSELDSKTGEKGFSDSNGELPETNLSTSN 268
Query: 246 VVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDE 305
+ + T + + D+ KV +E K+E +S S+DE
Sbjct: 269 ATETTESSGSDESGSSGKSTGYQQTKNEEDEKEKVQSSEEESKVKESGKNEKDASSSQDE 328
Query: 306 EDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEI 365
K+ KP+ KKK SS E + EP + +D + EE
Sbjct: 329 S--------KEEKPERKKKEESSSQG---------EGKEEEPEKREKEDSSSQEESKEEE 371
Query: 366 AEKLCESVAASLEGKKLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQR 423
E + ++S E ++ T I +++ ++ T ++S + R + E++
Sbjct: 372 PENKEKEASSSQEENEIKE-TEIKEKEESSSQEGNENKETEKKSSESQRKENTNSEKK 428
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.133 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,531,602
Number of Sequences: 26719
Number of extensions: 703960
Number of successful extensions: 5960
Number of sequences better than 10.0: 313
Number of HSP's better than 10.0 without gapping: 93
Number of HSP's successfully gapped in prelim test: 229
Number of HSP's that attempted gapping in prelim test: 4327
Number of HSP's gapped (non-prelim): 929
length of query: 630
length of database: 11,318,596
effective HSP length: 105
effective length of query: 525
effective length of database: 8,513,101
effective search space: 4469378025
effective search space used: 4469378025
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC136953.3


