

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.9 + phase: 0
(341 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
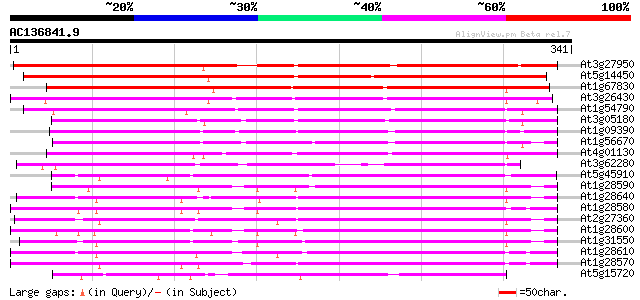
Score E
Sequences producing significant alignments: (bits) Value
At3g27950 early nodule-specific protein, putative 292 2e-79
At5g14450 early nodule-specific protein - like 279 1e-75
At1g67830 ENOD8-like protein 261 4e-70
At3g26430 nodulin, putative 256 2e-68
At1g54790 209 1e-54
At3g05180 putative nodulin 206 1e-53
At1g09390 putative lipase 196 2e-50
At1g56670 lipase homolog (Lip-4) 193 1e-49
At4g01130 putative acetyltransferase 174 8e-44
At3g62280 putative protein 164 8e-41
At5g45910 GDSL-motif lipase/hydrolase-like protein 127 9e-30
At1g28590 putative lipase (At1g28590) 120 1e-27
At1g28640 lipase, putative 119 2e-27
At1g28580 lipase like protein 116 2e-26
At2g27360 putative lipase 116 2e-26
At1g28600 lipase like protein 115 3e-26
At1g31550 unknown protein 114 8e-26
At1g28610 lipase, putative 111 5e-25
At1g28570 lipase like protein 105 3e-23
At5g15720 unknown protein 102 4e-22
>At3g27950 early nodule-specific protein, putative
Length = 361
Score = 292 bits (748), Expect = 2e-79
Identities = 157/334 (47%), Positives = 216/334 (64%), Gaps = 22/334 (6%)
Query: 3 TMTLIYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSIS 62
T+ +I +L F ++ S C +PA++NFGDSNSDTGA A + V PNG++FFG +
Sbjct: 7 TLAIIVLLLGFTEKLSALSSSCNFPAVFNFGDSNSDTGAISAAIGEVPPPNGVAFFGRSA 66
Query: 63 GRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLT--YLGF 120
GR DGRLI+DFI+E L LPYL+ YL+SVG+NYRHGANFA + IRP +A + +LG
Sbjct: 67 GRHSDGRLIIDFITENLTLPYLTPYLDSVGANYRHGANFATGGSCIRPTLACFSPFHLGT 126
Query: 121 QVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEE 180
QVSQFI FK+ T L++Q T DFSKA+YT+DIGQND+ G Q N +EE
Sbjct: 127 QVSQFIHFKTRTLSLYNQ------------TNDFSKALYTLDIGQNDLAIGFQ--NMTEE 172
Query: 181 EVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGC 240
+++ +IP I+ FT A++ LY E AR F IHNTGP C+PY +P D GC
Sbjct: 173 QLKATIPLIIENFTIALKLLYKEGARFFSIHNTGPTGCLPYLLKAFPAIPR----DPYGC 228
Query: 241 VKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC 300
+KP N +A E+N+QLK+++ QL++ P + FTYVDVY+ KY LI+ A+ GF++P ++CC
Sbjct: 229 LKPLNNVAIEFNKQLKNKITQLKKELPSSFFTYVDVYSAKYNLITKAKALGFIDPFDYCC 288
Query: 301 GSYQGNEIHYCGKKSIKNGT-VYGIACDDSSTYI 333
G + CGK NGT +Y +C + +I
Sbjct: 289 VGAIGRGMG-CGKTIFLNGTELYSSSCQNRKNFI 321
>At5g14450 early nodule-specific protein - like
Length = 389
Score = 279 bits (714), Expect = 1e-75
Identities = 145/327 (44%), Positives = 199/327 (60%), Gaps = 12/327 (3%)
Query: 9 ILCFFNLCVACPSKK-CVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCD 67
+LC F + + + C +PAIYNFGDSNSDTG A + P G FF +GR D
Sbjct: 21 LLCLFAVTTSVSVQPTCTFPAIYNFGDSNSDTGGISAAFEPIRDPYGQGFFHRPTGRDSD 80
Query: 68 GRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLG-------F 120
GRL +DFI+E L LPYLS+YLNS+GSN+RHGANFA + IR + G
Sbjct: 81 GRLTIDFIAERLGLPYLSAYLNSLGSNFRHGANFATGGSTIRRQNETIFQYGISPFSLDM 140
Query: 121 QVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEE 180
Q++QF FK+ + +LF Q R +PR E+F+KA+YT DIGQND+ G + S +
Sbjct: 141 QIAQFDQFKARSALLFTQIKSRYDREKLPRQEEFAKALYTFDIGQNDLSVGFR--TMSVD 198
Query: 181 EVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGC 240
+++ +IPDI++ AV+ +Y + R FW+HNTGP C+P FY G LD +GC
Sbjct: 199 QLKATIPDIVNHLASAVRNIYQQGGRTFWVHNTGPFGCLP-VNMFYMGTPAPGYLDKSGC 257
Query: 241 VKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC 300
VK NE+A E+NR+LK+ V LR+ A TYVDVYT KY ++SN + GF NPL+ CC
Sbjct: 258 VKAQNEMAMEFNRKLKETVINLRKELTQAAITYVDVYTAKYEMMSNPKKLGFANPLKVCC 317
Query: 301 GSYQGNEIHYCGKK-SIKNGTVYGIAC 326
G ++ + +CG K + N +YG +C
Sbjct: 318 GYHEKYDHIWCGNKGKVNNTEIYGGSC 344
>At1g67830 ENOD8-like protein
Length = 372
Score = 261 bits (667), Expect = 4e-70
Identities = 143/316 (45%), Positives = 195/316 (61%), Gaps = 13/316 (4%)
Query: 23 KCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELELP 82
+C +PAI+NFGDSNSDTG A P+G SFFGS +GR CDGRL++DFI+E L LP
Sbjct: 25 QCHFPAIFNFGDSNSDTGGLSAAFGQAGPPHGSSFFGSPAGRYCDGRLVIDFIAESLGLP 84
Query: 83 YLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGFQV----SQFILFKS-HTKILFD 137
YLS++L+SVGSN+ HGANFA A +PIR + + L GF QF+ F + H +
Sbjct: 85 YLSAFLDSVGSNFSHGANFATAGSPIRALNSTLRQSGFSPFSLDVQFVQFYNFHNRSQTV 144
Query: 138 QRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQAV 197
+ ++ +P ++ FSKA+YT DIGQND+ G N + E+V +P+I+SQF A+
Sbjct: 145 RSRGGVYKTMLPESDSFSKALYTFDIGQNDLTAG-YFANKTVEQVETEVPEIISQFMNAI 203
Query: 198 QKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKD 257
+ +Y + R FWIHNTGPI C+ Y +P N+ + D++GCV P N LAQ++N LK
Sbjct: 204 KNIYGQGGRYFWIHNTGPIGCLAYVIERFP--NKASDFDSHGCVSPLNHLAQQFNHALKQ 261
Query: 258 QVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC---GSYQGNEIHYCG-K 313
V +LR A TYVDVY++K+ L +A+ GF L CC G Y N+ CG K
Sbjct: 262 AVIELRSSLSEAAITYVDVYSLKHELFVHAQGHGFKGSLVSCCGHGGKYNYNKGIGCGMK 321
Query: 314 KSIKNGTVY-GIACDD 328
K +K VY G CD+
Sbjct: 322 KIVKGKEVYIGKPCDE 337
>At3g26430 nodulin, putative
Length = 380
Score = 256 bits (653), Expect = 2e-68
Identities = 143/345 (41%), Positives = 207/345 (59%), Gaps = 20/345 (5%)
Query: 1 MNTMTLIYILCFFNLCVACP---SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISF 57
M T L+ C+ P S C +PAI+NFGDSNSDTG A+ +PNG +F
Sbjct: 1 METNLLLVKCVLLASCLIHPRACSPSCNFPAIFNFGDSNSDTGGLSASFGQAPYPNGQTF 60
Query: 58 FGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTY 117
F S SGR DGRLI+DFI+EEL LPYL+++L+S+GSN+ HGANFA A + +RP A +
Sbjct: 61 FHSPSGRFSDGRLIIDFIAEELGLPYLNAFLDSIGSNFSHGANFATAGSTVRPPNATIAQ 120
Query: 118 LG-------FQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGY 170
G Q+ QF F + ++++ + + +P+ E FS+A+YT DIGQND+
Sbjct: 121 SGVSPISLDVQLVQFSDFITRSQLI--RNRGGVFKKLLPKKEYFSQALYTFDIGQNDLTA 178
Query: 171 GLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKN 230
GL K N + ++++ IPD+ Q + ++K+Y++ R FWIHNT P+ C+PY +P
Sbjct: 179 GL-KLNMTSDQIKAYIPDVHDQLSNVIRKVYSKGGRRFWIHNTAPLGCLPYVLDRFP--V 235
Query: 231 EKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQ 290
+D +GC P NE+A+ YN +LK +V +LR+ A FTYVD+Y++K TLI+ A+
Sbjct: 236 PASQIDNHGCAIPRNEIARYYNSELKRRVIELRKELSEAAFTYVDIYSIKLTLITQAKKL 295
Query: 291 GFVNPLEFCC---GSYQGNEIHYCGKKSIKNG--TVYGIACDDSS 330
GF PL CC G Y N++ CG K + G V +C+D S
Sbjct: 296 GFRYPLVACCGHGGKYNFNKLIKCGAKVMIKGKEIVLAKSCNDVS 340
>At1g54790
Length = 383
Score = 209 bits (533), Expect = 1e-54
Identities = 133/339 (39%), Positives = 187/339 (54%), Gaps = 20/339 (5%)
Query: 9 ILCFFNLCVACPSKKCV---YPAIYNFGDSNSDTGAGY-ATMAAVEHPNGISFFGSISGR 64
I+ F + + PS + YPAI NFGDSNSDTG A + V P G ++F SGR
Sbjct: 10 IVLFILISLFLPSSFSIILKYPAIINFGDSNSDTGNLISAGIENVNPPYGQTYFNLPSGR 69
Query: 65 CCDGRLILDFISEELELPYLSSYLNSVG-SNYRHGANFAVASA---PIRPIIAGLTYLGF 120
CDGRLI+DF+ +E++LP+L+ YL+S+G N++ G NFA A + P P
Sbjct: 70 YCDGRLIVDFLLDEMDLPFLNPYLDSLGLPNFKKGCNFAAAGSTILPANPTSVSPFSFDL 129
Query: 121 QVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEE 180
Q+SQFI FKS L +T +P + +SK +Y IDIGQNDI + + +
Sbjct: 130 QISQFIRFKSRAIELLS-KTGRKYEKYLPPIDYYSKGLYMIDIGQNDIAGAFY--SKTLD 186
Query: 181 EVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGC 240
+V SIP IL F +++LY E R WIHNTGP+ C+ + + K LD GC
Sbjct: 187 QVLASIPSILETFEAGLKRLYEEGGRNIWIHNTGPLGCLAQNIAKFGTDSTK--LDEFGC 244
Query: 241 VKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC 300
V HN+ A+ +N QL + + +P A TYVD++++K LI+N GF PL CC
Sbjct: 245 VSSHNQAAKLFNLQLHAMSNKFQAQYPDANVTYVDIFSIKSNLIANYSRFGFEKPLMACC 304
Query: 301 GSYQGNEIHY-----CGKKSIKNG-TVYGIACDDSSTYI 333
G G ++Y CG+ + +G +V AC+DSS YI
Sbjct: 305 G-VGGAPLNYDSRITCGQTKVLDGISVTAKACNDSSEYI 342
>At3g05180 putative nodulin
Length = 379
Score = 206 bits (524), Expect = 1e-53
Identities = 128/321 (39%), Positives = 182/321 (55%), Gaps = 21/321 (6%)
Query: 26 YPAIYNFGDSNSDTGAGYATMAAVEHPN-GISFFGS-ISGRCCDGRLILDFISEELELPY 83
+PA++NFGDSNSDTG + + + P+ I+FF S SGR C+GRLI+DF+ E ++ PY
Sbjct: 34 FPAVFNFGDSNSDTGELSSGLGFLPQPSYEITFFRSPTSGRFCNGRLIVDFLMEAIDRPY 93
Query: 84 LSSYLNSVG-SNYRHGANFAVASAPIRPIIAGLTY----LGFQVSQFILFKSHTKILFDQ 138
L YL+S+ YR G NFA A++ I+ A +Y G QVSQFI FKS L Q
Sbjct: 94 LRPYLDSISRQTYRRGCNFAAAASTIQKANAA-SYSPFGFGVQVSQFITFKSKVLQLIQQ 152
Query: 139 RTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQAVQ 198
E L+ +P FS +Y DIGQNDI + ++V +P IL F ++
Sbjct: 153 DEE--LQRYLPSEYFFSNGLYMFDIGQNDIAGAFY--TKTVDQVLALVPIILDIFQDGIK 208
Query: 199 KLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKDQ 258
+LY E AR +WIHNTGP+ C+ + +K LD GCV HN+ A+ +N QL
Sbjct: 209 RLYAEGARNYWIHNTGPLGCLAQVVSIFGE--DKSKLDEFGCVSDHNQAAKLFNLQLHGL 266
Query: 259 VFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHY-----CGK 313
+L + +P ++FTYVD++++K LI N GF + + CCG+ G ++Y CGK
Sbjct: 267 FKKLPQQYPNSRFTYVDIFSIKSDLILNHSKYGFDHSIMVCCGT-GGPPLNYDDQVGCGK 325
Query: 314 KSIKNGTVY-GIACDDSSTYI 333
+ NGT+ C DSS Y+
Sbjct: 326 TARSNGTIITAKPCYDSSKYV 346
>At1g09390 putative lipase
Length = 370
Score = 196 bits (497), Expect = 2e-50
Identities = 120/311 (38%), Positives = 166/311 (52%), Gaps = 13/311 (4%)
Query: 25 VYPAIYNFGDSNSDTGAGYATMA-AVEHPNGISFFGSISGRCCDGRLILDFISEELELPY 83
V P I+NFGDSNSDTG A + ++ PNG SFF +GR DGRL++DF+ + L
Sbjct: 34 VPPVIFNFGDSNSDTGGLVAGLGYSIGLPNGRSFFQRSTGRLSDGRLVIDFLCQSLNTSL 93
Query: 84 LSSYLNS-VGSNYRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILFDQRTEP 142
L+ YL+S VGS +++GANFA+ + P L Q+ QF+ FKS L
Sbjct: 94 LNPYLDSLVGSKFQNGANFAIVGSSTLPRYVPFA-LNIQLMQFLHFKSRALELAS--ISD 150
Query: 143 PLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYN 202
PL+ + F A+Y IDIGQNDI K S V + IP+++S+ A++ LY+
Sbjct: 151 PLKEMMIGESGFRNALYMIDIGQNDIADSFSK-GLSYSRVVKLIPNVISEIKSAIKILYD 209
Query: 203 EEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQL 262
E R FW+HNTGP+ C+P K D +GC+ +N A+ +N L L
Sbjct: 210 EGGRKFWVHNTGPLGCLPQKLSMVHSKG----FDKHGCLATYNAAAKLFNEGLDHMCRDL 265
Query: 263 RRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHYCGKKSIKNGTVY 322
R A YVD+Y +KY LI+N+ N GF PL CCG Y G +Y +I G
Sbjct: 266 RTELKEANIVYVDIYAIKYDLIANSNNYGFEKPLMACCG-YGGPPYNY--NVNITCGNGG 322
Query: 323 GIACDDSSTYI 333
+CD+ S +I
Sbjct: 323 SKSCDEGSRFI 333
>At1g56670 lipase homolog (Lip-4)
Length = 373
Score = 193 bits (490), Expect = 1e-49
Identities = 117/314 (37%), Positives = 168/314 (53%), Gaps = 23/314 (7%)
Query: 27 PAIYNFGDSNSDTGAGYATMA-AVEHPNGISFFGSISGRCCDGRLILDFISEELELPYLS 85
P I+NFGDSNSDTG A + + PNG FF +GR DGRL++DF+ + L L
Sbjct: 39 PVIFNFGDSNSDTGGLVAGLGYPIGFPNGRLFFRRSTGRLSDGRLLIDFLCQSLNTSLLR 98
Query: 86 SYLNSVG-SNYRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILFDQRTEPPL 144
YL+S+G + +++GANFA+A +P P + L QV QF FKS + L L
Sbjct: 99 PYLDSLGRTRFQNGANFAIAGSPTLPKNVPFS-LNIQVKQFSHFKSRSLELASSSNS--L 155
Query: 145 RSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEE 204
+ F A+Y IDIGQNDI + NS + V+ IP I+++ ++++LY+E
Sbjct: 156 KGMFISNNGFKNALYMIDIGQNDIARSFARGNSYSQTVKL-IPQIITEIKSSIKRLYDEG 214
Query: 205 ARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRR 264
R FWIHNTGP+ C+P K+ LD +GC+ +N A +N+ L +LR
Sbjct: 215 GRRFWIHNTGPLGCLPQKLSMVKSKD----LDQHGCLVSYNSAATLFNQGLDHMCEELRT 270
Query: 265 MFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHY-----CGKKSIKNG 319
A Y+D+Y +KY+LI+N+ GF +PL CCG Y G +Y CG K
Sbjct: 271 ELRDATIIYIDIYAIKYSLIANSNQYGFKSPLMACCG-YGGTPYNYNVKITCGHKGSN-- 327
Query: 320 TVYGIACDDSSTYI 333
C++ S +I
Sbjct: 328 -----VCEEGSRFI 336
>At4g01130 putative acetyltransferase
Length = 382
Score = 174 bits (440), Expect = 8e-44
Identities = 108/323 (33%), Positives = 167/323 (51%), Gaps = 18/323 (5%)
Query: 23 KCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELELP 82
KC + AI+NFGDSNSDTG +A A P G+++F +GR DGRLI+DF+++ L +P
Sbjct: 29 KCDFEAIFNFGDSNSDTGGFWAAFPAQSGPWGMTYFKKPAGRASDGRLIIDFLAKSLGMP 88
Query: 83 YLSSYLNSVGSNYRHGANFA-VASAPIRP----IIAGLT--YLGFQVSQFILFKSHTKIL 135
+LS YL S+GS++RHGANFA +AS + P ++G++ L Q++Q FK +
Sbjct: 89 FLSPYLQSIGSDFRHGANFATLASTVLLPNTSLFVSGISPFSLAIQLNQMKQFK--VNVD 146
Query: 136 FDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQ 195
+ P +P F K++YT IGQND L + E V+ +P ++ Q
Sbjct: 147 ESHSLDRPGLKILPSKIVFGKSLYTFYIGQNDFTSNL--ASIGVERVKLYLPQVIGQIAG 204
Query: 196 AVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQL 255
++++Y R F + N P+ C P Y H + +LD GC+ P N+ + YN L
Sbjct: 205 TIKEIYGIGGRTFLVLNLAPVGCYPAILTGYTHTD--ADLDKYGCLIPVNKAVKYYNTLL 262
Query: 256 KDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCG----SYQGNEIHYC 311
+ Q R A Y+D + + L + ++ G + ++ CCG Y N+ +C
Sbjct: 263 NKTLSQTRTELKNATVIYLDTHKILLDLFQHPKSYGMKHGIKACCGYGGRPYNFNQKLFC 322
Query: 312 GK-KSIKNGTVYGIACDDSSTYI 333
G K I N + AC D Y+
Sbjct: 323 GNTKVIGNFSTTAKACHDPHNYV 345
>At3g62280 putative protein
Length = 343
Score = 164 bits (414), Expect = 8e-41
Identities = 111/316 (35%), Positives = 162/316 (51%), Gaps = 42/316 (13%)
Query: 5 TLIYILCFFN-LCVA---CPSKKCVY-----PAIYNFGDSNSDTGAGYATMAA-VEHPNG 54
T+ + CFF LC++ C + + Y P + NFGDSNSDTG A + + P+G
Sbjct: 4 TVSSLQCFFLVLCLSLLVCSNSETSYKSNKKPILINFGDSNSDTGGVLAGVGLPIGLPHG 63
Query: 55 ISFFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAG 114
I+FF +GR DGRLI+DF E L++ YLS YL+S+ N++ G NFAV+ A PI +
Sbjct: 64 ITFFHRGTGRLGDGRLIVDFYCEHLKMTYLSPYLDSLSPNFKRGVNFAVSGATALPIFS- 122
Query: 115 LTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQK 174
L Q+ QF+ FK+ ++ L R + F A+Y IDIGQND+ L
Sbjct: 123 -FPLAIQIRQFVHFKNRSQELISSG-----RRDLIDDNGFRNALYMIDIGQNDLLLALYD 176
Query: 175 PNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGN 234
N + V IP +L + +A+Q G + H + +
Sbjct: 177 SNLTYAPVVEKIPSMLLEIKKAIQ---------------GELAI---------HLHNDSD 212
Query: 235 LDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVN 294
LD GC + HNE+A+ +N+ L +LR F A YVD+Y++KY L ++ + GFV+
Sbjct: 213 LDPIGCFRVHNEVAKAFNKGLLSLCNELRSQFKDATLVYVDIYSIKYKLSADFKLYGFVD 272
Query: 295 PLEFCCGSYQGNEIHY 310
PL CCG Y G +Y
Sbjct: 273 PLMACCG-YGGRPNNY 287
>At5g45910 GDSL-motif lipase/hydrolase-like protein
Length = 372
Score = 127 bits (319), Expect = 9e-30
Identities = 97/320 (30%), Positives = 150/320 (46%), Gaps = 19/320 (5%)
Query: 26 YPAIYNFGDSNSDTGAGYATMAAVEHPN------GISFFGSISGRCCDGRLILDFISEEL 79
Y +I+NFGDS SDTG + V+ PN G +FF +GRC DGRLI+DFI+E
Sbjct: 28 YESIFNFGDSLSDTG-NFLLSGDVDSPNIGRLPYGQTFFNRSTGRCSDGRLIIDFIAEAS 86
Query: 80 ELPYLSSYLNSVGSN----YRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKIL 135
LPY+ YL S+ +N ++ GANFAVA A + G V+ ++
Sbjct: 87 GLPYIPPYLQSLRTNDSVDFKRGANFAVAGATANE-FSFFKNRGLSVTLLTNKTLDIQLD 145
Query: 136 FDQRTEPPLRSGVPRTED-FSKAIYTI-DIGQNDIGYGLQKPNSSEEEVRRSIPDILSQF 193
+ ++ +P L P E F K+++ + +IG ND Y L S + + +P ++++
Sbjct: 146 WFKKLKPSLCKTKPECEQYFRKSLFLVGEIGGNDYNYPLLAFRSFKHAM-DLVPFVINKI 204
Query: 194 TQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNR 253
L E A + PI C + + N C P N LA+ +N
Sbjct: 205 MDVTSALIEEGAMTLIVPGNLPIGCSAALLERFNDNSGWLYDSRNQCYMPLNNLAKLHND 264
Query: 254 QLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFV-NPLEFCCGSYQGNEIHYCG 312
+LK + LR+ +P AK Y D Y+ ++ GF + L+ CCG G + Y
Sbjct: 265 KLKKGLAALRKKYPYAKIIYADYYSSAMQFFNSPSKYGFTGSVLKACCG---GGDGRYNV 321
Query: 313 KKSIKNGTVYGIACDDSSTY 332
+ +++ G C+D STY
Sbjct: 322 QPNVRCGEKGSTTCEDPSTY 341
>At1g28590 putative lipase (At1g28590)
Length = 403
Score = 120 bits (300), Expect = 1e-27
Identities = 98/327 (29%), Positives = 148/327 (44%), Gaps = 36/327 (11%)
Query: 26 YPAIYNFGDSNSDTGAGYATM------AAVEHPNGISFFGSISGRCCDGRLILDFISEEL 79
+ +I +FGDS +DTG A+ P G +FF +GR DGRLI+DFI+E L
Sbjct: 34 FKSIISFGDSIADTGNLLGLSDPNDLPASAFPPYGETFFHHPTGRYSDGRLIIDFIAEFL 93
Query: 80 ELPYLSSYLNSVGSNYRHGANFAVASAP-IRPIIA---GLTYLGFQVSQFILFKSHTKIL 135
P + + +N++ G NFAVA A + P G+ VS + +S T+ L
Sbjct: 94 GFPLVPPFYGCQNANFKKGVNFAVAGATALEPSFLEERGIHSTITNVSLSVQLRSFTESL 153
Query: 136 FDQRTEPPLRSGVP---RTEDFSKAIYTIDIGQNDIGYGL--QKPNSSEEEVRRSIPDIL 190
P G P R + I +IG ND + L +KP EE+ +P ++
Sbjct: 154 -------PNLCGSPSDCRDMIENALILMGEIGGNDYNFALFQRKPVKEVEEL---VPFVI 203
Query: 191 SQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQE 250
+ + A+ +L R F + PI Y Y N++ GC+K N+ ++
Sbjct: 204 ATISSAITELVCMGGRTFLVPGNFPIGYSASYLTLYKTSNKEEYDPLTGCLKWLNDFSEY 263
Query: 251 YNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVN-PLEFCC---GSYQGN 306
YN+QL++++ LR+++P Y D Y L GF+N PL CC GSY N
Sbjct: 264 YNKQLQEELNGLRKLYPHVNIIYADYYNALLRLFQEPAKFGFMNRPLPACCGVGGSYNFN 323
Query: 307 EIHYCGKKSIKNGTVYGIACDDSSTYI 333
CG ++ CDD S Y+
Sbjct: 324 FSRRCGSVGVE-------YCDDPSQYV 343
>At1g28640 lipase, putative
Length = 390
Score = 119 bits (299), Expect = 2e-27
Identities = 98/346 (28%), Positives = 156/346 (44%), Gaps = 30/346 (8%)
Query: 5 TLIYILCFFNLCVACPSKKCV-YPAIYNFGDSNSDTGAGYATMAAVEH-------PNGIS 56
+ + +L + VA +C + +I +FGDS +DTG Y ++ V H P G S
Sbjct: 11 SFLLVLYSTTIIVASSESRCRRFKSIISFGDSIADTG-NYLHLSDVNHLPQSAFLPYGES 69
Query: 57 FFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAV--ASAPIRPIIAG 114
FF SGR DGRLI+DFI+E L LPY+ SY S ++ G NFAV A+A R + G
Sbjct: 70 FFHPPSGRYSDGRLIIDFIAEFLGLPYVPSYFGSQNVSFDQGINFAVYGATALDRVFLVG 129
Query: 115 LTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPR--TEDFSKAIYTI-DIGQNDIGYG 171
G + S F ++ ++ P L + R E ++ + +IG ND Y
Sbjct: 130 ---KGIE-SDFTNVSLSVQLNIFKQILPNLCTSSSRDCREMLGDSLILMGEIGVNDYNYP 185
Query: 172 LQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNE 231
+ S E+++ +P ++ + A+ L + + F + P+ C P Y + E
Sbjct: 186 FFE-GKSINEIKQLVPLVIKAISSAIVDLIDLGGKTFLVPGNFPLGCYPAYLTLFQTAAE 244
Query: 232 KGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQG 291
+ + GC+ NE + +N QLK ++ +L+ ++ Y D Y + L G
Sbjct: 245 EDHDPFTGCIPRLNEFGEYHNEQLKTELKRLQELYDHVNIIYADYYNSLFRLYQEPVKYG 304
Query: 292 FVN-PLEFCC---GSYQGNEIHYCGKKSIKNGTVYGIACDDSSTYI 333
F N PL CC G Y CG + + C + S Y+
Sbjct: 305 FKNRPLAACCGVGGQYNFTIGKECGHRGVS-------CCQNPSEYV 343
>At1g28580 lipase like protein
Length = 390
Score = 116 bits (291), Expect = 2e-26
Identities = 98/347 (28%), Positives = 159/347 (45%), Gaps = 26/347 (7%)
Query: 1 MNTMTLIYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTG--AGYATMAAVEH----PNG 54
M + I++ F V+ +K + +I +FGDS +DTG G + + H P G
Sbjct: 10 MKLLVFIFLSTFVVTNVSSETKCREFKSIISFGDSIADTGNLLGLSDPKDLPHMAFPPYG 69
Query: 55 ISFFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAV--ASAPIRPII 112
+FF +GR +GRLI+DFI+E L LP + + S +N+ G NFAV A+A R +
Sbjct: 70 ENFFHHPTGRFSNGRLIIDFIAEFLGLPLVPPFYGSHNANFEKGVNFAVGGATALERSFL 129
Query: 113 A--GLTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVP---RTEDFSKAIYTIDIGQND 167
G+ + VS + S + L P G P R + I +IG ND
Sbjct: 130 EDRGIHFPYTNVSLGVQLNSFKESL-------PSICGSPSDCRDMIENALILMGEIGGND 182
Query: 168 IGYGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYP 227
Y + EE++ +P +++ + A+ +L R F + P+ C Y +
Sbjct: 183 YNYAFFV-DKGIEEIKELMPLVITTISSAITELIGMGGRTFLVPGEFPVGCSVLYLTSHQ 241
Query: 228 HKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNA 287
N + GC+K N+ + + QL+ ++ +L++++P Y D Y + L
Sbjct: 242 TSNMEEYDPLTGCLKWLNKFGENHGEQLRAELNRLQKLYPHVNIIYADYYNALFHLYQEP 301
Query: 288 RNQGFVN-PLEFCCGSYQGNEIHYCGKKSIKNGTVYGIACDDSSTYI 333
GF+N PL CCG+ G +Y + K GT +CDD S Y+
Sbjct: 302 AKFGFMNRPLSACCGA--GGPYNYTVGR--KCGTDIVESCDDPSKYV 344
>At2g27360 putative lipase
Length = 394
Score = 116 bits (290), Expect = 2e-26
Identities = 94/347 (27%), Positives = 150/347 (43%), Gaps = 30/347 (8%)
Query: 4 MTLIYILCFFNLCVACPSKKCV-YPAIYNFGDSNSDTGAGYATMAAVEHPN--------- 53
M L + + F + V +C + +I +FGDS +DTG + + PN
Sbjct: 8 MLLSFFISTFLITVVTSQTRCRNFKSIISFGDSITDTG----NLLGLSSPNDLPESAFPP 63
Query: 54 -GISFFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPII 112
G +FF SGR DGRLI+DFI+E L +P++ + S N+ G NFAV A
Sbjct: 64 YGETFFHHPSGRFSDGRLIIDFIAEFLGIPHVPPFYGSKNGNFEKGVNFAVGGATALECS 123
Query: 113 AGLTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTI--DIGQNDIGY 170
L G SQ + + F + S P D + + + +IG ND +
Sbjct: 124 V-LEEKGTHCSQSNISLGNQLKSFKESLPYLCGSSSPDCRDMIENAFILIGEIGGNDYNF 182
Query: 171 GLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKN 230
L + EEV+ +P +++ + A+ +L + AR F + P+ C Y Y N
Sbjct: 183 PLFD-RKNIEEVKELVPLVITTISSAISELVDMGARTFLVPGNFPLGCSVAYLTLYETPN 241
Query: 231 EKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQ 290
++ GC+ N+ + +N QL+ ++ +LR ++P Y D Y L+
Sbjct: 242 KEEYNPLTGCLTWLNDFSVYHNEQLQAELKRLRNLYPHVNIIYGDYYNTLLRLMQEPSKF 301
Query: 291 GFVN-PLEFCC---GSYQGNEIHYCGKKSIKNGTVYGIACDDSSTYI 333
G ++ PL CC G Y CG K ++ C D S Y+
Sbjct: 302 GLMDRPLPACCGLGGPYNFTFSIKCGSKGVE-------YCSDPSKYV 341
>At1g28600 lipase like protein
Length = 393
Score = 115 bits (289), Expect = 3e-26
Identities = 102/355 (28%), Positives = 154/355 (42%), Gaps = 40/355 (11%)
Query: 1 MNTMTLIYILCFFNLCVACPSKKCVYP---AIYNFGDSNSDTG--AGYATMAAVE----H 51
M ++ + I F L V S + P +I +FGDS +DTG G + +
Sbjct: 1 MASLDSLVIFLFSTLFVTIVSSETPCPNFKSIISFGDSIADTGNLVGLSDRNQLPVTAFP 60
Query: 52 PNGISFFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPI 111
P G +FF +GR CDGR+I+DFI+E + LPY+ Y S N+ G NFAVA A
Sbjct: 61 PYGETFFHHPTGRSCDGRIIMDFIAEFVGLPYVPPYFGSKNRNFDKGVNFAVAGATALK- 119
Query: 112 IAGLTYLGFQ----VSQFILFKSHTKILFDQRTEPPLRSGVP---RTEDFSKAIYTIDIG 164
+ L G Q VS + KS K L P G P R + I +IG
Sbjct: 120 SSFLKKRGIQPHTNVSLGVQLKSFKKSL-------PNLCGSPSDCRDMIGNALILMGEIG 172
Query: 165 QNDIGYGL--QKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYY 222
ND + +KP +EV +P +++ + + +L + F + PI C Y
Sbjct: 173 GNDYNFPFFNRKP---VKEVEELVPFVIASISSTITELIGMGGKTFLVPGEFPIGCSVVY 229
Query: 223 YFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYT 282
Y N+ + GC+K N+ + ++ +LK ++ +LR+++P Y D Y
Sbjct: 230 LTLYKTSNKDEYDPSTGCLKWLNKFGEYHSEKLKVELNRLRKLYPHVNIIYADYYNSLLR 289
Query: 283 LISNARNQGFV-NPLEFCC---GSYQGNEIHYCGKKSIKNGTVYGIACDDSSTYI 333
+ GF+ P CC G Y N CG +K +C D S Y+
Sbjct: 290 IFKEPAKFGFMERPFPACCGIGGPYNFNFTRKCGSVGVK-------SCKDPSKYV 337
>At1g31550 unknown protein
Length = 394
Score = 114 bits (285), Expect = 8e-26
Identities = 95/343 (27%), Positives = 149/343 (42%), Gaps = 31/343 (9%)
Query: 7 IYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEH---------PNGISF 57
+++ F V+ S+ + +I +FGDS +DTG + +H P G +F
Sbjct: 15 LFLSTLFVSIVSSESQCRNFESIISFGDSIADTGN---LLGLSDHNNLPMSAFPPYGETF 71
Query: 58 FGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTY 117
F +GR DGRLI+DFI+E L LPY+ Y S N+ G NFAVASA + L
Sbjct: 72 FHHPTGRFSDGRLIIDFIAEFLGLPYVPPYFGSTNGNFEKGVNFAVASATALE-SSFLEE 130
Query: 118 LGFQVSQFILFKSHTKILFDQRTEPPLRSGVP---RTEDFSKAIYTIDIGQNDIGYGLQK 174
G+ KI + P G+P R + I +IG ND + +
Sbjct: 131 KGYHCPHNFSLGVQLKIF---KQSLPNLCGLPSDCRDMIGNALILMGEIGANDYNFPFFQ 187
Query: 175 PNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGN 234
+EV+ +P ++S + A+ +L R F + P+ C + + N +
Sbjct: 188 LRPL-DEVKELVPLVISTISSAITELIGMGGRTFLVPGGFPLGCSVAFLTLHQTSNMEEY 246
Query: 235 LDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVN 294
GC+K N+ + ++ QL++++ +LR++ P Y D Y L GF+N
Sbjct: 247 DPLTGCLKWLNKFGEYHSEQLQEELNRLRKLNPHVNIIYADYYNASLRLGREPSKYGFIN 306
Query: 295 -PLEFCC---GSYQGNEIHYCGKKSIKNGTVYGIACDDSSTYI 333
L CC G Y N CG ++ AC D S Y+
Sbjct: 307 RHLSACCGVGGPYNFNLSRSCGSVGVE-------ACSDPSKYV 342
>At1g28610 lipase, putative
Length = 383
Score = 111 bits (278), Expect = 5e-25
Identities = 91/347 (26%), Positives = 156/347 (44%), Gaps = 27/347 (7%)
Query: 1 MNTMTLIYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEH-------PN 53
++++ ++ F V+ ++ +I +FGDS +DTG ++ H P
Sbjct: 4 LDSLVSFFLSTLFVTIVSSQTQCRNLESIISFGDSITDTG-NLVGLSDRNHLPVTAFLPY 62
Query: 54 GISFFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPII- 112
G +FF +GR C+GR+I+DFI+E L LP++ + S N+ G NFAVA A
Sbjct: 63 GETFFHHPTGRSCNGRIIIDFIAEFLGLPHVPPFYGSKNGNFEKGVNFAVAGATALETSI 122
Query: 113 ---AGLTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTI--DIGQND 167
G+ Y +S I K+ + L P L D + I +IG ND
Sbjct: 123 LEKRGIYYPHSNISLGIQLKTFKESL------PNLCGSPTDCRDMIGNAFIIMGEIGGND 176
Query: 168 IGYGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYP 227
+ +S EV+ +P ++++ + A+ +L + R F + P+ C Y Y
Sbjct: 177 FNFAFFVNKTS--EVKELVPLVITKISSAIVELVDMGGRTFLVPGNFPLGCSATYLTLYQ 234
Query: 228 HKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNA 287
N++ GC+ N+ ++ YN +L+ ++ +L +++P Y D + L
Sbjct: 235 TSNKEEYDPLTGCLTWLNDFSEYYNEKLQAELNRLSKLYPHVNIIYGDYFNALLRLYQEP 294
Query: 288 RNQGFVN-PLEFCCGSYQGNEIHYCGKKSIKNGTVYGIACDDSSTYI 333
GF++ PL CCG G ++ K K G+V C D S Y+
Sbjct: 295 SKFGFMDRPLPACCG--LGGPYNFTLSK--KCGSVGVKYCSDPSKYV 337
>At1g28570 lipase like protein
Length = 389
Score = 105 bits (263), Expect = 3e-23
Identities = 92/345 (26%), Positives = 150/345 (42%), Gaps = 21/345 (6%)
Query: 1 MNTMTLIYILCFFNLCVACPSKKCV-YPAIYNFGDSNSDTGAGYATMAAVEHPN------ 53
M ++ IL F L +C + +I +FGDS +DTG A P
Sbjct: 6 MKLVSFFLILSTFCLTTVNSEPQCHNFKSIISFGDSIADTGNLLALSDPTNLPKVAFLPY 65
Query: 54 GISFFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAV--ASAPIRPI 111
G +FF +GR +GRLI+DFI+E L P + + S +N+ G NFAV A+A R
Sbjct: 66 GETFFHHPTGRFSNGRLIIDFIAEFLGFPLVPPFYGSQNANFEKGVNFAVGGATALERSF 125
Query: 112 IA--GLTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIG 169
+ G+ + VS + S + L + P R + I +IG ND
Sbjct: 126 LEERGIHFPYTNVSLAVQLSSFKESLPNLCVSP----SDCRDMIENSLILMGEIGGNDYN 181
Query: 170 YGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHK 229
Y + EE++ +P ++ + A+ +L + F + P+ C Y Y
Sbjct: 182 YAFFV-GKNIEEIKELVPLVIETISSAITELIGMGGKTFLVPGEFPLGCSVAYLSLYQTS 240
Query: 230 NEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARN 289
N + GC+K N+ ++ ++ QL+ ++ +L++++P Y D Y L
Sbjct: 241 NIEEYDPLTGCLKWLNKFSEYHDEQLQAELNRLQKLYPHVNIIYADYYNTLLRLAQEPAK 300
Query: 290 QGFVN-PLEFCCGSYQGNEIHYCGKKSIKNGTVYGIACDDSSTYI 333
GF++ PL CC G ++ + K GT CDD S Y+
Sbjct: 301 FGFISRPLPACCA--LGGPFNFTLGR--KRGTQVPECCDDPSKYV 341
>At5g15720 unknown protein
Length = 364
Score = 102 bits (253), Expect = 4e-22
Identities = 88/290 (30%), Positives = 135/290 (46%), Gaps = 29/290 (10%)
Query: 27 PAIYNFGDSNSDTGAG--YATMAAVEH-PNGISFFGSISGRCCDGRLILDFISEELELPY 83
PA + FGDS D+G T+A + P GI F G +GR C+GR ++D+ + L LP
Sbjct: 29 PAFFVFGDSLVDSGNNNYIPTLARANYFPYGIDF-GFPTGRFCNGRTVVDYGATYLGLPL 87
Query: 84 LSSYLN--SVGSNYRHGANFAVASAPI-----RPIIAGLTYLGFQVSQFILFKSHTKILF 136
+ YL+ S+G N G N+A A+A I R A T+ G Q+SQF +I
Sbjct: 88 VPPYLSPLSIGQNALRGVNYASAAAGILDETGRHYGARTTFNG-QISQF-------EITI 139
Query: 137 DQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKP---NSSEEEVRRSIPDILSQF 193
+ R ++ + +K+I I+IG ND P ++S+ D+L +
Sbjct: 140 ELRLRRFFQNPADLRKYLAKSIIGINIGSNDYINNYLMPERYSTSQTYSGEDYADLLIKT 199
Query: 194 TQA-VQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYN 252
A + +LYN AR + +GP+ CIP N +GCV N + +N
Sbjct: 200 LSAQISRLYNLGARKMVLAGSGPLGCIPSQLSMVTGNN------TSGCVTKINNMVSMFN 253
Query: 253 RQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGS 302
+LKD L P + F Y +V+ + + ++ N G V E CCG+
Sbjct: 254 SRLKDLANTLNTTLPGSFFVYQNVFDLFHDMVVNPSRYGLVVSNEACCGN 303
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,130,959
Number of Sequences: 26719
Number of extensions: 357564
Number of successful extensions: 1097
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 96
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 752
Number of HSP's gapped (non-prelim): 126
length of query: 341
length of database: 11,318,596
effective HSP length: 100
effective length of query: 241
effective length of database: 8,646,696
effective search space: 2083853736
effective search space used: 2083853736
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC136841.9


