

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.8 + phase: 0 /pseudo
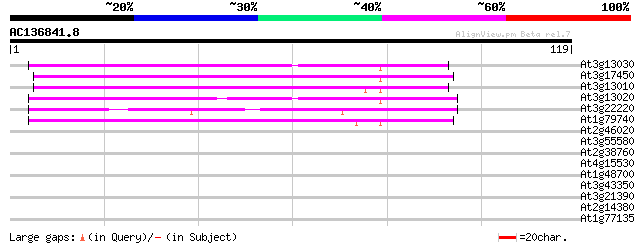
(119 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g13030 unknown protein (At3g13030) 58 9e-10
At3g17450 unknown protein 55 4e-09
At3g13010 hypothetical protein 50 2e-07
At3g13020 hypothetical protein 49 5e-07
At3g22220 hypothetical protein 45 4e-06
At1g79740 hypothetical protein 43 3e-05
At2g46020 putative SNF2 subfamily transcriptional activator 28 0.56
At3g55580 regulator of chromosome condensation-like protein 28 0.95
At2g38760 putative annexin 27 2.1
At4g15530 pyruvate,orthophosphate dikinase 26 2.8
At1g48700 hypothetical protein 25 4.7
At3g43350 putative protein 25 6.2
At3g21390 unknown protein 25 8.1
At2g14380 putative retroelement pol polyprotein 25 8.1
At1g77135 hypothetical protein 25 8.1
>At3g13030 unknown protein (At3g13030)
Length = 544
Score = 57.8 bits (138), Expect = 9e-10
Identities = 35/93 (37%), Positives = 46/93 (48%), Gaps = 5/93 (5%)
Query: 5 LHGPLHAAGYFLNPQFHSSHGFRDDIEGKGGLHDCITRMVADPEERAKIEIQLDDFDKWA 64
LH PLHAAGYFLNP S F DIE GL + MV D + KI Q+D + +
Sbjct: 384 LHNPLHAAGYFLNPTAFYSTNFHLDIEVVTGLISSLIHMVEDCHVQFKISTQIDMY-RLG 442
Query: 65 NDMGYSVAAGNEV----PSVWWSPFGKGLPELQ 93
D + +++ P+ WW+ PELQ
Sbjct: 443 KDCFNEASQADQITGISPAEWWAHKASQYPELQ 475
>At3g17450 unknown protein
Length = 877
Score = 55.5 bits (132), Expect = 4e-09
Identities = 30/92 (32%), Positives = 41/92 (43%), Gaps = 3/92 (3%)
Query: 6 HGPLHAAGYFLNPQFHSSHGFRDDIEGKGGLHDCITRMVADPEERAKIEIQLDDFDKWAN 65
H PL+ A YF NP + F E G+++CI R+ D R +Q+ D+
Sbjct: 646 HHPLYVAAYFFNPAYKYRPDFMAQSEVVRGVNECIVRLEPDNTRRITALMQIPDYTCAKA 705
Query: 66 DMGYSVAAGNEV---PSVWWSPFGKGLPELQK 94
D G +A G PS WW G ELQ+
Sbjct: 706 DFGTDIAIGTRTELDPSAWWQQHGISCLELQR 737
>At3g13010 hypothetical protein
Length = 572
Score = 49.7 bits (117), Expect = 2e-07
Identities = 34/91 (37%), Positives = 42/91 (45%), Gaps = 3/91 (3%)
Query: 6 HGPLHAAGYFLNPQFHSSHGFRDDIEGKGGLHDCITRMVADPEERAKIEIQLDDFDKWAN 65
H PLHAAGYFLNP + S F GL + +V +P + KI QLD +
Sbjct: 424 HNPLHAAGYFLNPMAYYSDDFHTYQHVYTGLAFSLVHLVKEPHLQVKIGTQLDVYRYGRG 483
Query: 66 DMGYSVAAG--NEV-PSVWWSPFGKGLPELQ 93
+ AG N V P WW+ PELQ
Sbjct: 484 CFMKASQAGQLNGVSPVNWWTQKANQYPELQ 514
>At3g13020 hypothetical protein
Length = 605
Score = 48.5 bits (114), Expect = 5e-07
Identities = 33/95 (34%), Positives = 44/95 (45%), Gaps = 7/95 (7%)
Query: 5 LHGPLHAAGYFLNPQFHSSHGFRDDIEGKGGLHDCITRMVADPEERAKIEIQLDDFDKWA 64
LH PLHAAGY+LNP S F D E GL + + E + KI QLD + +
Sbjct: 458 LHNPLHAAGYYLNPTSFYSTDFHLDPEVSSGLTHSLVHVA--KEGQIKIASQLDRY-RLG 514
Query: 65 NDMGYSVAAGNEV----PSVWWSPFGKGLPELQKF 95
D + +++ P WW+ PELQ F
Sbjct: 515 KDCFNEASQPDQISGISPIDWWTEKASQHPELQSF 549
>At3g22220 hypothetical protein
Length = 759
Score = 45.4 bits (106), Expect = 4e-06
Identities = 30/101 (29%), Positives = 50/101 (48%), Gaps = 17/101 (16%)
Query: 5 LHGPLHAAGYFLNPQFHSSHGFRDDIEGKGGLH----DCITRMVADPEERAKIEIQLDDF 60
L PL+AAG++LNP+F + D E + +H DCI ++V D + +I + D
Sbjct: 526 LQQPLYAAGFYLNPKFF----YSIDEEMRSEIHLAVVDCIEKLVPDVNIQ---DIVIKDI 578
Query: 61 DKWANDMGY------SVAAGNEVPSVWWSPFGKGLPELQKF 95
+ + N +G A +P+ WWS +G+ L +F
Sbjct: 579 NSYKNAVGIFGRNLAIRARDTMLPAEWWSTYGESCLNLSRF 619
>At1g79740 hypothetical protein
Length = 518
Score = 42.7 bits (99), Expect = 3e-05
Identities = 28/93 (30%), Positives = 42/93 (45%), Gaps = 3/93 (3%)
Query: 5 LHGPLHAAGYFLNPQFHSSHGFRDDIEGKGGLHDCITRMVADPEERAKIEIQLDDFDKWA 64
LH PLHAA FLNP + + K + +++ + R I Q+ F +
Sbjct: 309 LHSPLHAAAAFLNPSIQYNPEIKFLTSLKEDFFKVLEKLLPTSDLRRDITNQIFTFTRAK 368
Query: 65 NDMGYSVA--AGNEV-PSVWWSPFGKGLPELQK 94
G ++A A + V P +WW FG P LQ+
Sbjct: 369 GMFGCNLAMEARDSVSPGLWWEQFGDSAPVLQR 401
>At2g46020 putative SNF2 subfamily transcriptional activator
Length = 1245
Score = 28.5 bits (62), Expect = 0.56
Identities = 13/52 (25%), Positives = 25/52 (48%), Gaps = 1/52 (1%)
Query: 16 LNPQFHSSHGFRDDIEGKGGLHDCITRMVADPEERAKIEIQLDDFDKWANDM 67
L H +++ + LH+ + RM+A EE ++ Q+D+ W +M
Sbjct: 560 LETLLHDEERYQETVHDVPSLHE-VNRMIARSEEEVELFDQMDEEFDWTEEM 610
>At3g55580 regulator of chromosome condensation-like protein
Length = 488
Score = 27.7 bits (60), Expect = 0.95
Identities = 18/59 (30%), Positives = 28/59 (46%)
Query: 22 SSHGFRDDIEGKGGLHDCITRMVADPEERAKIEIQLDDFDKWANDMGYSVAAGNEVPSV 80
SS G +EG+GG R ++ ++ A+ Q D+ D A S+A G + SV
Sbjct: 181 SSGGTSSQVEGRGGGEPTKKRRISPSKQAAENSSQSDNIDLSALPCLVSLAPGVRIVSV 239
>At2g38760 putative annexin
Length = 321
Score = 26.6 bits (57), Expect = 2.1
Identities = 14/54 (25%), Positives = 27/54 (49%)
Query: 18 PQFHSSHGFRDDIEGKGGLHDCITRMVADPEERAKIEIQLDDFDKWANDMGYSV 71
P+ H + RD IEG G D +TR + E ++++ + F+ + M ++
Sbjct: 248 PEKHFAKVVRDSIEGFGTDEDSLTRAIVTRAEIDLMKVRGEYFNMYNTSMDNAI 301
>At4g15530 pyruvate,orthophosphate dikinase
Length = 960
Score = 26.2 bits (56), Expect = 2.8
Identities = 9/19 (47%), Positives = 13/19 (68%)
Query: 16 LNPQFHSSHGFRDDIEGKG 34
L+PQFH G+R+ + KG
Sbjct: 448 LHPQFHDPSGYREKVVAKG 466
>At1g48700 hypothetical protein
Length = 286
Score = 25.4 bits (54), Expect = 4.7
Identities = 7/25 (28%), Positives = 18/25 (72%)
Query: 47 PEERAKIEIQLDDFDKWANDMGYSV 71
P+ K+ +++++F KWAN+ +++
Sbjct: 90 PDFSEKLLLEVENFRKWANETNFTI 114
>At3g43350 putative protein
Length = 830
Score = 25.0 bits (53), Expect = 6.2
Identities = 12/27 (44%), Positives = 17/27 (62%), Gaps = 2/27 (7%)
Query: 42 RMVADPEERAKIEIQLDDFDKWANDMG 68
R++AD E K +I+ DF KW D+G
Sbjct: 362 RLLADISEHEKRDIE--DFSKWILDVG 386
>At3g21390 unknown protein
Length = 335
Score = 24.6 bits (52), Expect = 8.1
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 2/33 (6%)
Query: 34 GGLHDCITRMVADPEERAKI--EIQLDDFDKWA 64
GG+ I+RMV P + KI ++QL+ WA
Sbjct: 22 GGVAGAISRMVTSPLDVIKIRFQVQLEPTATWA 54
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 24.6 bits (52), Expect = 8.1
Identities = 13/43 (30%), Positives = 24/43 (55%)
Query: 51 AKIEIQLDDFDKWANDMGYSVAAGNEVPSVWWSPFGKGLPELQ 93
A++E +LD + + G+S AA + S+ + + KG E+Q
Sbjct: 177 AEVENRLDQYCDFHKRSGHSTAACRHLQSILLNKYKKGDIEVQ 219
>At1g77135 hypothetical protein
Length = 585
Score = 24.6 bits (52), Expect = 8.1
Identities = 12/37 (32%), Positives = 17/37 (45%)
Query: 59 DFDKWANDMGYSVAAGNEVPSVWWSPFGKGLPELQKF 95
D+D N +GY A +E WW +LQ+F
Sbjct: 478 DYDCNWNVVGYHQFASDEAHKTWWRVHDAMPKKLQRF 514
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.333 0.149 0.525
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,093,276
Number of Sequences: 26719
Number of extensions: 127501
Number of successful extensions: 271
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 255
Number of HSP's gapped (non-prelim): 15
length of query: 119
length of database: 11,318,596
effective HSP length: 95
effective length of query: 24
effective length of database: 8,780,291
effective search space: 210726984
effective search space used: 210726984
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC136841.8


