

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.7 + phase: 0 /pseudo
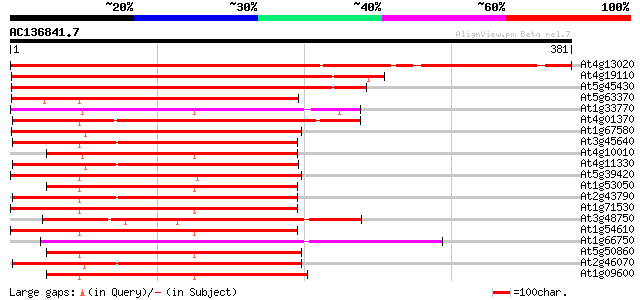
(381 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g13020 serine/threonine-specific protein kinase MHK 417 e-117
At4g19110 putative serine/threonine-protein kinase 295 3e-80
At5g45430 serine/threonine-protein kinase Mak (male germ cell-as... 292 2e-79
At5g63370 protein kinase 184 7e-47
At1g33770 protein kinase, putative 170 1e-42
At4g01370 MAP kinase 4 163 2e-40
At1g67580 putative protein kinase 163 2e-40
At3g45640 mitogen-activated protein kinase 3 161 6e-40
At4g10010 protein kinase like protein 160 8e-40
At4g11330 MAP kinase (ATMPK5) 159 2e-39
At5g39420 cyclin-dependent kinase CDC2C (CDC2CAt) 157 1e-38
At1g53050 cell division-related protein, putative 157 1e-38
At2g43790 MAP kinase (ATMPK6) 156 2e-38
At1g71530 protein kinase-like 156 2e-38
At3g48750 protein kinase (cdc2) 156 2e-38
At1g54610 Unknown protein (T22H22.5) 155 3e-38
At1g66750 unknown protein 155 3e-38
At5g50860 cyclin-dependent protein kinase-like 154 1e-37
At2g46070 mitogen-activated protein kinase like protein 154 1e-37
At1g09600 putative protein kinase 154 1e-37
>At4g13020 serine/threonine-specific protein kinase MHK
Length = 435
Score = 417 bits (1072), Expect = e-117
Identities = 218/382 (57%), Positives = 275/382 (71%), Gaps = 15/382 (3%)
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMD 60
+ T+E+VAVK++KRKF +WEE NLRE+KALRK+NH +IIKL+E+VRE+NELFFIFE MD
Sbjct: 24 LETYEVVAVKKMKRKFYYWEECVNLREVKALRKLNHPHIIKLKEIVREHNELFFIFECMD 83
Query: 61 CNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADF 120
NLY ++KERE+PFSE EIR FM QMLQGL+HMHK G+FHRDLKPENLLVTN++LKIADF
Sbjct: 84 HNLYHIMKERERPFSEGEIRSFMSQMLQGLAHMHKNGYFHRDLKPENLLVTNNILKIADF 143
Query: 121 GLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGES 180
GLAREV+SMPPYT+YVSTRWYRAPEVLLQS YTPAVDMWA+GAILAEL+ LTP+FPGES
Sbjct: 144 GLAREVASMPPYTEYVSTRWYRAPEVLLQSSLYTPAVDMWAVGAILAELYALTPLFPGES 203
Query: 181 EIDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITA 240
EIDQ+YKI C+LG PD T F + SR++ + H +++D++PNA+ EAIDLI
Sbjct: 204 EIDQLYKICCVLGKPDWTTFPEAKSISRIMS-ISHTEFPQTRIADLLPNAAPEAIDLINR 262
Query: 241 RCGSVTATSFFPSCSILHKVSNFLTYTEQVNTRVPRSLSDPLELKLSNKRVKPNLELKLH 300
C ++ H F + Q + + LEL+L N PNLEL L
Sbjct: 263 LCSWDPLKRPTADEALNHP---FFSMATQASYPI-----HDLELRLDNMAALPNLELNLW 314
Query: 301 DFGPDPDDCFLGLTLAVKPSVSNLDVVQNARQGMGENLV*LDAL*KCRICYFARILMTIQ 360
DF +P++CFLGLTLAVKPS L++++N Q M EN + + R L++
Sbjct: 315 DFNREPEECFLGLTLAVKPSAPKLEMLRNVSQDMSENFLFCPGVNNDREPSVFWSLLS-- 372
Query: 361 TNQPDQNGVHSSAETS-LSLSF 381
PD+NG+H+ E+S LSLSF
Sbjct: 373 ---PDENGLHAPVESSPLSLSF 391
>At4g19110 putative serine/threonine-protein kinase
Length = 461
Score = 295 bits (755), Expect = 3e-80
Identities = 143/263 (54%), Positives = 198/263 (74%), Gaps = 11/263 (4%)
Query: 2 RTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDC 61
+T E+VA+K++K+K+ W+E NLRE+K+LR+MNH NI+KL+EV+REN+ L+F+FEYM+C
Sbjct: 25 QTGEVVAIKKMKKKYYSWDECINLREVKSLRRMNHPNIVKLKEVIRENDILYFVFEYMEC 84
Query: 62 NLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFG 121
NLYQL+K+R+K F+E +I+ + Q+ QGLS+MH++G+FHRDLKPENLLV+ D++KIADFG
Sbjct: 85 NLYQLMKDRQKLFAEADIKNWCFQVFQGLSYMHQRGYFHRDLKPENLLVSKDIIKIADFG 144
Query: 122 LAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESE 181
LAREV+S PP+T+YVSTRWYRAPEVLLQS YT VDMWA+GAI+AEL +L PIFPG SE
Sbjct: 145 LAREVNSSPPFTEYVSTRWYRAPEVLLQSYVYTSKVDMWAMGAIMAELLSLRPIFPGASE 204
Query: 182 IDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITAR 241
D++YKI ++G P + G N + +++ ++ V LS ++P+AS +AI+LI
Sbjct: 205 ADEIYKICSVIGTPTEETWLEGLNLANTINYQFPQLPG-VPLSSLMPSASEDAINLIERL 263
Query: 242 C----------GSVTATSFFPSC 254
C V FF SC
Sbjct: 264 CSWDPSSRPTAAEVLQHPFFQSC 286
>At5g45430 serine/threonine-protein kinase Mak (male germ
cell-associated kinase)-like protein
Length = 499
Score = 292 bits (748), Expect = 2e-79
Identities = 138/241 (57%), Positives = 188/241 (77%), Gaps = 1/241 (0%)
Query: 2 RTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDC 61
+T E+VA+KR+K+K+ WEE NLRE+K+L +MNH NI+KL+EV+REN+ L+F+FEYM+C
Sbjct: 25 QTNEVVAIKRMKKKYFSWEECVNLREVKSLSRMNHPNIVKLKEVIRENDILYFVFEYMEC 84
Query: 62 NLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFG 121
NLYQL+K+R K F+E +IR + Q+ QGLS+MH++G+FHRDLKPENLLV+ DV+KIAD G
Sbjct: 85 NLYQLMKDRPKHFAESDIRNWCFQVFQGLSYMHQRGYFHRDLKPENLLVSKDVIKIADLG 144
Query: 122 LAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESE 181
LARE+ S PPYT+YVSTRWYRAPEVLLQS YT VDMWA+GAI+AEL +L P+FPG SE
Sbjct: 145 LAREIDSSPPYTEYVSTRWYRAPEVLLQSYVYTSKVDMWAMGAIMAELLSLRPLFPGASE 204
Query: 182 IDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITAR 241
D++YKI ++G P + G N + ++++ + V LS ++P AS +A++LI
Sbjct: 205 ADEIYKICSVIGSPTEETWLEGLNLASVINYQFPQFPG-VHLSSVMPYASADAVNLIERL 263
Query: 242 C 242
C
Sbjct: 264 C 264
>At5g63370 protein kinase
Length = 612
Score = 184 bits (467), Expect = 7e-47
Identities = 95/205 (46%), Positives = 141/205 (68%), Gaps = 10/205 (4%)
Query: 2 RTFEIVAVKRLKRKFC-FWEEY----TNLREIKALRKMNHQNIIKLREVV---RENNELF 53
+T EIVA+K++K K F EEY T+LREI L NH I+ ++EVV + +N+++
Sbjct: 318 KTKEIVALKKIKMKEDRFEEEYGFPLTSLREINILLSCNHPAIVNVKEVVVGGKNDNDVY 377
Query: 54 FIFEYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN- 112
+ E+++ +L ++ R++PFS E++C M Q+L GL ++H HRDLKP NLL+ N
Sbjct: 378 MVMEHLEHDLRGVMDRRKEPFSTSEVKCLMMQLLDGLKYLHTNWIIHRDLKPSNLLMNNC 437
Query: 113 DVLKIADFGLAREVSS-MPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFT 171
LKI DFG+AR+ S + PYTQ V T+WYR PE+LL + Y+ AVDMW++G I+AEL +
Sbjct: 438 GELKICDFGMARQYGSPIKPYTQMVITQWYRPPELLLGAKEYSTAVDMWSVGCIMAELLS 497
Query: 172 LTPIFPGESEIDQMYKIYCILGMPD 196
P+FPG+SE+DQ+ KI+ +LG P+
Sbjct: 498 QKPLFPGKSELDQLQKIFAVLGTPN 522
>At1g33770 protein kinase, putative
Length = 614
Score = 170 bits (430), Expect = 1e-42
Identities = 101/246 (41%), Positives = 145/246 (58%), Gaps = 12/246 (4%)
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNL-REIKALRKMNHQNIIKLREVVRE--NNELFFIFE 57
+ T +IVA+K+++ E + REI LRK++H N++KL+ +V + L +FE
Sbjct: 161 LETGKIVAMKKVRFANMDPESVRFMAREINILRKLDHPNVMKLQCLVTSKLSGSLHLVFE 220
Query: 58 YMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLK 116
YM+ +L L F+E +I+CFMKQ+L GL H H +G HRD+K NLLV ND VLK
Sbjct: 221 YMEHDLSGLALRPGVKFTEPQIKCFMKQLLCGLEHCHSRGILHRDIKGSNLLVNNDGVLK 280
Query: 117 IADFGLAR--EVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTP 174
I DFGLA + P T V T WYRAPE+LL S Y PA+D+W++G ILAELF P
Sbjct: 281 IGDFGLASFYKPDQDQPLTSRVVTLWYRAPELLLGSTEYGPAIDLWSVGCILAELFVCKP 340
Query: 175 IFPGESEIDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVK--LSDIIPNASM 232
I PG +E++QM+KI+ + G P + N ++ ++ P K L + N S
Sbjct: 341 IMPGRTEVEQMHKIFKLCGSPSEEFW----NTTKFPQATSYKPQHPYKRVLLETFKNLSS 396
Query: 233 EAIDLI 238
++DL+
Sbjct: 397 SSLDLL 402
>At4g01370 MAP kinase 4
Length = 376
Score = 163 bits (412), Expect = 2e-40
Identities = 96/245 (39%), Positives = 150/245 (61%), Gaps = 12/245 (4%)
Query: 3 TFEIVAVKRLKRKFC-FWEEYTNLREIKALRKMNHQNIIKLREVV----REN-NELFFIF 56
T E VA+K++ F + LREIK L+ M+H+N+I +++++ REN N+++ ++
Sbjct: 65 TGEEVAIKKIGNAFDNIIDAKRTLREIKLLKHMDHENVIAVKDIIKPPQRENFNDVYIVY 124
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVL 115
E MD +L+Q+I+ + P +++ R F+ Q+L+GL ++H HRDLKP NLL+ N L
Sbjct: 125 ELMDTDLHQIIRSNQ-PLTDDHCRFFLYQLLRGLKYVHSANVLHRDLKPSNLLLNANCDL 183
Query: 116 KIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPI 175
K+ DFGLAR S T+YV TRWYRAPE+LL YT A+D+W++G IL E T P+
Sbjct: 184 KLGDFGLARTKSETDFMTEYVVTRWYRAPELLLNCSEYTAAIDIWSVGCILGETMTREPL 243
Query: 176 FPGESEIDQMYKIYCILGMP-DSTCFTIGANNSRLLDFVGHEVVAP-VKLSDIIPNASME 233
FPG+ + Q+ I ++G P DS+ + ++N+R +V P + PN S
Sbjct: 244 FPGKDYVHQLRLITELIGSPDDSSLGFLRSDNAR--RYVRQLPQYPRQNFAARFPNMSAG 301
Query: 234 AIDLI 238
A+DL+
Sbjct: 302 AVDLL 306
>At1g67580 putative protein kinase
Length = 752
Score = 163 bits (412), Expect = 2e-40
Identities = 88/202 (43%), Positives = 133/202 (65%), Gaps = 5/202 (2%)
Query: 2 RTFEIVAVKRLK-RKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENN--ELFFIFEY 58
+T EIVA+K++K K T+LREI L +H +I+ ++EVV ++ +F + EY
Sbjct: 427 KTGEIVALKKVKMEKEREGFPLTSLREINILLSFHHPSIVDVKEVVVGSSLDSIFMVMEY 486
Query: 59 MDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKI 117
M+ +L L++ ++ FS+ E++C M Q+L+G+ ++H HRDLK NLL+ N LKI
Sbjct: 487 MEHDLKALMETMKQRFSQSEVKCLMLQLLEGVKYLHDNWVLHRDLKTSNLLLNNRGELKI 546
Query: 118 ADFGLAREVSS-MPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIF 176
DFGLAR+ S + PYT V T WYRAPE+LL + Y+ A+DMW++G I+AEL P+F
Sbjct: 547 CDFGLARQYGSPLKPYTHLVVTLWYRAPELLLGAKQYSTAIDMWSLGCIMAELLMKAPLF 606
Query: 177 PGESEIDQMYKIYCILGMPDST 198
G++E DQ+ KI+ ILG P+ +
Sbjct: 607 NGKTEFDQLDKIFRILGTPNES 628
>At3g45640 mitogen-activated protein kinase 3
Length = 370
Score = 161 bits (407), Expect = 6e-40
Identities = 88/200 (44%), Positives = 126/200 (63%), Gaps = 8/200 (4%)
Query: 3 TFEIVAVKRLKRKFC-FWEEYTNLREIKALRKMNHQNIIKLREVV-----RENNELFFIF 56
T E+VA+K++ F + LREIK LR ++H+NII +R+VV R+ ++++
Sbjct: 60 TNELVAMKKIANAFDNHMDAKRTLREIKLLRHLDHENIIAIRDVVPPPLRRQFSDVYIST 119
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVL 115
E MD +L+Q+I+ + SEE + F+ Q+L+GL ++H HRDLKP NLL+ N L
Sbjct: 120 ELMDTDLHQIIRSNQS-LSEEHCQYFLYQLLRGLKYIHSANIIHRDLKPSNLLLNANCDL 178
Query: 116 KIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPI 175
KI DFGLAR S T+YV TRWYRAPE+LL S YT A+D+W++G I EL P+
Sbjct: 179 KICDFGLARPTSENDFMTEYVVTRWYRAPELLLNSSDYTAAIDVWSVGCIFMELMNRKPL 238
Query: 176 FPGESEIDQMYKIYCILGMP 195
FPG+ + QM + +LG P
Sbjct: 239 FPGKDHVHQMRLLTELLGTP 258
>At4g10010 protein kinase like protein
Length = 649
Score = 160 bits (406), Expect = 8e-40
Identities = 83/175 (47%), Positives = 111/175 (63%), Gaps = 5/175 (2%)
Query: 26 REIKALRKMNHQNIIKLREVVRE--NNELFFIFEYMDCNLYQLIKEREKPFSEEEIRCFM 83
REI LRK++H N++KL +V + L+ +FEYM+ +L L F+E +I+C+M
Sbjct: 202 REINILRKLDHPNVMKLECLVTSKLSGSLYLVFEYMEHDLSGLALRPGVKFTESQIKCYM 261
Query: 84 KQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLKIADFGLAR--EVSSMPPYTQYVSTRW 140
KQ+L GL H H +G HRD+K NLLV ND VLKI DFGLA P T V T W
Sbjct: 262 KQLLSGLEHCHSRGILHRDIKGPNLLVNNDGVLKIGDFGLANIYHPEQDQPLTSRVVTLW 321
Query: 141 YRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMP 195
YRAPE+LL + Y P +D+W++G IL ELF PI PG +E++QM+KI+ G P
Sbjct: 322 YRAPELLLGATEYGPGIDLWSVGCILTELFLGKPIMPGRTEVEQMHKIFKFCGSP 376
>At4g11330 MAP kinase (ATMPK5)
Length = 373
Score = 159 bits (403), Expect = 2e-39
Identities = 80/201 (39%), Positives = 129/201 (63%), Gaps = 8/201 (3%)
Query: 3 TFEIVAVKRLKRKFCFW-EEYTNLREIKALRKMNHQNIIKLREVVRENN-----ELFFIF 56
T E +A+K++ + F + LREIK LR + H+N++ +++++R +++ +F
Sbjct: 62 THEEIAIKKIGKAFDNKVDAKRTLREIKLLRHLEHENVVVIKDIIRPPKKEDFVDVYIVF 121
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDV-L 115
E MD +L+Q+I+ + +++ + F+ Q+L+GL ++H HRDLKP NLL+ ++ L
Sbjct: 122 ELMDTDLHQIIRSNQS-LNDDHCQYFLYQILRGLKYIHSANVLHRDLKPSNLLLNSNCDL 180
Query: 116 KIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPI 175
KI DFGLAR S T+YV TRWYRAPE+LL S YT A+D+W++G I AE+ T P+
Sbjct: 181 KITDFGLARTTSETEYMTEYVVTRWYRAPELLLNSSEYTSAIDVWSVGCIFAEIMTREPL 240
Query: 176 FPGESEIDQMYKIYCILGMPD 196
FPG+ + Q+ I ++G PD
Sbjct: 241 FPGKDYVHQLKLITELIGSPD 261
>At5g39420 cyclin-dependent kinase CDC2C (CDC2CAt)
Length = 644
Score = 157 bits (396), Expect = 1e-38
Identities = 86/204 (42%), Positives = 128/204 (62%), Gaps = 6/204 (2%)
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNL-REIKALRKMNHQNIIKLREVV--RENNELFFIFE 57
+ T ++VA+K++K E + REI LRK+NH NI+KL +V R ++ ++ +FE
Sbjct: 125 VETGKMVALKKVKFDNLQPESIRFMAREILILRKLNHPNIMKLEGIVTSRASSSIYLVFE 184
Query: 58 YMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLK 116
YM+ +L L + F+E +I+C+MKQ+L GL H H +G HRD+K N+LV N VLK
Sbjct: 185 YMEHDLAGLSSNPDIRFTEPQIKCYMKQLLWGLEHCHMRGVIHRDIKASNILVNNKGVLK 244
Query: 117 IADFGLAREV--SSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTP 174
+ DFGLA V S+ T V T WYRAPE+L+ S Y +VD+W++G + AE+ P
Sbjct: 245 LGDFGLANVVTPSNKNQLTSRVVTLWYRAPELLMGSTSYGVSVDLWSVGCVFAEILMGKP 304
Query: 175 IFPGESEIDQMYKIYCILGMPDST 198
I G +EI+Q++KIY + G P +
Sbjct: 305 ILKGRTEIEQLHKIYKLCGSPQDS 328
>At1g53050 cell division-related protein, putative
Length = 694
Score = 157 bits (396), Expect = 1e-38
Identities = 81/175 (46%), Positives = 114/175 (64%), Gaps = 5/175 (2%)
Query: 26 REIKALRKMNHQNIIKLREVV--RENNELFFIFEYMDCNLYQLIKEREKPFSEEEIRCFM 83
REI+ LR+++H NIIKL +V R + L+ +FEYM+ +L L FSE +++C++
Sbjct: 180 REIQILRRLDHPNIIKLEGLVTSRMSCSLYLVFEYMEHDLAGLASHPAIKFSESQVKCYL 239
Query: 84 KQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLKIADFGLAR--EVSSMPPYTQYVSTRW 140
+Q+L GL H H +G HRD+K NLL+ N VLKIADFGLA + P T V T W
Sbjct: 240 QQLLHGLDHCHSRGVLHRDIKGSNLLIDNSGVLKIADFGLASFFDPRQTQPLTSRVVTLW 299
Query: 141 YRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMP 195
YR PE+LL + Y AVD+W+ G ILAEL+ PI PG +E++Q++KI+ + G P
Sbjct: 300 YRPPELLLGATRYGAAVDLWSAGCILAELYAGKPIMPGRTEVEQLHKIFKLCGSP 354
>At2g43790 MAP kinase (ATMPK6)
Length = 395
Score = 156 bits (395), Expect = 2e-38
Identities = 84/200 (42%), Positives = 123/200 (61%), Gaps = 8/200 (4%)
Query: 3 TFEIVAVKRLKRKFCFW-EEYTNLREIKALRKMNHQNIIKLREVV-----RENNELFFIF 56
T E VA+K++ F + LREIK LR M+H+NI+ +R+++ N+++ +
Sbjct: 85 TNESVAIKKIANAFDNKIDAKRTLREIKLLRHMDHENIVAIRDIIPPPLRNAFNDVYIAY 144
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVL 115
E MD +L+Q+I+ + SEE + F+ Q+L+GL ++H HRDLKP NLL+ N L
Sbjct: 145 ELMDTDLHQIIRSNQA-LSEEHCQYFLYQILRGLKYIHSANVLHRDLKPSNLLLNANCDL 203
Query: 116 KIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPI 175
KI DFGLAR S T+YV TRWYRAPE+LL S YT A+D+W++G I EL P+
Sbjct: 204 KICDFGLARVTSESDFMTEYVVTRWYRAPELLLNSSDYTAAIDVWSVGCIFMELMDRKPL 263
Query: 176 FPGESEIDQMYKIYCILGMP 195
FPG + Q+ + ++G P
Sbjct: 264 FPGRDHVHQLRLLMELIGTP 283
>At1g71530 protein kinase-like
Length = 655
Score = 156 bits (395), Expect = 2e-38
Identities = 87/201 (43%), Positives = 126/201 (62%), Gaps = 6/201 (2%)
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNL-REIKALRKMNHQNIIKLREVV--RENNELFFIFE 57
+ T +IVA+K+++ E + REI LRK++H N++KL +V R + L+ +FE
Sbjct: 167 LETGKIVAMKKVRFVNMDPESVRFMAREILILRKLDHPNVMKLEGLVTSRLSGSLYLVFE 226
Query: 58 YMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLK 116
YM+ +L L FSE +I+C+M+Q+ +GL H H++G HRD+K NLL+ N+ VLK
Sbjct: 227 YMEHDLAGLAATPGIKFSEPQIKCYMQQLFRGLEHCHRRGILHRDIKGSNLLINNEGVLK 286
Query: 117 IADFGLAR--EVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTP 174
I DFGLA T V T WYRAPE+LL + Y PA+D+W+ G IL ELF P
Sbjct: 287 IGDFGLANFYRGDGDLQLTSRVVTLWYRAPELLLGATEYGPAIDLWSAGCILTELFAGKP 346
Query: 175 IFPGESEIDQMYKIYCILGMP 195
I PG +E++QM+KI+ + G P
Sbjct: 347 IMPGRTEVEQMHKIFKLCGSP 367
>At3g48750 protein kinase (cdc2)
Length = 294
Score = 156 bits (394), Expect = 2e-38
Identities = 86/223 (38%), Positives = 136/223 (60%), Gaps = 10/223 (4%)
Query: 23 TNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLYQLIKEREKPFSEE--EIR 80
T +REI L++M H NI+KL++VV L+ +FEY+D +L + + + FS++ I+
Sbjct: 47 TAIREISLLKEMQHSNIVKLQDVVHSEKRLYLVFEYLDLDLKKHM-DSTPDFSKDLHMIK 105
Query: 81 CFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN--DVLKIADFGLAREVS-SMPPYTQYVS 137
++ Q+L+G+++ H HRDLKP+NLL+ + LK+ADFGLAR + +T V
Sbjct: 106 TYLYQILRGIAYCHSHRVLHRDLKPQNLLIDRRTNSLKLADFGLARAFGIPVRTFTHEVV 165
Query: 138 TRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPDS 197
T WYRAPE+LL S Y+ VD+W++G I AE+ + P+FPG+SEIDQ++KI+ I+G P
Sbjct: 166 TLWYRAPEILLGSHHYSTPVDIWSVGCIFAEMISQKPLFPGDSEIDQLFKIFRIMGTPYE 225
Query: 198 TCFTIGANNSRLLDF-VGHEVVAPVKLSDIIPNASMEAIDLIT 239
+ + L D+ P L +PN + +DL++
Sbjct: 226 DTW---RGVTSLPDYKSAFPKWKPTDLETFVPNLDPDGVDLLS 265
>At1g54610 Unknown protein (T22H22.5)
Length = 572
Score = 155 bits (393), Expect = 3e-38
Identities = 86/201 (42%), Positives = 125/201 (61%), Gaps = 6/201 (2%)
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNL-REIKALRKMNHQNIIKLREVV--RENNELFFIFE 57
M T +IVA+K+++ E + REI LR+++H N++KL +V R + L+ +F+
Sbjct: 138 MLTGKIVALKKVRFDNLEPESVKFMAREILVLRRLDHPNVVKLEGLVTSRMSCSLYLVFQ 197
Query: 58 YMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLK 116
YMD +L L FSE E++C M+Q++ GL H H +G HRD+K NLL+ + VLK
Sbjct: 198 YMDHDLAGLASSPVVKFSESEVKCLMRQLISGLEHCHSRGVLHRDIKGSNLLIDDGGVLK 257
Query: 117 IADFGLAR--EVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTP 174
IADFGLA + + P T V T WYRAPE+LL + Y +D+W+ G ILAEL P
Sbjct: 258 IADFGLATIFDPNHKRPMTSRVVTLWYRAPELLLGATDYGVGIDLWSAGCILAELLAGRP 317
Query: 175 IFPGESEIDQMYKIYCILGMP 195
I PG +E++Q++KIY + G P
Sbjct: 318 IMPGRTEVEQLHKIYKLCGSP 338
>At1g66750 unknown protein
Length = 348
Score = 155 bits (392), Expect = 3e-38
Identities = 100/276 (36%), Positives = 147/276 (53%), Gaps = 6/276 (2%)
Query: 22 YTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLYQLIKEREKPFSEEEIRC 81
+T LREIK L+++NH +I++L + + L +FEYM +L +I++R S +I+
Sbjct: 55 FTALREIKLLKELNHPHIVELIDAFPHDGSLHLVFEYMQTDLEAVIRDRNIFLSPGDIKS 114
Query: 82 FMKQMLQGLSHMHKKGFFHRDLKPENLLV-TNDVLKIADFGLAREVSS-MPPYTQYVSTR 139
+M L+GL++ HKK HRD+KP NLL+ N +LK+ADFGLAR S +T V
Sbjct: 115 YMLMTLKGLAYCHKKWVLHRDMKPNNLLIGENGLLKLADFGLARLFGSPNRRFTHQVFAT 174
Query: 140 WYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPDSTC 199
WYRAPE+L S Y VD+WA G I AEL P PG +EIDQ+ KI+ G P +
Sbjct: 175 WYRAPELLFGSRQYGAGVDVWAAGCIFAELLLRRPFLPGSTEIDQLGKIFQAFGTPVPSQ 234
Query: 200 FTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITARCGSVTATSFFPSCSILHK 259
+ ++ L D++ L I P AS +A+DL+ ++ H+
Sbjct: 235 W---SDMIYLPDYMEFSYTPAPPLRTIFPMASDDALDLLAKMFIYDPRQRITIQQALDHR 291
Query: 260 -VSNFLTYTEQVNTRVPRSLSDPLELKLSNKRVKPN 294
S+ + TE ++P S D LE K S + N
Sbjct: 292 YFSSSPSPTEPGKLQIPASKGDALEPKASEQNQHGN 327
>At5g50860 cyclin-dependent protein kinase-like
Length = 580
Score = 154 bits (388), Expect = 1e-37
Identities = 78/178 (43%), Positives = 112/178 (62%), Gaps = 5/178 (2%)
Query: 26 REIKALRKMNHQNIIKLREVV--RENNELFFIFEYMDCNLYQLIKEREKPFSEEEIRCFM 83
REI LR++NH N+IKL+ +V R + L+ +FEYM+ +L L + F +++CFM
Sbjct: 160 REILVLRRLNHPNVIKLQGLVTSRVSCSLYLVFEYMEHDLSGLAATQGLKFDLPQVKCFM 219
Query: 84 KQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLKIADFGLAR--EVSSMPPYTQYVSTRW 140
KQ+L GL H H +G HRD+K NLL+ ND +LKIADFGLA + T V T W
Sbjct: 220 KQLLSGLEHCHSRGVLHRDIKGSNLLIDNDGILKIADFGLATFYDPKQKQTMTSRVVTLW 279
Query: 141 YRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPDST 198
YR PE+LL + Y VD+W+ G I+AEL P+ PG +E++Q++KI+ + G P +
Sbjct: 280 YRPPELLLGATSYGTGVDLWSAGCIMAELLAGKPVMPGRTEVEQLHKIFKLCGSPSDS 337
>At2g46070 mitogen-activated protein kinase like protein
Length = 372
Score = 154 bits (388), Expect = 1e-37
Identities = 79/203 (38%), Positives = 129/203 (62%), Gaps = 8/203 (3%)
Query: 3 TFEIVAVKRLKRKFC-FWEEYTNLREIKALRKMNHQNIIKLREVVREN-----NELFFIF 56
T E VA+K++ F + LREIK LR M+H+N+I ++++VR N+++ ++
Sbjct: 63 TGEKVAIKKIGNAFDNIIDAKRTLREIKLLRHMDHENVITIKDIVRPPQRDIFNDVYIVY 122
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVL 115
E MD +L ++++ + + ++ R + Q+L+GL ++H HRDL+P N+L+ + + L
Sbjct: 123 ELMDTDLQRILRSNQT-LTSDQCRFLVYQLLRGLKYVHSANILHRDLRPSNVLLNSKNEL 181
Query: 116 KIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPI 175
KI DFGLAR S T+YV TRWYRAPE+LL YT A+D+W++G IL E+ T P+
Sbjct: 182 KIGDFGLARTTSDTDFMTEYVVTRWYRAPELLLNCSEYTAAIDIWSVGCILGEIMTGQPL 241
Query: 176 FPGESEIDQMYKIYCILGMPDST 198
FPG+ + Q+ I ++G PD++
Sbjct: 242 FPGKDYVHQLRLITELVGSPDNS 264
>At1g09600 putative protein kinase
Length = 714
Score = 154 bits (388), Expect = 1e-37
Identities = 82/182 (45%), Positives = 114/182 (62%), Gaps = 5/182 (2%)
Query: 26 REIKALRKMNHQNIIKLREVV--RENNELFFIFEYMDCNLYQLIKEREKPFSEEEIRCFM 83
REI LR+++H N++KL ++ R + ++ IFEYM+ +L L FSE +I+C+M
Sbjct: 209 REIIILRRLDHPNVMKLEGLITSRVSGSMYLIFEYMEHDLAGLASTPGINFSEAQIKCYM 268
Query: 84 KQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVLKIADFGLAR--EVSSMPPYTQYVSTRW 140
KQ+L GL H H +G HRD+K NLL+ N+ LKI DFGLA + P T V T W
Sbjct: 269 KQLLHGLEHCHSRGVLHRDIKGSNLLLDHNNNLKIGDFGLANFYQGHQKQPLTSRVVTLW 328
Query: 141 YRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPDSTCF 200
YR PE+LL S Y VD+W+ G ILAELFT PI PG +E++Q++KI+ + G P +
Sbjct: 329 YRPPELLLGSTDYGVTVDLWSTGCILAELFTGKPIMPGRTEVEQLHKIFKLCGSPSEEYW 388
Query: 201 TI 202
I
Sbjct: 389 KI 390
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,308,037
Number of Sequences: 26719
Number of extensions: 348154
Number of successful extensions: 3099
Number of sequences better than 10.0: 924
Number of HSP's better than 10.0 without gapping: 463
Number of HSP's successfully gapped in prelim test: 461
Number of HSP's that attempted gapping in prelim test: 1470
Number of HSP's gapped (non-prelim): 1086
length of query: 381
length of database: 11,318,596
effective HSP length: 101
effective length of query: 280
effective length of database: 8,619,977
effective search space: 2413593560
effective search space used: 2413593560
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136841.7


