

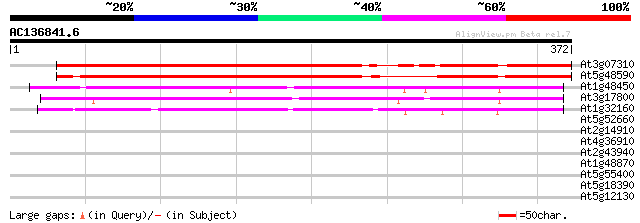
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.6 + phase: 0
(372 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g07310 unknown protein 328 2e-90
At5g48590 unknown protein 292 2e-79
At1g48450 unknown protein 180 1e-45
At3g17800 unknown protein 171 6e-43
At1g32160 unknown protein 157 9e-39
At5g52660 unknown protein 32 0.56
At2g14910 unknown protein 30 1.6
At4g36910 unknown protein 29 3.6
At2g43940 unknown protein 29 4.7
At1g48870 hypothetical protein 29 4.7
At5g55400 fimbrin 28 8.1
At5g18390 unknown protein 28 8.1
At5g12130 unknown protein 28 8.1
>At3g07310 unknown protein
Length = 368
Score = 328 bits (842), Expect = 2e-90
Identities = 182/343 (53%), Positives = 238/343 (69%), Gaps = 30/343 (8%)
Query: 32 LIKASAGASSHCE-SSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLADDRDA 90
++ +A S CE SSLN PL PR+ G+FL VL N R LFH A +ELK LADDR+A
Sbjct: 42 MVVVAAAGQSRCEPGSSLNAPLEPRSAQGRFLRSVLLNKRQLFHYAAADELKQLADDREA 101
Query: 91 ANSRMLLASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLSRCLY 150
A +RM L+S SDEA LHRRIAE+KE C+ AV+DIM +LIF+K+SEIR PLVPKLSRC+Y
Sbjct: 102 ALARMSLSSGSDEASLHRRIAELKERYCKTAVQDIMYMLIFYKYSEIRVPLVPKLSRCIY 161
Query: 151 NGRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTESWATTKVRQFLLGRIY 210
NGRLEI PSKDWELESI++ + L++I+EHV+ V GL+ VT++WATT++++ L ++Y
Sbjct: 162 NGRLEIWPSKDWELESIYSCDTLEIIKEHVSAVIGLRVNSCVTDNWATTQIQKLHLRKVY 221
Query: 211 VASILYGYFLKSVSLRYHLERNLNLANHDVH-PGHRTNLSFKDMCPYGFEDDIFGHLSNM 269
ASILYGYFLKS SLR+ LE +L+ D+H G+ + IFG
Sbjct: 222 AASILYGYFLKSASLRHQLECSLS----DIHGSGY-------------LKSPIFGCSFT- 263
Query: 270 KPIGQGLIRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALFNSEGFDSV 329
G I +++ L+ Y+ F P +LQRCAK R++EA NL+ S ALF +E
Sbjct: 264 --TGTAQISNKQQ---LRHYISDFDPETLQRCAKPRTEEARNLIEKQSLALFGTE----- 313
Query: 330 DSDDVILTSFSSLKRLVLEAVAFGSFLWETEDYIDNVYKLKDH 372
+SD+ I+TSFSSLKRLVLEAVAFG+FLW+TE Y+D YKLK++
Sbjct: 314 ESDETIVTSFSSLKRLVLEAVAFGTFLWDTELYVDGAYKLKEN 356
>At5g48590 unknown protein
Length = 344
Score = 292 bits (747), Expect = 2e-79
Identities = 163/341 (47%), Positives = 217/341 (62%), Gaps = 50/341 (14%)
Query: 32 LIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLADDRDAA 91
L+ SA +S S++ PL+PR+ G+FLS VL R LFH AV + LK LADD++A+
Sbjct: 43 LVVVSAASSGQ----SIDAPLVPRSPQGRFLSSVLVKKRQLFHFAVADLLKQLADDKEAS 98
Query: 92 NSRMLLASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLSRCLYN 151
SRM L+ SDEA LHRRIA++KE+ C++A+EDIM +LI +KFSEIR PLVPKL C+YN
Sbjct: 99 LSRMFLSYGSDEASLHRRIAQLKESDCQIAIEDIMYMLILYKFSEIRVPLVPKLPSCIYN 158
Query: 152 GRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTESWATTKVRQFLLGRIYV 211
GRLEI PSKDWELESIH+ +VL++I+EH V L+ S+T+ ATT++ + L ++Y
Sbjct: 159 GRLEISPSKDWELESIHSFDVLELIKEHSNAVISLRVNSSLTDDCATTEIDKNRLSKVYT 218
Query: 212 ASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNLSFKDMCPYGFEDDIFGHLSNMKP 271
AS+LYGYFLKS SLR+ LE +L+ H G T
Sbjct: 219 ASVLYGYFLKSASLRHQLECSLS-----QHHGSFT------------------------- 248
Query: 272 IGQGLIRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALFNSEGFDSVDS 331
+ L+ Y+ F P L+RCAK RS EA +L+ S ALF E S
Sbjct: 249 ------------KQLRHYISEFDPKILRRCAKPRSHEAKSLIEKQSLALFGPE----ESS 292
Query: 332 DDVILTSFSSLKRLVLEAVAFGSFLWETEDYIDNVYKLKDH 372
+ I+TSFSSLKRL+LEAVAFG+FLW+TE+Y+D +KLK++
Sbjct: 293 KESIVTSFSSLKRLLLEAVAFGTFLWDTEEYVDGAFKLKEN 333
>At1g48450 unknown protein
Length = 423
Score = 180 bits (457), Expect = 1e-45
Identities = 121/375 (32%), Positives = 203/375 (53%), Gaps = 28/375 (7%)
Query: 14 FTRPISFSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLF 73
F+RP ++ R ++KASA + ES + PL ++ VG+FLS +L +H +L
Sbjct: 51 FSRPFQSGTTCVKSRRSFVVKASASGDASTESIA---PLQLKSPVGQFLSQILVSHPHLV 107
Query: 74 HVAVQEELKLLADDRDAANSRMLLASE-SDEALLHRRIAEMKENQCEVAVEDIMSLLIFH 132
AV+++L+ L DRDA +S + +L+RRIAE+KE + A+E+I+ L+
Sbjct: 108 PAAVEQQLEQLQIDRDAEEQSKDASSVLGTDIVLYRRIAEVKEKERRRALEEILYALVVQ 167
Query: 133 KFSEIRAPLVPKL--SRCLYNGRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKP 190
KF + LVP + S +GR++ P+ D ELE +H+ EV +MI+ H++ + K
Sbjct: 168 KFMDANVTLVPSITSSSADPSGRVDTWPTLDGELERLHSPEVYEMIQNHLSIIL----KN 223
Query: 191 SVTESWATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRT-NLS 249
+ A ++ + +G++Y AS++YGYFLK + R+ LE+ + + G + +
Sbjct: 224 RTDDLTAVAQISKLGVGQVYAASVMYGYFLKRIDQRFQLEKTMRILPGGSDEGETSIEQA 283
Query: 250 FKDMCPYGFED--DIFGHLSNMKPIGQ--------GLIRQEEEIEDLKCYVMRFHPGSLQ 299
+D+ +E+ + + +S+ + +G G + + LK YVM F +LQ
Sbjct: 284 GRDVERNFYEEAEETYQAVSSNQDVGSFVGGINASGGFSSDMKQSRLKTYVMSFDGETLQ 343
Query: 300 RCAKLRSKEAVNLVRSYSSALFNS-------EGFDSVDSDDVILTSFSSLKRLVLEAVAF 352
R A +RS+E+V ++ ++ ALF +G D+ I SF LKRLVLEAV F
Sbjct: 344 RYATIRSRESVGIIEKHTEALFGRPEIVITPQGTIDSSKDEHIKISFKGLKRLVLEAVTF 403
Query: 353 GSFLWETEDYIDNVY 367
GSFLW+ E ++D+ Y
Sbjct: 404 GSFLWDVESHVDSRY 418
>At3g17800 unknown protein
Length = 421
Score = 171 bits (433), Expect = 6e-43
Identities = 113/363 (31%), Positives = 189/363 (51%), Gaps = 23/363 (6%)
Query: 21 SSSLSFRSRVPLIKASAGASSHCESSSLNTPLLP---RTQVGKFLSGVLQNHRNLFHVAV 77
S + + ++R + ++ AS+ S S P+ P ++ G+FLS +L +H +L AV
Sbjct: 61 SLTTTAKTRRSFVVRASSASNDASSGSSPKPIAPLQLQSPAGQFLSQILVSHPHLVPAAV 120
Query: 78 QEELKLLADDRDAANSRMLLAS-ESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSE 136
+++L+ L DRD+ AS + +L+RRIAE+KEN+ +E+I+ L+ KF E
Sbjct: 121 EQQLEQLQTDRDSQGQNKDSASVPGTDIVLYRRIAELKENERRRTLEEILYALVVQKFME 180
Query: 137 IRAPLVPKLSRCL-YNGRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTES 195
LVP +S +GR++ P+K +LE +H+ E+ +MI H+ + G + + +
Sbjct: 181 ANVSLVPSVSPSSDPSGRVDTWPTKVEKLERLHSPEMYEMIHNHLALILGSR----MGDL 236
Query: 196 WATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNLSFKDMCP 255
+ ++ + +G++Y AS++YGYFLK V R+ LE+ + + + +
Sbjct: 237 NSVAQISKLRVGQVYAASVMYGYFLKRVDQRFQLEKTMKILPGGSDESKTSVEQAEGTAT 296
Query: 256 Y----GFEDDIFGHLSNMKPIGQGLIRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVN 311
Y ++ + G G E + L+ YVM F +LQR A +RS+EAV
Sbjct: 297 YQAAVSSHPEVGAFAGGVSAKGFG---SEIKPSRLRSYVMSFDAETLQRYATIRSREAVG 353
Query: 312 LVRSYSSALFNS-------EGFDSVDSDDVILTSFSSLKRLVLEAVAFGSFLWETEDYID 364
++ ++ ALF EG D+ I SF +KRLVLEAV FGSFLW+ E ++D
Sbjct: 354 IIEKHTEALFGKPEIVITPEGTVDSSKDEQIKISFGGMKRLVLEAVTFGSFLWDVESHVD 413
Query: 365 NVY 367
Y
Sbjct: 414 ARY 416
>At1g32160 unknown protein
Length = 406
Score = 157 bits (397), Expect = 9e-39
Identities = 111/363 (30%), Positives = 192/363 (52%), Gaps = 24/363 (6%)
Query: 19 SFSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQ 78
S S++ + R R ++AS S+ E+ + P+ + VG+ L +L+ H +L V V
Sbjct: 49 SSSTNENGRGRSVTVRASGDEDSN-ENFAPLAPVELESPVGQLLEQILRTHPHLLPVTVD 107
Query: 79 EELKLLADDRDAANSRMLLASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSEIR 138
E+L+ A + ++ + S S + +L +RI+E+++ + + +I+ L+ H+F E
Sbjct: 108 EQLEKFAAESESRKAD----SSSTQDILQKRISEVRDKERRKTLAEIIYCLVVHRFVEKG 163
Query: 139 APLVPKLSRCLYN-GRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTESWA 197
++P++ GR+++ P+++ +LE IH+ + +MI+ H+++V G P+V +
Sbjct: 164 ISMIPRIKPTSDPAGRIDLWPNQEEKLEVIHSADAFEMIQSHLSSVLG--DGPAVGPLSS 221
Query: 198 TTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNLSFKDMCP-Y 256
++ + LG++Y AS +YGYFL+ V RY LER +N +T F++ P Y
Sbjct: 222 IVQIGKIKLGKLYAASAMYGYFLRRVDQRYQLERTMNTLPKRPE---KTRERFEEPSPPY 278
Query: 257 GFEDD---IFGHLSNMKPIGQGLIRQEEEIED--LKCYVMRFHPGSLQRCAKLRSKEAVN 311
D I P + R E+E L+ YV +LQR A +RSKEA+
Sbjct: 279 PLWDPDSLIRIQPEEYDPDEYAIQRNEDESSSYGLRSYVTYLDSDTLQRYATIRSKEAMT 338
Query: 312 LVRSYSSALFN-------SEGFDSVDSDDVILTSFSSLKRLVLEAVAFGSFLWETEDYID 364
L+ + ALF +G +D+V+ S S L LVLEAVAFGSFLW++E Y++
Sbjct: 339 LIEKQTQALFGRPDIRILEDGKLDTSNDEVLSLSVSGLAMLVLEAVAFGSFLWDSESYVE 398
Query: 365 NVY 367
+ Y
Sbjct: 399 SKY 401
>At5g52660 unknown protein
Length = 331
Score = 32.0 bits (71), Expect = 0.56
Identities = 15/24 (62%), Positives = 18/24 (74%)
Query: 28 SRVPLIKASAGASSHCESSSLNTP 51
S PL KA AGA+++C SSS NTP
Sbjct: 199 SFTPLPKAGAGANNNCSSSSENTP 222
>At2g14910 unknown protein
Length = 386
Score = 30.4 bits (67), Expect = 1.6
Identities = 30/140 (21%), Positives = 61/140 (43%), Gaps = 19/140 (13%)
Query: 123 EDIMSLLIFHKFSEIRAPLVPKLSRCLYNGRLEILPSKDWEL-ESIHTLEVLDMIREHVT 181
+D SL F S+ R+P +C+ + ++ + D L + + LD ++ ++
Sbjct: 59 DDGFSLDDFTLHSDSRSP-----KKCVLSDLIQEIEPLDVSLIQKDVPVTTLDAMKRTIS 113
Query: 182 TVTGL----KAKPSVTESWATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNLAN 237
+ GL + + + W L ++ V+S++ GY L++ R LE+NL+++
Sbjct: 114 GMLGLLPSDRFQVHIESLWEP-------LSKLLVSSMMTGYTLRNAEYRLFLEKNLDMSG 166
Query: 238 H--DVHPGHRTNLSFKDMCP 255
D H T + P
Sbjct: 167 GGLDSHASENTEYDMEGTFP 186
>At4g36910 unknown protein
Length = 236
Score = 29.3 bits (64), Expect = 3.6
Identities = 22/70 (31%), Positives = 35/70 (49%), Gaps = 3/70 (4%)
Query: 19 SFSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQ 78
+FS S +SR+P ++AG++ SSS P VG+F++ H V
Sbjct: 41 TFSRSFPSKSRIPSASSAAGSTLMTNSSS---PRSGVYTVGEFMTKKEDLHVVKPTTTVD 97
Query: 79 EELKLLADDR 88
E L+LL ++R
Sbjct: 98 EALELLVENR 107
>At2g43940 unknown protein
Length = 383
Score = 28.9 bits (63), Expect = 4.7
Identities = 30/116 (25%), Positives = 48/116 (40%), Gaps = 21/116 (18%)
Query: 17 PISFSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLF--- 73
P SF + SFRS+ P K +SS C S S + SG +++
Sbjct: 25 PDSFHAPSSFRSKNPNFKRRLSSSSCCSSQS------------QLFSGRCRSYSRCVTMC 72
Query: 74 ---HVAVQEELKLLADDRDAANSRMLLASESDEAL-LHRRIAEMKENQCEVAVEDI 125
H+ QE ++L D D + +L S+ D L + + + E VAV ++
Sbjct: 73 LPEHMRNQENTEILTDKDD--HIECVLESDEDSGLRIPTQAQAIVEGSGSVAVSEL 126
>At1g48870 hypothetical protein
Length = 593
Score = 28.9 bits (63), Expect = 4.7
Identities = 20/78 (25%), Positives = 37/78 (46%), Gaps = 8/78 (10%)
Query: 260 DDIFGHLSNMKPIGQGLIRQEEEIEDLKCYVMRFHPGS--------LQRCAKLRSKEAVN 311
DD+F S++ IG+ ++ +EEE+E+ + V P S L++ L +
Sbjct: 12 DDLFFDSSDVLSIGEQVVVEEEEVEEAEFQVWSGEPISVEERRVRFLKKMGLLEERCLER 71
Query: 312 LVRSYSSALFNSEGFDSV 329
+V YS + +S S+
Sbjct: 72 MVSDYSDEVTSSSSDSSL 89
>At5g55400 fimbrin
Length = 714
Score = 28.1 bits (61), Expect = 8.1
Identities = 21/73 (28%), Positives = 33/73 (44%), Gaps = 5/73 (6%)
Query: 136 EIRAPLVPKLSRCLYNGRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTES 195
E+R LV + G +++ W+L H L++L +R + G S S
Sbjct: 466 EMRFSLVNVAGNDIVQGNKKLILGFLWQLMRTHMLQLLKSLR---SRTRGKDMTDSEIIS 522
Query: 196 WATTKVRQFLLGR 208
WA KVR ++GR
Sbjct: 523 WANRKVR--IMGR 533
>At5g18390 unknown protein
Length = 459
Score = 28.1 bits (61), Expect = 8.1
Identities = 19/60 (31%), Positives = 26/60 (42%), Gaps = 5/60 (8%)
Query: 216 YGYFLKSVSLRYHLERNLNLANHDVHPGHRTNLSF-----KDMCPYGFEDDIFGHLSNMK 270
Y + +VS ++ L N+ V GH+ S K MC G DD F S+MK
Sbjct: 325 YKTLIPAVSKIGKIDEAFRLLNNCVEDGHKPFPSLYAPIIKGMCRNGMFDDAFSFFSDMK 384
>At5g12130 unknown protein
Length = 384
Score = 28.1 bits (61), Expect = 8.1
Identities = 25/90 (27%), Positives = 39/90 (42%), Gaps = 17/90 (18%)
Query: 266 LSNMKPIG--QGLIRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALFNS 323
LS++ PI +G+ +++EE E + L K N S SS+ +S
Sbjct: 57 LSHLPPIACSRGIDQEDEEKESREL---------------LPHKNDENATTSRSSSSVDS 101
Query: 324 EGFDSVDSDDVILTSFSSLKRLVLEAVAFG 353
G ++ TSF ++ V AVAFG
Sbjct: 102 GGLKDYQQEETYKTSFKTVALCVGTAVAFG 131
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,209,489
Number of Sequences: 26719
Number of extensions: 343538
Number of successful extensions: 946
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 919
Number of HSP's gapped (non-prelim): 15
length of query: 372
length of database: 11,318,596
effective HSP length: 101
effective length of query: 271
effective length of database: 8,619,977
effective search space: 2336013767
effective search space used: 2336013767
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136841.6


