

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.5 + phase: 0
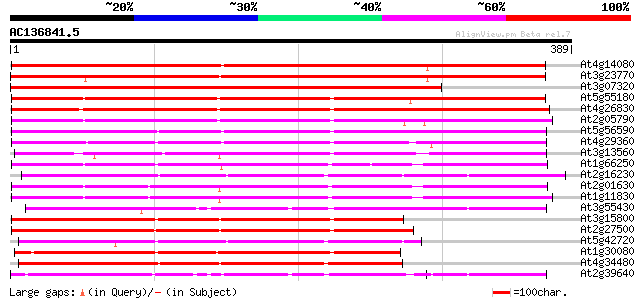
(389 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g14080 A6 anther-specific protein 442 e-124
At3g23770 beta-1,3-glucanase, putative 440 e-124
At3g07320 putative beta-1,3-glucanase precursor 400 e-112
At5g55180 beta-1,3-glucanase-like protein 299 2e-81
At4g26830 putative beta-1,3-glucanase 295 4e-80
At2g05790 putative beta-1,3-glucanase 291 3e-79
At5g56590 beta-1,3-glucanase-like protein 285 3e-77
At4g29360 beta-1,3-glucanase-like protein 262 2e-70
At3g13560 glucan endo-1,3-beta-glucosidase precursor, putative 248 4e-66
At1g66250 beta-1,3-glucanase precursor, putative 247 9e-66
At2g16230 putative beta-1,3-glucanase 245 3e-65
At2g01630 beta-1,3-glucanase like protein 244 4e-65
At1g11830 putative beta-1,3-glucanase, predicted GPI-anchored pr... 244 8e-65
At3g55430 beta-1,3-glucanase - like protein 217 8e-57
At3g15800 putative beta-1,3-glucanase precursor 217 8e-57
At2g27500 beta-1,3-glucanase like protein 209 3e-54
At5g42720 beta-1,3-glucanase-like protein 199 2e-51
At1g30080 beta-1,3-glucanase precursor, putative 197 6e-51
At4g34480 putative protein (fragment) 196 2e-50
At2g39640 putative beta-1,3-glucanase, predicted GPI-anchored pr... 196 2e-50
>At4g14080 A6 anther-specific protein
Length = 478
Score = 442 bits (1136), Expect = e-124
Identities = 201/373 (53%), Positives = 284/373 (75%), Gaps = 4/373 (1%)
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYR 61
+PN +T +SSNQT+A++WV+TN++P+Y +T IR++LVGNE++S + ++VPAM +
Sbjct: 95 VPNHQITALSSNQTIADEWVRTNILPYYPQTQIRFVLVGNEILSYNSGNVSVNLVPAMRK 154
Query: 62 MKHSLTIFGLHKVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFFL 121
+ +SL + G+H +KVGTPLAMD L++SFPPSNGTFR +I VM P+L+FL+ TNS+FFL
Sbjct: 155 IVNSLRLHGIHNIKVGTPLAMDSLRSSFPPSNGTFREEITGPVMLPLLKFLNGTNSYFFL 214
Query: 122 DVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYPD 181
+V+P+F W+ +P+N +LD+ALF+ + T TD TGLVY NL DQM+D+V FAM +LGYP
Sbjct: 215 NVHPYFRWSRNPMNTSLDFALFQGHS-TYTDPQTGLVYRNLLDQMLDSVLFAMTKLGYPH 273
Query: 182 IQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFAL 241
+++ I+ETGWPN GD+D+ GANI NA TYNRN +KK++ PP+GTP+RPG +P+F+F+L
Sbjct: 274 MRLAISETGWPNFGDIDETGANILNAATYNRNLIKKMSASPPIGTPSRPGLPIPTFVFSL 333
Query: 242 YNENLKTGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPPPDD---YKGKAWCVVA 298
+NEN K+G GT+RH+G+L+P+GS IY++D +G+TP + PLP P + YKG+ WCV
Sbjct: 334 FNENQKSGSGTQRHWGILHPDGSPIYDVDFTGQTPLTGFNPLPKPTNNVPYKGQVWCVPV 393
Query: 299 EGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTC 358
EGAN+T + E L AC+Q N TC + PG+ C+EP S+ HASYA +SYWAQFR C
Sbjct: 394 EGANETELEETLRMACAQSNTTCAALAPGRECYEPVSIYWHASYALNSYWAQFRNQSIQC 453
Query: 359 NFNGLATQIAEDP 371
FNGLA + +P
Sbjct: 454 FFNGLAHETTTNP 466
>At3g23770 beta-1,3-glucanase, putative
Length = 476
Score = 440 bits (1131), Expect = e-124
Identities = 205/377 (54%), Positives = 282/377 (74%), Gaps = 7/377 (1%)
Query: 1 MLPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQT---WPHIVP 57
M+PN + ++ ++Q A+ WV TN++PF+ +T IR++LVGNE++S +++Q W ++VP
Sbjct: 89 MVPNNQIISIGADQAAADNWVATNVLPFHPQTRIRFVLVGNEVLSYSSDQDKQIWANLVP 148
Query: 58 AMYRMKHSLTIFGLHKVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNS 117
AM ++ +SL G+H +KVGTPLAMD L++SFPPS+GTFR DIA+ VM P+L+FL+ TNS
Sbjct: 149 AMRKVVNSLRARGIHNIKVGTPLAMDALRSSFPPSSGTFREDIAVPVMLPLLKFLNGTNS 208
Query: 118 FFFLDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERL 177
FFFLDVYP+F W++DP+N +LD+ALFES N T TD TGLVYTNL DQM+D+V FAM +L
Sbjct: 209 FFFLDVYPYFPWSTDPVNNHLDFALFES-NSTYTDPQTGLVYTNLLDQMLDSVIFAMTKL 267
Query: 178 GYPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSF 237
GYP+I + I+ETGWPNDGD+ + GANI NA TYNRN +KK+T PP+GTPAR G+ +P+F
Sbjct: 268 GYPNISLAISETGWPNDGDIHETGANIVNAATYNRNLIKKMTANPPLGTPARRGAPIPTF 327
Query: 238 IFALYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPPPDD---YKGKAW 294
+F+L+NEN K G GTERH+G+L P+G+ IY+ID SG+ + LP P + +KG W
Sbjct: 328 LFSLFNENQKPGSGTERHWGILNPDGTPIYDIDFSGRRSFSGFDSLPKPSNNVPFKGNVW 387
Query: 295 CVVAEGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRV 354
CV +GA++ + +AL++AC + N TC + PG C+ P +V HASYAFSSYWAQFR
Sbjct: 388 CVAVDGADEAELGQALNFACGRSNATCAALAPGGECYAPVTVTWHASYAFSSYWAQFRNQ 447
Query: 355 GGTCNFNGLATQIAEDP 371
C FNGLA + +P
Sbjct: 448 SSQCYFNGLARETTTNP 464
>At3g07320 putative beta-1,3-glucanase precursor
Length = 384
Score = 400 bits (1028), Expect = e-112
Identities = 187/300 (62%), Positives = 240/300 (79%), Gaps = 1/300 (0%)
Query: 1 MLPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMY 60
M+PNEL+ N+S + +L++ W+++N++PFY T IRYLLVGNE++S ++ +VPAM
Sbjct: 77 MVPNELLVNISKSASLSDDWIRSNILPFYPTTKIRYLLVGNEILSLPDSELKSSLVPAMR 136
Query: 61 RMKHSLTIFGLHKVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFF 120
+++ SL G+ KVKVGT LA DVLQ+SFPPS+G FR DI+ +MKPML+FL+ T SF F
Sbjct: 137 KIQRSLKSLGVKKVKVGTTLATDVLQSSFPPSSGEFREDISGLIMKPMLQFLNRTKSFLF 196
Query: 121 LDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYP 180
+DVYP+FAW DP +++LDYA+FES N+TVTD + L Y NLFDQM+DA FAM+R+GYP
Sbjct: 197 VDVYPYFAWAQDPTHVDLDYAIFESTNVTVTDPVSNLTYHNLFDQMIDAFVFAMKRVGYP 256
Query: 181 DIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFA 240
DI+I++AETGWPN+GD DQIGANI+NA TYNRN VKK+ PPVGTPARPG +LP+F+FA
Sbjct: 257 DIRIWVAETGWPNNGDYDQIGANIYNAATYNRNVVKKLAADPPVGTPARPGKVLPAFVFA 316
Query: 241 LYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPPPDD-YKGKAWCVVAE 299
LYNEN KTG GTERHFGLL+PNG+++Y IDLSGKT E P P +D YKGK WCVVA+
Sbjct: 317 LYNENQKTGPGTERHFGLLHPNGTQVYGIDLSGKTEYKESLPAPENNDLYKGKIWCVVAK 376
>At5g55180 beta-1,3-glucanase-like protein
Length = 460
Score = 299 bits (765), Expect = 2e-81
Identities = 153/374 (40%), Positives = 232/374 (61%), Gaps = 8/374 (2%)
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYR 61
LPNE + + +++Q+ + WVQ N+ + T I + VGNE+ N T ++VPAM
Sbjct: 80 LPNENLASAAADQSYTDTWVQDNIKKYIPATDIEAIAVGNEVFVDPRNTT-TYLVPAMKN 138
Query: 62 MKHSLTIFGLHK-VKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFF 120
++ SL F L K +K+ +P+A+ L +S+PPS G+F+ ++ V+KPML+ L T+S
Sbjct: 139 VQSSLVKFNLDKSIKISSPIALSALASSYPPSAGSFKPELIEPVIKPMLDLLRKTSSHLM 198
Query: 121 LDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYP 180
++ YPFFA+ ++ I+LDYALF+ +N DSG GL Y +L D +DAV+ AM +G+
Sbjct: 199 VNAYPFFAYAANADKISLDYALFK-ENAGNVDSGNGLKYNSLLDAQIDAVFAAMSAVGFN 257
Query: 181 DIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFA 240
D+++ + ETGWP+ GD ++IGA NA YN VK+V GTP +P L ++FA
Sbjct: 258 DVKLVVTETGWPSAGDENEIGAGSANAAAYNGGLVKRVLTGN--GTPLKPKEPLNVYLFA 315
Query: 241 LYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGK-TP--EYEYKPLPPPDDYKGKAWCVV 297
L+NEN KTG +ER++GL YPN +++Y++ L+GK TP + + K +P G+ WCV
Sbjct: 316 LFNENQKTGPTSERNYGLFYPNENKVYDVSLNGKSTPVNDNKEKVVPVKPSLVGQTWCVA 375
Query: 298 AEGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGT 357
K + E L YAC +G C +QPG C+ P+S+ HASYAF+SY+ + R GT
Sbjct: 376 NGKTTKEKLQEGLDYACGEGGADCRPIQPGATCYNPESLEAHASYAFNSYYQKNARGVGT 435
Query: 358 CNFNGLATQIAEDP 371
CNF G A +++ P
Sbjct: 436 CNFGGAAYVVSQPP 449
>At4g26830 putative beta-1,3-glucanase
Length = 448
Score = 295 bits (754), Expect = 4e-80
Identities = 151/375 (40%), Positives = 230/375 (61%), Gaps = 7/375 (1%)
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYR 61
LPNEL+++ +S+Q+ A+ W++T+++P++ T I + VGNE+ T P++V AM
Sbjct: 78 LPNELLSSAASHQSFADNWIKTHIMPYFPATEIEAIAVGNEVFVDPT--ITPYLVNAMKN 135
Query: 62 MKHSLTIFGLHK-VKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFF 120
+ SL + L K +K+ +P+A+ L S+PPS+G+F+ ++ V+KPML L T+S+
Sbjct: 136 IHTSLVKYKLDKAIKISSPIALSALANSYPPSSGSFKPELIEPVVKPMLALLQQTSSYLM 195
Query: 121 LDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYP 180
++ YPFFA+ ++ I+LDYALF+ +N DSGTGL Y +LFD +DAVY A+ +G+
Sbjct: 196 VNAYPFFAYAANADKISLDYALFK-ENAGNIDSGTGLKYNSLFDAQIDAVYAALSAVGFK 254
Query: 181 DIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFA 240
+++ + ETGWP+ GD ++IGA+ NA YN VK+V GTP RP L ++FA
Sbjct: 255 GVKVMVTETGWPSVGDENEIGASESNAAAYNAGLVKRVLTGK--GTPLRPTEPLNVYLFA 312
Query: 241 LYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGK-TPEYEYKPLPPPDDYKGKAWCVVAE 299
L+NEN K G +ER++GL YPN ++Y + + K T P ++G WCV
Sbjct: 313 LFNENQKPGPTSERNYGLFYPNEGKVYNVPFTKKSTTPVNGNRGKVPVTHEGHTWCVSNG 372
Query: 300 GANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTCN 359
K + EAL YAC +G C +QPG C+ P+S+ HASYAF+SY+ + R GTC
Sbjct: 373 EVAKEKLQEALDYACGEGGADCRPIQPGATCYHPESLEAHASYAFNSYYQKNSRRVGTCF 432
Query: 360 FNGLATQIAEDPSKL 374
F G A + + P L
Sbjct: 433 FGGAAHVVTQPPRTL 447
>At2g05790 putative beta-1,3-glucanase
Length = 473
Score = 291 bits (746), Expect = 3e-79
Identities = 154/393 (39%), Positives = 229/393 (58%), Gaps = 22/393 (5%)
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYR 61
LPNEL+ + + + A WV+ N+ ++ T I + VGNE+ T N T ++PAM
Sbjct: 78 LPNELLFSAAKRTSFAVSWVKRNVAAYHPSTQIESIAVGNEVFVDTHNTT-SFLIPAMRN 136
Query: 62 MKHSLTIFGLHK-VKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFF 120
+ +L F LH +K+ +PLA+ LQ S+P S+G+FR ++ SV+KPML+FL T S
Sbjct: 137 IHKALMSFNLHSDIKISSPLALSALQNSYPSSSGSFRPELIDSVIKPMLDFLRETGSRLM 196
Query: 121 LDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYP 180
++VYPFFA+ + I LDYAL +N + DSG GL Y NLFD +DAV+ AM L Y
Sbjct: 197 INVYPFFAYEGNSDVIPLDYALLR-ENPGMVDSGNGLRYFNLFDAQIDAVFAAMSALKYD 255
Query: 181 DIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFA 240
DI+I + ETGWP+ GD +++GA + NA +YN N ++++ + GTP RP + L ++FA
Sbjct: 256 DIEIIVTETGWPSKGDENEVGATLANAASYNGNLIRRILTRG--GTPLRPKADLTVYLFA 313
Query: 241 LYNENLKTGLGTERHFGLLYPNGSRIYEIDLS----------GKTPEYEYKPLPPP---- 286
L+NEN K G +ER++GL +P+ ++Y+I + G TP + P
Sbjct: 314 LFNENKKLGPTSERNYGLFFPDEKKVYDIPFTTEGLKHYRDGGHTPVTGGDQVTKPPMSG 373
Query: 287 ---DDYKGKAWCVVAEGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYA 343
G WCV A + + L YAC +G C +QPG C+ PD++ HAS+A
Sbjct: 374 GVSKSLNGYTWCVANGDAGEERLQGGLDYACGEGGADCRPIQPGANCYSPDTLEAHASFA 433
Query: 344 FSSYWAQFRRVGGTCNFNGLATQIAEDPSKLFF 376
F+SY+ + R GG+C F G A +++ PSK F
Sbjct: 434 FNSYYQKKGRAGGSCYFGGAAYVVSQPPSKYNF 466
>At5g56590 beta-1,3-glucanase-like protein
Length = 506
Score = 285 bits (729), Expect = 3e-77
Identities = 142/373 (38%), Positives = 221/373 (59%), Gaps = 6/373 (1%)
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYR 61
+PN + S +Q+ + W++ +++P+Y T I Y+ VG E +VPAM
Sbjct: 79 VPNSDLNAFSQSQSNVDTWLKNSVLPYYPTTKITYITVGAESTDDPHINASSFVVPAMQN 138
Query: 62 MKHSLTIFGL-HKVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFF 120
+ +L GL ++KV T L++ +L SFPPS G F + A ++PMLEFL S F
Sbjct: 139 VLTALRKVGLSRRIKVSTTLSLGILSRSFPPSAGAFNSSYAY-FLRPMLEFLAENKSPFM 197
Query: 121 LDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYP 180
+D+YP++A+ P N++LDY LFES + V D TGL+Y N+FD VDA+Y+A+ L +
Sbjct: 198 IDLYPYYAYRDSPNNVSLDYVLFESSS-EVIDPNTGLLYKNMFDAQVDALYYALTALNFR 256
Query: 181 DIQIFIAETGWPNDGD-LDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIF 239
I+I + ETGWP G ++ A+ NA TYN N ++ V GTPA+PG + +IF
Sbjct: 257 TIKIMVTETGWPTKGSPKEKAAASSDNAETYNSNIIRHVVTNQ--GTPAKPGEAMNVYIF 314
Query: 240 ALYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPPPDDYKGKAWCVVAE 299
+L+NEN K GL +ER++GL YP+ + +Y++D +GK+ + +WC+ +
Sbjct: 315 SLFNENRKAGLDSERNWGLFYPDQTSVYQLDFTGKSNGFHSNSSGTNSSGSSNSWCIASS 374
Query: 300 GANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTCN 359
A++ + AL +AC GN C +QP +PCF+PD++V HAS+ F+SY+ Q R C+
Sbjct: 375 KASERDLKGALDWACGPGNVDCTAIQPSQPCFQPDTLVSHASFVFNSYFQQNRATDVACS 434
Query: 360 FNGLATQIAEDPS 372
F G ++ +DPS
Sbjct: 435 FGGAGVKVNKDPS 447
>At4g29360 beta-1,3-glucanase-like protein
Length = 534
Score = 262 bits (670), Expect = 2e-70
Identities = 138/399 (34%), Positives = 224/399 (55%), Gaps = 37/399 (9%)
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYR 61
+PN + + Q+ + W+ N++P+Y T I + VG E+ + N T ++PAM
Sbjct: 80 VPNADLLAFAQFQSNVDTWLSNNILPYYPSTKITSISVGLEVTEAPDNATGL-VLPAMRN 138
Query: 62 MKHSLTIFGLHK-VKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFF 120
+ +L GL K +K+ + ++ +L SFPPS+ +F + + +KPMLEFL S F
Sbjct: 139 IHTALKKSGLDKKIKISSSHSLAILSRSFPPSSASFSKKHS-AFLKPMLEFLVENESPFM 197
Query: 121 LDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYP 180
+D+YP++A+ + L+YALFES + V D TGL+Y+N+FD +DA+YFA+ + +
Sbjct: 198 IDLYPYYAYRDSTEKVPLEYALFESSS-QVVDPATGLLYSNMFDAQLDAIYFALTAMSFK 256
Query: 181 DIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFA 240
+++ + E+GWP+ G + A NA YN N ++ V P GTPA+PG + ++F+
Sbjct: 257 TVKVMVTESGWPSKGSPKETAATPENALAYNTNLIRHVIGDP--GTPAKPGEEIDVYLFS 314
Query: 241 LYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPPPDDYKG--------- 291
L+NEN K G+ +ER++G+ Y NG+ +Y +D +G+ P+ P + G
Sbjct: 315 LFNENRKPGIESERNWGMFYANGTNVYALDFTGENTT----PVSPTNSTTGTSPSPSSSP 370
Query: 292 ------------------KAWCVVAEGANKTAVVEALSYACSQGNRTCELVQPGKPCFEP 333
K WC+ + A+ T + AL +AC GN C VQP +PCFEP
Sbjct: 371 IINGNSTVTIGGGGGGGTKKWCIASSQASVTELQTALDWACGPGNVDCSAVQPDQPCFEP 430
Query: 334 DSVVGHASYAFSSYWAQFRRVGGTCNFNGLATQIAEDPS 372
D+V+ HASYAF++Y+ Q C+FNG + ++ +DPS
Sbjct: 431 DTVLSHASYAFNTYYQQSGASSIDCSFNGASVEVDKDPS 469
>At3g13560 glucan endo-1,3-beta-glucosidase precursor, putative
Length = 505
Score = 248 bits (633), Expect = 4e-66
Identities = 133/376 (35%), Positives = 212/376 (56%), Gaps = 23/376 (6%)
Query: 4 NELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVP----AM 59
NE + + + A WV N+ + T I + VG+E+++ T PH+ P A+
Sbjct: 81 NEEILKIGRFPSAAAAWVNKNVAAYIPSTNITAIAVGSEVLT-----TIPHVAPILASAL 135
Query: 60 YRMKHSLTIFGLH-KVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSF 118
+ +L L+ KVKV +P++MD++ FPPS TF +V + +L+FL T SF
Sbjct: 136 NNIHKALVASNLNFKVKVSSPMSMDIMPKPFPPSTSTFSPSWNTTVYQ-LLQFLKNTGSF 194
Query: 119 FFLDVYPFFAWTSDPININLDYALFE--SDNITVTDSGTGLVYTNLFDQMVDAVYFAMER 176
F L+ YP++ +T+ LDYALF+ S + D T L Y ++FD MVDA Y++ME
Sbjct: 195 FMLNAYPYYGYTTANGIFPLDYALFKQLSPVKQIVDPNTLLHYNSMFDAMVDAAYYSMEA 254
Query: 177 LGYPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPS 236
L + I + + ETGWP+ G D+ A + NA T+N N +K+V G P++P + +
Sbjct: 255 LNFSKIPVVVTETGWPSSGGSDEAAATVANAETFNTNLIKRVLNNS--GPPSQPDIPINT 312
Query: 237 FIFALYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPPPDDYKGKAWCV 296
+I+ LYNE+ ++G +ER++G+L+PNG+ +Y + LSG + +CV
Sbjct: 313 YIYELYNEDKRSGPVSERNWGILFPNGTSVYPLSLSGGSSSAALNG--------SSMFCV 364
Query: 297 VAEGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGG 356
A+ +V+ L++AC QG C +QPG+PC+ P+ V HAS+AF+ Y+ + + GG
Sbjct: 365 AKADADDDKLVDGLNWACGQGRANCAAIQPGQPCYLPNDVKSHASFAFNDYYQKMKSAGG 424
Query: 357 TCNFNGLATQIAEDPS 372
TC+F+G A DPS
Sbjct: 425 TCDFDGTAITTTRDPS 440
>At1g66250 beta-1,3-glucanase precursor, putative
Length = 418
Score = 247 bits (630), Expect = 9e-66
Identities = 135/375 (36%), Positives = 212/375 (56%), Gaps = 15/375 (4%)
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYR 61
+PN+ + + + + A WV+ N++ Y T+I + VG+E+++S +N P +V A+
Sbjct: 43 IPNDQLLGIGQSNSTAANWVKRNVIAHYPATMITAVSVGSEVLTSLSNAA-PVLVSAIKN 101
Query: 62 MKHSLTIFGLHK-VKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFF 120
+ +L L K +KV TPL+ ++ FPPS F + +V+ P+L FL TNS+
Sbjct: 102 VHAALLSANLDKLIKVSTPLSTSLILDPFPPSQAFFNRSLN-AVIVPLLSFLQSTNSYLM 160
Query: 121 LDVYPFFAWTSDPININLDYALFES--DNITVTDSGTGLVYTNLFDQMVDAVYFAMERLG 178
++VYP+ + I LDYALF+ N D+ T + Y+N FD MVDA YFAM L
Sbjct: 161 VNVYPYIDYMQSNGVIPLDYALFKPIPPNKEAVDANTLVRYSNAFDAMVDATYFAMAFLN 220
Query: 179 YPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFI 238
+ +I + + E+GWP+ G+ ++ A + NA TYN N ++ V K GTP RPG + ++I
Sbjct: 221 FTNIPVLVTESGWPSKGETNEPDATLDNANTYNSNLIRHVLNK--TGTPKRPGIAVSTYI 278
Query: 239 FALYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPPPDDYKGKAWCVVA 298
+ LYNE+ K GL +E+++GL NG +Y + L+ +D + +C
Sbjct: 279 YELYNEDTKAGL-SEKNWGLFNANGEPVYVLRLTNSGSVLA-------NDTTNQTYCTAR 330
Query: 299 EGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTC 358
EGA+ + AL +AC G C ++ G+ C+EPD+VV HA+YAF +Y+ Q C
Sbjct: 331 EGADTKMLQAALDWACGPGKIDCSPIKQGETCYEPDNVVAHANYAFDTYYHQTGNNPDAC 390
Query: 359 NFNGLATQIAEDPSK 373
NFNG+A+ DPSK
Sbjct: 391 NFNGVASITTTDPSK 405
>At2g16230 putative beta-1,3-glucanase
Length = 456
Score = 245 bits (626), Expect = 3e-65
Identities = 138/378 (36%), Positives = 218/378 (57%), Gaps = 7/378 (1%)
Query: 9 NVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYRMKHSLTI 68
+++S+ +A+QW+ +N++PFY + I + VGNE++ S ++PAM ++ +L
Sbjct: 85 SIASDLNIASQWINSNVLPFYPASNIILINVGNEVLLSNDLNLVNQLLPAMQNVQKALEA 144
Query: 69 FGLH-KVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFFLDVYPFF 127
L K+KV T AM VL S PPS G+F +K +L+FL T S F ++ YPFF
Sbjct: 145 VSLGGKIKVSTVHAMTVLGNSEPPSAGSFAPSYQAG-LKGILQFLSDTGSPFAINPYPFF 203
Query: 128 AWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYPDIQIFIA 187
A+ SDP L + LF+ + V DS TG+ Y N+FD VDAV+ A++ +G+ +++ +A
Sbjct: 204 AYQSDPRPETLAFCLFQPNPGRV-DSNTGIKYMNMFDAQVDAVHSALKSIGFEKVEVLVA 262
Query: 188 ETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFALYNENLK 247
ETGWP+ GD +++G ++ NA YN N + + + VGTP PG + ++IFAL++ENLK
Sbjct: 263 ETGWPSTGDSNEVGPSVENAKAYNGNLIAHL--RSMVGTPLMPGKSIDTYIFALFDENLK 320
Query: 248 TGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPPPDDYKGKAWCVVAEGANKTAVV 307
G E+ FGL P+ S Y+I L+ KT + P WCV E A + +
Sbjct: 321 PGPSFEQSFGLFKPDLSMAYDIGLT-KTTSSQTSQSPQLGKVTSMGWCVPKEDATQEQLQ 379
Query: 308 EALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTCNFNGLATQI 367
++L + C QG C + PG CFEP++V H +YA + Y+ + C+F+ A
Sbjct: 380 DSLDWVCGQG-IDCGPIMPGGVCFEPNNVASHTAYAMNLYFQKSPENPTDCDFSKTARIT 438
Query: 368 AEDPSKLFFLIALRTFIL 385
+E+PSKLF + +++L
Sbjct: 439 SENPSKLFSSSSFISYLL 456
>At2g01630 beta-1,3-glucanase like protein
Length = 501
Score = 244 bits (624), Expect = 4e-65
Identities = 133/375 (35%), Positives = 211/375 (55%), Gaps = 14/375 (3%)
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYR 61
+PN+ + +S + A WV N+ +Y T I + VG+E+++S TN +V A+
Sbjct: 76 VPNDQLLGISQSNATAANWVTRNVAAYYPATNITTIAVGSEVLTSLTNAA-SVLVSALKY 134
Query: 62 MKHSLTIFGLHK-VKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFF 120
++ +L L + +KV TP + ++ SFPPS F N V+ P+L+FL T S
Sbjct: 135 IQAALVTANLDRQIKVSTPHSSTIILDSFPPSQAFF-NKTWDPVIVPLLKFLQSTGSPLL 193
Query: 121 LDVYPFFAWTSDPININLDYALFE--SDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLG 178
L+VYP+F + I LDYALF+ N D+ T L YTN+FD +VDA YFAM L
Sbjct: 194 LNVYPYFDYVQSNGVIPLDYALFQPLQANKEAVDANTLLHYTNVFDAIVDAAYFAMSYLN 253
Query: 179 YPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFI 238
+ +I I + E+GWP+ G + A + NA TYN N ++ V K GTP PG+ + ++I
Sbjct: 254 FTNIPIVVTESGWPSKGGPSEHDATVENANTYNSNLIQHVINK--TGTPKHPGTAVTTYI 311
Query: 239 FALYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPPPDDYKGKAWCVVA 298
+ LYNE+ + G +E+++GL Y NG+ +Y + L+G +D + +C+
Sbjct: 312 YELYNEDTRPGPVSEKNWGLFYTNGTPVYTLRLAGAGAILA-------NDTTNQTFCIAK 364
Query: 299 EGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTC 358
E ++ + AL +AC G C + G+ C+EPD VV H++YAF++Y+ + + G+C
Sbjct: 365 EKVDRKMLQAALDWACGPGKVDCSALMQGESCYEPDDVVAHSTYAFNAYYQKMGKASGSC 424
Query: 359 NFNGLATQIAEDPSK 373
+F G+AT DPS+
Sbjct: 425 DFKGVATVTTTDPSR 439
>At1g11830 putative beta-1,3-glucanase, predicted GPI-anchored
protein
Length = 477
Score = 244 bits (622), Expect = 8e-65
Identities = 130/378 (34%), Positives = 214/378 (56%), Gaps = 14/378 (3%)
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYR 61
+PN + + S+ + A W+ N+V +Y +TLI + VG+E++++ + P ++PA+
Sbjct: 97 VPNNQLLAIGSSNSTAASWIGRNVVAYYPETLITAISVGDEVLTTVPSSA-PLLLPAIES 155
Query: 62 MKHSLTIFGLH-KVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFF 120
+ ++L LH ++KV TP A ++ +FPPS F N S+M P+L+FL T S
Sbjct: 156 LYNALVASNLHTQIKVSTPHAASIMLDTFPPSQAYF-NQTWHSIMVPLLQFLSKTGSPLM 214
Query: 121 LDVYPFFAWTSDPININLDYALFE--SDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLG 178
+++YP++ + + + LD LFE + + + D T L YTN+ D MVDA Y +M+ L
Sbjct: 215 MNLYPYYVYMQNKGVVPLDNCLFEPLTPSKEMVDPNTLLHYTNVLDAMVDAAYVSMKNLN 274
Query: 179 YPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFI 238
D+ + + E+GWP+ GD + A I NA TYN N +K V + GTP P +I
Sbjct: 275 VSDVAVLVTESGWPSKGDSKEPYATIDNADTYNSNLIKHVFDR--TGTPLHPEMTSSVYI 332
Query: 239 FALYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPPPDDYKGKAWCVVA 298
+ L+NE+L+ +E +GL Y N + +Y + +SG +D + +C+
Sbjct: 333 YELFNEDLRAPPVSEASWGLFYGNSTPVYLLHVSGSGTFLA-------NDTTNQTYCIAM 385
Query: 299 EGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTC 358
+G + + AL +AC G C +QPG+ C++P++V GHAS+AF+SY+ + R G+C
Sbjct: 386 DGVDAKTLQAALDWACGPGRSNCSEIQPGESCYQPNNVKGHASFAFNSYYQKEGRASGSC 445
Query: 359 NFNGLATQIAEDPSKLFF 376
+F G+A DPSKLFF
Sbjct: 446 DFKGVAMITTTDPSKLFF 463
>At3g55430 beta-1,3-glucanase - like protein
Length = 449
Score = 217 bits (553), Expect = 8e-57
Identities = 121/363 (33%), Positives = 196/363 (53%), Gaps = 12/363 (3%)
Query: 12 SNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYRMKHSLTIFGL 71
+N A +WV N++PF+ +T I+Y+ VGNE++ + N +++PAM + ++L G+
Sbjct: 91 ANGRQARRWVSVNILPFHPQTKIKYISVGNEILLTGDNNMINNLLPAMRNLNNALVRAGV 150
Query: 72 HKVKVGTPLAMDVLQTSFP--PSNGTFRNDIALSVMKPMLEFLHVTNSFFFLDVYPFFAW 129
VKV T +++++ PS+G FR ++ P+L + T S F ++ YP+F +
Sbjct: 151 RDVKVTTAHSLNIIAYDLTGAPSSGRFRPGWDKGILAPILAYHRRTKSPFMVNPYPYFGF 210
Query: 130 TSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYPDIQIFIAET 189
DP N+N +A+F + V D T VYTN+FD ++D+ Y AM+ LGY D+ I + ET
Sbjct: 211 --DPKNVN--FAIFRTPYKAVRDPFTRHVYTNMFDALMDSTYSAMKALGYGDVNIVVGET 266
Query: 190 GWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFALYNENLKTG 249
GWP+ D + NA +N N +K+ + GTP P ++IF L+NE K G
Sbjct: 267 GWPS--ACDAPWCSPANAAWFNLNIIKRAQGQ---GTPLMPNRRFETYIFGLFNEEGKPG 321
Query: 250 LGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPPPDDYKGKAWCVVAEGANKTAVVEA 309
ER++GL + S +Y++ L +P P G WCV GA T + ++
Sbjct: 322 PTAERNWGLFRADFSPVYDVGLLRNGQGGGGRPALPAPSTAGGKWCVARSGATNTQLQDS 381
Query: 310 LSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTCNFNGLATQIAE 369
+++ C QG C+ +Q G CF P S+ HAS+ ++Y+ R G CNF+G +
Sbjct: 382 INWVCGQG-VDCKPIQAGGSCFNPSSLRTHASFVMNAYFQSHGRTDGACNFSGTGMIVGN 440
Query: 370 DPS 372
+PS
Sbjct: 441 NPS 443
>At3g15800 putative beta-1,3-glucanase precursor
Length = 399
Score = 217 bits (553), Expect = 8e-57
Identities = 116/274 (42%), Positives = 172/274 (62%), Gaps = 6/274 (2%)
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFY-SKTLIRYLLVGNELISSTTNQTWPHIVPAMY 60
L NE + ++S + A W++ N+ PF T I + VGNE++ T W ++PA
Sbjct: 96 LGNEFLKDISVGEDRAMNWIKENVEPFIRGGTKISGIAVGNEILGGTDIGLWEALLPAAK 155
Query: 61 RMKHSLTIFGLHKV-KVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFF 119
+ +L GLH V +V +P + V S+PPS+ TFR+D+A MKP+L F S F
Sbjct: 156 NVYSALRRLGLHNVVEVSSPHSEAVFANSYPPSSCTFRDDVA-PFMKPLLAFFWQIQSPF 214
Query: 120 FLDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGY 179
+++ YPF A+ SDPI I+++YALFE N + D T L Y N+FD MVDA Y A+E+ GY
Sbjct: 215 YINAYPFLAYKSDPITIDINYALFEH-NKGILDPKTKLHYDNMFDAMVDASYAALEKAGY 273
Query: 180 PDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIF 239
+ + ++ETGW + GD D+ GA++ NA TYNRN K++ K+ GTP RP ++ +++F
Sbjct: 274 TKVPVIVSETGWASKGDADEPGASVKNARTYNRNLRKRLQKRK--GTPYRPDMVVRAYVF 331
Query: 240 ALYNENLKTGLGTERHFGLLYPNGSRIYEIDLSG 273
AL+NEN K G +ER+FGL P+G+ Y+I L+G
Sbjct: 332 ALFNENSKPGPTSERNFGLFKPDGTIAYDIGLTG 365
>At2g27500 beta-1,3-glucanase like protein
Length = 392
Score = 209 bits (531), Expect = 3e-54
Identities = 109/280 (38%), Positives = 173/280 (60%), Gaps = 5/280 (1%)
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYR 61
L NE + N+S++ T A W+Q L P SKT I ++VGNE+ + + ++PAM
Sbjct: 82 LGNEYLQNMSTDPTKAQDWLQQRLEPHISKTRITSIVVGNEIFKTNDHVLIQSLLPAMKS 141
Query: 62 MKHSLTIFGLHK-VKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFF 120
+ +LT GL K V V + ++D+L TS+PPS+G+F+ + + ++P+L+F S F
Sbjct: 142 VYAALTNLGLEKQVTVTSAHSLDILSTSYPPSSGSFKEEF-IQYLQPLLDFHSQIESPFL 200
Query: 121 LDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYP 180
++ YPFFA+ P + L+Y LF+ N + D T L Y N+ VDA+Y A++ LG+
Sbjct: 201 INAYPFFAYKDSPKEVPLEYVLFQP-NQGMVDPNTNLHYDNMLFAQVDALYSAIKTLGHT 259
Query: 181 DIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFA 240
DI++ I+ETGWP+ GD ++IGA+ NA YN N +K + ++ GTPA+ + ++FA
Sbjct: 260 DIEVRISETGWPSKGDENEIGASPENAALYNGNLLKLIQQRK--GTPAKQSVPIDVYVFA 317
Query: 241 LYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEY 280
L+NENLK G +ER++GL YP+G +Y + + G P+ Y
Sbjct: 318 LFNENLKPGPVSERNYGLFYPDGKPVYNVGMQGYLPDIIY 357
>At5g42720 beta-1,3-glucanase-like protein
Length = 438
Score = 199 bits (506), Expect = 2e-51
Identities = 112/281 (39%), Positives = 165/281 (57%), Gaps = 7/281 (2%)
Query: 7 VTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYRMKHSL 66
V ++S+ + A WV+TN+VP+Y + I + VGNE+ S N ++PAM ++ +L
Sbjct: 84 VPGLASDPSFARSWVETNVVPYYPASKIVLIAVGNEITSFGDNSLMSQLLPAMKNVQTAL 143
Query: 67 TIFGLH--KVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFFLDVY 124
L K+KV T M VL S PPS F+ + A ++K +LEF T S F ++ Y
Sbjct: 144 EAASLGGGKIKVSTVHIMSVLAGSDPPSTAVFKPEHA-DILKGLLEFNSETGSPFAVNPY 202
Query: 125 PFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYPDIQI 184
PFFA+ D L Y LF+++ V D + L Y N+FD VDAVY A+ +G+ D++I
Sbjct: 203 PFFAYQDDRRPETLAYCLFQANPGRV-DPNSNLKYMNMFDAQVDAVYSALNSMGFKDVEI 261
Query: 185 FIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFALYNE 244
+AETGWP GD ++ GA + NA YN+N + + K GTP PG ++ +++FALY+E
Sbjct: 262 MVAETGWPYKGDPEEAGATVENARAYNKNLIAHL--KSGSGTPLMPGRVIDTYLFALYDE 319
Query: 245 NLKTGLGTERHFGLLYPNGSRIYEIDLSGKTPEYEYKPLPP 285
NLK G G+ER FGL P+ + Y+I L+ KT Y + P
Sbjct: 320 NLKPGKGSERAFGLFRPDLTMTYDIGLT-KTTNYNQTSMAP 359
>At1g30080 beta-1,3-glucanase precursor, putative
Length = 408
Score = 197 bits (502), Expect = 6e-51
Identities = 109/269 (40%), Positives = 164/269 (60%), Gaps = 6/269 (2%)
Query: 4 NELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYRMK 63
NE++ ++ Q A QWV T + P++ T I + VGNEL + + +++PAM +
Sbjct: 91 NEMLPSLVDPQQ-ALQWVTTRIKPYFPATKIGGIAVGNELYTDDDSSLIGYLMPAMMSIH 149
Query: 64 HSLTIFGLHK-VKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFFLD 122
+L GL K ++V TP ++ VLQ S+PPS G FR ++A VM +L FL TNS F+++
Sbjct: 150 GALVQTGLDKYIQVSTPNSLSVLQESYPPSAGCFRPEVA-GVMTQLLGFLRNTNSPFWIN 208
Query: 123 VYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYPDI 182
YP+FA+ P I LDY LF + N + D T Y N+ VDAV FAM RLG+ DI
Sbjct: 209 AYPYFAYKDSPTKIPLDYVLF-NPNPGMVDPYTKYRYDNMLYAQVDAVIFAMARLGFKDI 267
Query: 183 QIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFALY 242
++ ++ETGWP+ GD D++GA + NA YN+N +++ + GTP RP ++FAL+
Sbjct: 268 EVGVSETGWPSKGDGDEVGATVANAAVYNKNILRRQLQNE--GTPLRPNLSFDVYLFALF 325
Query: 243 NENLKTGLGTERHFGLLYPNGSRIYEIDL 271
NE+LK G +ER++GL P+ + Y + L
Sbjct: 326 NEDLKPGPTSERNYGLYQPDETMTYNVGL 354
>At4g34480 putative protein (fragment)
Length = 356
Score = 196 bits (498), Expect = 2e-50
Identities = 107/267 (40%), Positives = 165/267 (61%), Gaps = 5/267 (1%)
Query: 7 VTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYRMKHSL 66
V +++S+ A QW+ +N++PFY + I + VGNE++ S ++PAM ++ +L
Sbjct: 84 VPSLASDPNAATQWINSNVLPFYPASKIMLITVGNEILMSNDPNLVNQLLPAMQNVQKAL 143
Query: 67 TIFGLH-KVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFFLDVYP 125
L K+KV T +M VL +S PPS+G+F +K +L+FL T S F ++ YP
Sbjct: 144 EAVSLGGKIKVSTVNSMTVLGSSDPPSSGSFAAGYQTG-LKGILQFLSDTGSPFAINPYP 202
Query: 126 FFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYPDIQIF 185
FFA+ SDP L + LFE N DS TG+ YTN+FD VDAV+ A++ +G+ ++I
Sbjct: 203 FFAYQSDPRPETLAFCLFEP-NAGRVDSKTGIKYTNMFDAQVDAVHSALKSMGFEKVEIV 261
Query: 186 IAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFALYNEN 245
+AETGW + GD +++GA++ NA YN N + + + VGTP PG + ++IFALY+EN
Sbjct: 262 VAETGWASRGDANEVGASVDNAKAYNGNLIAHL--RSMVGTPLMPGKPVDTYIFALYDEN 319
Query: 246 LKTGLGTERHFGLLYPNGSRIYEIDLS 272
LK G +ER FGL + S +Y++ L+
Sbjct: 320 LKPGPSSERAFGLFKTDLSMVYDVGLA 346
>At2g39640 putative beta-1,3-glucanase, predicted GPI-anchored
protein
Length = 549
Score = 196 bits (497), Expect = 2e-50
Identities = 128/374 (34%), Positives = 191/374 (50%), Gaps = 21/374 (5%)
Query: 1 MLPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMY 60
M+PN + + N A QWV N++PF + +Y+ VGNE+++S N ++VPAM
Sbjct: 82 MVPNGNIPAMV-NVANARQWVAANVLPFQQQIKFKYVCVGNEILASNDNNLISNLVPAMQ 140
Query: 61 RMKHSLTIFGLHKVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFF 120
+ +L L +KV TP A + PS F ND + +LEF S F
Sbjct: 141 SLNEALKASNLTYIKVTTPHAFTISYNRNTPSESRFTND-QKDIFTKILEFHRQAKSPFM 199
Query: 121 LDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYP 180
++ Y FF T D N+N YA+F N +TD+ T YTN+FD ++DA Y AM+ LGY
Sbjct: 200 INAYTFF--TMDTNNVN--YAIFGPSN-AITDTNTQQTYTNMFDAVMDATYSAMKALGYG 254
Query: 181 DIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFA 240
D+ I + ETGWP D + NA YN N +K+ +GTP P + FIFA
Sbjct: 255 DVDIAVGETGWPT--ACDASWCSPQNAENYNLNIIKRA---QVIGTPLMPNRHIDIFIFA 309
Query: 241 LYNENLKTGLGTERHFGLLYPNGSRIYEID-LSGKTPEYEYKPLP-PPDDYKGKAWCVVA 298
L+NE+ K G ER++G+ P+ S +Y++ L G PLP PP + GK WCV
Sbjct: 310 LFNEDGKPGPTRERNWGIFKPDFSPMYDVGVLKGGG-----SPLPFPPINNNGK-WCVGK 363
Query: 299 EGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTC 358
A + + + CS G C + PG CF+ +++ +S+ ++Y+ V C
Sbjct: 364 PEATLMQLQANIDWVCSHG-IDCTPISPGGICFDNNNMTTRSSFIMNAYYQSKGCVDVVC 422
Query: 359 NFNGLATQIAEDPS 372
+F+G + +PS
Sbjct: 423 DFSGTGIVTSTNPS 436
Score = 43.1 bits (100), Expect = 3e-04
Identities = 20/83 (24%), Positives = 40/83 (48%), Gaps = 1/83 (1%)
Query: 290 KGKAWCVVAEGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWA 349
K WC+ + A +T + + + CSQG C+ + PG CF+ +++ +++ ++Y+
Sbjct: 456 KSANWCMAKQEATETQLQANIDWVCSQGI-DCKPISPGGICFDNNNMKTRSTFIMNAYYE 514
Query: 350 QFRRVGGTCNFNGLATQIAEDPS 372
C+F G +PS
Sbjct: 515 SKGYSKDACDFRGSGIVTTTNPS 537
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,293,781
Number of Sequences: 26719
Number of extensions: 427522
Number of successful extensions: 1303
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 77
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1011
Number of HSP's gapped (non-prelim): 87
length of query: 389
length of database: 11,318,596
effective HSP length: 101
effective length of query: 288
effective length of database: 8,619,977
effective search space: 2482553376
effective search space used: 2482553376
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136841.5


