

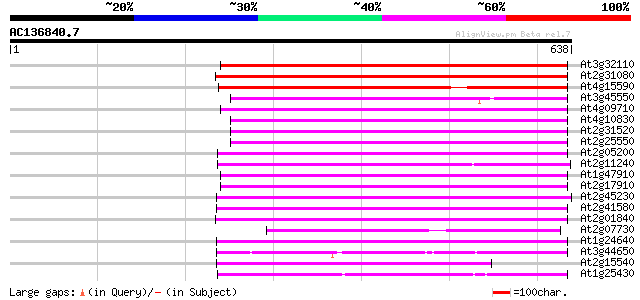
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136840.7 - phase: 0 /pseudo
(638 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g32110 non-LTR reverse transcriptase, putative 363 e-100
At2g31080 putative non-LTR retroelement reverse transcriptase 354 1e-97
At4g15590 reverse transcriptase like protein 331 9e-91
At3g45550 putative protein 296 2e-80
At4g09710 RNA-directed DNA polymerase -like protein 295 4e-80
At4g10830 putative protein 291 8e-79
At2g31520 putative non-LTR retroelement reverse transcriptase 291 1e-78
At2g25550 putative non-LTR retroelement reverse transcriptase 291 1e-78
At2g05200 putative non-LTR retroelement reverse transcriptase 288 5e-78
At2g11240 pseudogene 278 7e-75
At1g47910 reverse transcriptase, putative 275 7e-74
At2g17910 putative non-LTR retroelement reverse transcriptase 265 8e-71
At2g45230 putative non-LTR retroelement reverse transcriptase 264 1e-70
At2g41580 putative non-LTR retroelement reverse transcriptase 263 3e-70
At2g01840 putative non-LTR retroelement reverse transcriptase 254 8e-68
At2g07730 putative non-LTR retroelement reverse transcriptase 252 4e-67
At1g24640 hypothetical protein 244 8e-65
At3g44650 putative protein 236 2e-62
At2g15540 putative non-LTR retroelement reverse transcriptase 236 3e-62
At1g25430 hypothetical protein 236 4e-62
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 363 bits (932), Expect = e-100
Identities = 175/395 (44%), Positives = 252/395 (63%)
Query: 240 KVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIAL 299
+V A+ S YKAPGPDGF +F++Q +VG+ V F + F +D L+ L
Sbjct: 972 EVEGAIRSMGKYKAPGPDGFQPVFYQQGWEVVGESVTKFVMDFFSSGSFPQETNDVLVVL 1031
Query: 300 IPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAI 359
I KV P FRPISLCN+++K ITKV+V RL+ ++N +IGP Q+SF+PGR +TDN +
Sbjct: 1032 IAKVLKPEKITQFRPISLCNVLFKTITKVMVGRLKGVINKLIGPAQTSFIPGRLSTDNIV 1091
Query: 360 VLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTS 419
V+QEV+H MR K KG++ K+DLEKA+D + WD L L G P T ++ IM CV
Sbjct: 1092 VVQEVVHSMRRKKGVKGWMLLKLDLEKAYDRIRWDLLEDTLKAAGLPGTWVQWIMKCVEG 1151
Query: 420 SSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNP 479
S +LWNG + F+P GLRQGDPLSPYLF+LC+E+L I +++ W+PI +S
Sbjct: 1152 PSMRLLWNGEKTDAFKPLRGLRQGDPLSPYLFVLCIERLCHLIESSIAAKKWKPIKISQS 1211
Query: 480 GPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDKI 539
GP+LSH+ FADD++LF +A+ Q+R +R + ++F SG K++L KS+ ++S V +D
Sbjct: 1212 GPRLSHICFADDLILFAEASIDQIRVLRGVLEKFCGASGQKVSLEKSKIYFSKNVLRDLG 1271
Query: 540 NTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKPGRLAL 599
++ SG++ T L KYLG PI++ R K F +I + RLA W+ ++L+ GRL L
Sbjct: 1272 KRISEESGMKATKDLGKYLGVPILQKRINKETFGEVIKRFSSRLAGWKGRMLSFAGRLTL 1331
Query: 600 ASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
VL+SI + M LPQS D +D+V+R F+W
Sbjct: 1332 TKAVLTSILVHTMSTIKLPQSTLDGLDKVSRAFLW 1366
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 354 bits (908), Expect = 1e-97
Identities = 176/400 (44%), Positives = 253/400 (63%)
Query: 235 ALNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISD 294
A K +V +A+ S +KAPGPDG+ +F++Q VG V F T S +D
Sbjct: 291 AFTKAEVVSAVKSMGRFKAPGPDGYQPVFYQQCWETVGPSVTRFVLEFFETGVLPASTND 350
Query: 295 TLIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRST 354
L+ LI KV P + FRP+SLCN+++KIITK++V RL+ +++ +IGP Q+SF+PGR +
Sbjct: 351 ALLVLIAKVAKPERIQQFRPVSLCNVLFKIITKMMVTRLKNVISKLIGPAQASFIPGRLS 410
Query: 355 TDNAIVLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIM 414
DN +++QE +H MR K +KG++ K+DLEKA+D V WDFL+ L G + IM
Sbjct: 411 IDNIVLVQEAVHSMRRKKGRKGWMLLKLDLEKAYDRVRWDFLQETLEAAGLSEGWTSRIM 470
Query: 415 HCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPI 474
VT S S+LWNG R F P GLRQGDPLSPYLF+LC+E+L I +V + W+PI
Sbjct: 471 AGVTDPSMSVLWNGERTDSFVPARGLRQGDPLSPYLFVLCLERLCHLIEASVGKREWKPI 530
Query: 475 HVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGV 534
VS G KLSH+ FADD++LF +A+ Q+R IR + +RF + SG K++L KS+ F+S V
Sbjct: 531 AVSCGGSKLSHVCFADDLILFAEASVAQIRIIRRVLERFCEASGQKVSLEKSKIFFSHNV 590
Query: 535 TQDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKP 594
+++ ++ SGI CT L KYLG PI++ R K F +++++ RLA W+ + L+
Sbjct: 591 SREMEQLISEESGIGCTKELGKYLGMPILQKRMNKETFGEVLERVSARLAGWKGRSLSLA 650
Query: 595 GRLALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
GR+ L VLSSI + M LP S D +D+ +R F+W
Sbjct: 651 GRITLTKAVLSSIPVHVMSAILLPVSTLDTLDRYSRTFLW 690
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 331 bits (848), Expect = 9e-91
Identities = 162/397 (40%), Positives = 245/397 (60%), Gaps = 17/397 (4%)
Query: 238 KKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLI 297
+ +V A+ S +KAPGPDG+ +F++Q VG+ V F + S +D L+
Sbjct: 246 RDEVVVAVRSMGRFKAPGPDGYQPVFYQQCWETVGESVSKFVMEFFESGVLPKSTNDVLL 305
Query: 298 ALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDN 357
L+ KV P FRP+SLCN+++KIITK++V RL+ +++ +IGP Q+SF+PGR + DN
Sbjct: 306 VLLAKVAKPERITQFRPVSLCNVLFKIITKMMVIRLKNVISKLIGPAQASFIPGRLSFDN 365
Query: 358 AIVLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCV 417
+V+QE +H MR K +KG++ K+DLEKA+D + WDFL L G + IK IM CV
Sbjct: 366 IVVVQEAVHSMRRKKGRKGWMLLKLDLEKAYDRIRWDFLAETLEAAGLSEGWIKRIMECV 425
Query: 418 TSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVS 477
S+LWNG + F P GLRQGDP+SPYLF+LC+E+L I AV +G W+ I +S
Sbjct: 426 AGPEMSLLWNGEKTDSFTPERGLRQGDPISPYLFVLCIERLCHQIETAVGRGDWKSISIS 485
Query: 478 NPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQD 537
GPK+SH+ FADD++LF +A+ Q K++L KS+ F+S+ V++D
Sbjct: 486 QGGPKVSHVCFADDLILFAEASVAQ-----------------KVSLEKSKIFFSNNVSRD 528
Query: 538 KINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKPGRL 597
+T+ +GI T L KYLG P+++ R K F +++++ RL+ W+++ L+ GR+
Sbjct: 529 LEGLITAETGIGSTRELGKYLGMPVLQKRINKDTFGEVLERVSSRLSGWKSRSLSLAGRI 588
Query: 598 ALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
L VL SI + M LP S+ + +D+V+RNF+W
Sbjct: 589 TLTKAVLMSIPIHTMSSILLPASLLEQLDKVSRNFLW 625
>At3g45550 putative protein
Length = 851
Score = 296 bits (758), Expect = 2e-80
Identities = 160/387 (41%), Positives = 227/387 (58%), Gaps = 7/387 (1%)
Query: 252 KAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIALIPKVDPPLTYKD 311
KAPGPDG F+KQ IVG+DV F +SH S++ T I +IPK+ P T D
Sbjct: 65 KAPGPDGLTARFYKQCWDIVGNDVIKEVKLFFESSHMKTSVNHTNICMIPKIQNPQTLSD 124
Query: 312 FRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAIVLQEVIHYMRIS 371
+RPI+LCN++YK+I+K +V+RL+ LN+I+ Q++F+PGR DN ++ E++H +++
Sbjct: 125 YRPIALCNVLYKVISKCMVNRLKAHLNSIVSDSQAAFIPGRIINDNVMIAHEIMHSLKVR 184
Query: 372 KK-KKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTSSSFSILWNGNR 430
K+ K Y+A K D+ KA+D V WDFL + + FGF D I IM V S +S+L NG+
Sbjct: 185 KRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCDKWIGWIMAAVKSVHYSVLINGSP 244
Query: 431 LPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNPGPKLSHLLFAD 490
PT G+RQGDPLSPYLFILC + LS I G + + N P ++HL FAD
Sbjct: 245 HGYISPTRGIRQGDPLSPYLFILCGDILSHLIKVKASSGDIRGVRIGNGAPAITHLQFAD 304
Query: 491 DVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSS---GVTQDKINTLTSISG 547
D L F +AN + ++D+FD + +SG KIN+ KS + S G TQ ++ TL +I
Sbjct: 305 DSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSLITFGSRVYGSTQTRLKTLLNIPN 364
Query: 548 IRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKPGRLALASFVLSSI 607
KYLG P GR KK F +IID++++R ASW K L+ G+ L V ++
Sbjct: 365 ---QGGGGKYLGLPEQFGRKKKEMFNYIIDRVKERTASWSAKFLSPAGKEILLKSVALAM 421
Query: 608 STYYMQINWLPQSVCDNIDQVTRNFIW 634
Y M LPQ + I+ + NF W
Sbjct: 422 PVYAMSCFKLPQGIVSEIESLLMNFWW 448
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 295 bits (756), Expect = 4e-80
Identities = 159/396 (40%), Positives = 226/396 (56%), Gaps = 1/396 (0%)
Query: 240 KVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIAL 299
++ AL S KAPGPDGF FF Y I+ DV S F+ S P +++T + L
Sbjct: 373 EIKEALFSISADKAPGPDGFSASFFHAYWDIIEADVSRDIRSFFVDSCLSPRLNETHVTL 432
Query: 300 IPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAI 359
IPK+ P D+RPI+LCN+ YKI+ K+L RL+P L+ +I +QS+F+PGR+ DN +
Sbjct: 433 IPKISAPRKVSDYRPIALCNVQYKIVAKILTRRLQPWLSELISLHQSAFVPGRAIADNVL 492
Query: 360 VLQEVIHYMRISKKKKGY-VAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVT 418
+ E++H++R+S KK +A K D+ KA+D + W+FL+ L GF D I+ +M CV
Sbjct: 493 ITHEILHFLRVSGAKKYCSMAIKTDMSKAYDRIKWNFLQEVLMRLGFHDKWIRWVMQCVC 552
Query: 419 SSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSN 478
+ S+S L NG+ P+ GLRQGDPLSPYLFILC E LS A ++G I V+
Sbjct: 553 TVSYSFLINGSPQGSVVPSRGLRQGDPLSPYLFILCTEVLSGLCRKAQEKGVMVGIRVAR 612
Query: 479 PGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDK 538
P+++HLLFADD + F K N T + ++ ++ SG INL KS +SS QD
Sbjct: 613 GSPQVNHLLFADDTMFFCKTNPTCCGALSNILKKYELASGQSINLAKSAITFSSKTPQDI 672
Query: 539 INTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKPGRLA 598
+ I + KYLG P GR K+ F I+D++++R SW + L+ G+
Sbjct: 673 KRRVKLSLRIDNEGGIGKYLGLPEHFGRRKRDIFSSIVDRIRQRSHSWSIRFLSSAGKQI 732
Query: 599 LASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
L VLSS+ +Y M LP S+C I V F W
Sbjct: 733 LLKAVLSSMPSYAMMCFKLPASLCKQIQSVLTRFWW 768
>At4g10830 putative protein
Length = 1294
Score = 291 bits (745), Expect = 8e-79
Identities = 157/384 (40%), Positives = 226/384 (57%), Gaps = 1/384 (0%)
Query: 252 KAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIALIPKVDPPLTYKD 311
KAPGPDG F+K IVG DV F TS+ SI+ T I +IPK+ P T D
Sbjct: 806 KAPGPDGLTARFYKSCWEIVGPDVIKEVKIFFRTSYMKQSINHTNICMIPKITNPETLSD 865
Query: 312 FRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAIVLQEVIHYMRIS 371
+RPI+LCN++YKII+K LV RL+ L+ I+ Q++F+PGR DN ++ E++H ++
Sbjct: 866 YRPIALCNVLYKIISKCLVERLKGHLDAIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTR 925
Query: 372 KK-KKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTSSSFSILWNGNR 430
K+ + Y+A K D+ KA+D V W+FL + + FGF +T IK IM V S ++S+L NG
Sbjct: 926 KRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGFSETWIKWIMGAVKSVNYSVLVNGIP 985
Query: 431 LPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNPGPKLSHLLFAD 490
+P G+RQGDPLSPYLFILC + L+ I N V +G I + N P ++HL FAD
Sbjct: 986 HGTIQPQRGIRQGDPLSPYLFILCADILNHLIKNRVAEGDIRGIRIGNGVPGVTHLQFAD 1045
Query: 491 DVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDKINTLTSISGIRC 550
D L F ++N + ++D+FD + +SG KIN+ KS + S V N L +I GI+
Sbjct: 1046 DSLFFCQSNVRNCQALKDVFDVYEYYSGQKINMSKSMITFGSRVHGTTQNRLKNILGIQS 1105
Query: 551 TTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKPGRLALASFVLSSISTY 610
KYLG P GR K+ F +II++++KR +SW K L+ G+ + V S+ Y
Sbjct: 1106 HGGGGKYLGLPEQFGRKKRDMFNYIIERVKKRTSSWSAKYLSPAGKEIMLKSVAMSMPVY 1165
Query: 611 YMQINWLPQSVCDNIDQVTRNFIW 634
M LP ++ I+ + NF W
Sbjct: 1166 AMSCFKLPLNIVSEIEALLMNFWW 1189
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 291 bits (744), Expect = 1e-78
Identities = 157/384 (40%), Positives = 222/384 (56%), Gaps = 1/384 (0%)
Query: 252 KAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIALIPKVDPPLTYKD 311
KAPGPDG F+K IVG DV F TS PSI+ T I +IPK+ P T D
Sbjct: 600 KAPGPDGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSD 659
Query: 312 FRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAIVLQEVIHYMRIS 371
+RPI+LCN++YK+I+K LV+RL+ LN+I+ Q++F+PGR DN ++ EV+H +++
Sbjct: 660 YRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVR 719
Query: 372 KK-KKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTSSSFSILWNGNR 430
K+ K Y+A K D+ KA+D V WDFL + + FGF + I IM V S +S+L NG+
Sbjct: 720 KRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSP 779
Query: 431 LPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNPGPKLSHLLFAD 490
PT G+RQGDPLSPYLFILC + LS IN G + + N P ++HL FAD
Sbjct: 780 HGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGNGAPAITHLQFAD 839
Query: 491 DVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDKINTLTSISGIRC 550
D L F +AN + ++D+FD + +SG KIN+ KS + S V + L I I
Sbjct: 840 DSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSKLKQILEIPN 899
Query: 551 TTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKPGRLALASFVLSSISTY 610
KYLG P GR KK F +IID+++KR ++W + L+ G+ + V ++ Y
Sbjct: 900 QGGGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVY 959
Query: 611 YMQINWLPQSVCDNIDQVTRNFIW 634
M LP+ + I+ + NF W
Sbjct: 960 AMSCFKLPKGIVSEIESLLMNFWW 983
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 291 bits (744), Expect = 1e-78
Identities = 157/384 (40%), Positives = 222/384 (56%), Gaps = 1/384 (0%)
Query: 252 KAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIALIPKVDPPLTYKD 311
KAPGPDG F+K IVG DV F TS PSI+ T I +IPK+ P T D
Sbjct: 826 KAPGPDGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSD 885
Query: 312 FRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAIVLQEVIHYMRIS 371
+RPI+LCN++YK+I+K LV+RL+ LN+I+ Q++F+PGR DN ++ EV+H +++
Sbjct: 886 YRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVR 945
Query: 372 KK-KKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTSSSFSILWNGNR 430
K+ K Y+A K D+ KA+D V WDFL + + FGF + I IM V S +S+L NG+
Sbjct: 946 KRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSP 1005
Query: 431 LPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNPGPKLSHLLFAD 490
PT G+RQGDPLSPYLFILC + LS IN G + + N P ++HL FAD
Sbjct: 1006 HGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGNGAPAITHLQFAD 1065
Query: 491 DVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDKINTLTSISGIRC 550
D L F +AN + ++D+FD + +SG KIN+ KS + S V + L I I
Sbjct: 1066 DSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSRLKQILEIPN 1125
Query: 551 TTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKPGRLALASFVLSSISTY 610
KYLG P GR KK F +IID+++KR ++W + L+ G+ + V ++ Y
Sbjct: 1126 QGGGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVY 1185
Query: 611 YMQINWLPQSVCDNIDQVTRNFIW 634
M LP+ + I+ + NF W
Sbjct: 1186 AMSCFKLPKGIVSEIESLLMNFWW 1209
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 288 bits (738), Expect = 5e-78
Identities = 159/399 (39%), Positives = 224/399 (55%), Gaps = 1/399 (0%)
Query: 237 NKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTL 296
N ++V A+ S KAPGPDGF F+ Y HI+ DV F + +F +++T
Sbjct: 324 NDEEVKDAVFSINASKAPGPDGFTAGFYHSYWHIISTDVGREIRLFFTSKNFPRRMNETH 383
Query: 297 IALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTD 356
I LIPK P D+RPI+LCNI YKI+ K++ R++ IL +I QS+F+PGR +D
Sbjct: 384 IRLIPKDLGPRKVADYRPIALCNIFYKIVAKIMTKRMQLILPKLISENQSAFVPGRVISD 443
Query: 357 NAIVLQEVIHYMRISKKKKGY-VAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMH 415
N ++ EV+H++R S KK +A K D+ KA+D V WDFL+ L FGF I ++
Sbjct: 444 NVLITHEVLHFLRTSSAKKHCSMAVKTDMSKAYDRVEWDFLKKVLQRFGFHSIWIDWVLE 503
Query: 416 CVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIH 475
CVTS S+S L NG PT GLRQGDPLSP LFILC E LS A + +
Sbjct: 504 CVTSVSYSFLINGTPQGKVVPTRGLRQGDPLSPCLFILCTEVLSGLCTRAQRLRQLPGVR 563
Query: 476 VSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVT 535
VS GP+++HLLFADD + F+K++ + ++ R+ K SG IN HKS +SS
Sbjct: 564 VSINGPRVNHLLFADDTMFFSKSDPESCNKLSEILSRYGKASGQSINFHKSSVTFSSKTP 623
Query: 536 QDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKPG 595
+ + I IR KYLG P GR K+ F IIDK++++ SW ++ L++ G
Sbjct: 624 RSVKGQVKRILKIRKEGGTGKYLGLPEHFGRRKRDIFGAIIDKIRQKSHSWASRFLSQAG 683
Query: 596 RLALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
+ + VL+S+ Y M LP ++C I + F W
Sbjct: 684 KQVMLKAVLASMPLYSMSCFKLPSALCRKIQSLLTRFWW 722
>At2g11240 pseudogene
Length = 1044
Score = 278 bits (711), Expect = 7e-75
Identities = 153/403 (37%), Positives = 229/403 (55%), Gaps = 3/403 (0%)
Query: 237 NKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTL 296
N +++ +A S KAPGPDGF FF+ VG ++ S F +S P+I+ T
Sbjct: 255 NPEEIKSACFSIHADKAPGPDGFSASFFQSNWMTVGPNIVLEIQSFFSSSTLQPTINKTH 314
Query: 297 IALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTD 356
I LIPK+ D+RPI+LC + YKII+K+L RL+PIL II QS+F+P R++ D
Sbjct: 315 ITLIPKIQSLKRMVDYRPIALCTVFYKIISKLLSRRLQPILQEIISENQSAFVPKRASND 374
Query: 357 NAIVLQEVIHYMR-ISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMH 415
N ++ E +HY++ + +K+ ++A K ++ KA+D + WDF++ + + GF T I I+
Sbjct: 375 NVLITHEALHYLKSLGAEKRCFMAVKTNMSKAYDRIEWDFIKLVMQEMGFHQTWISWILQ 434
Query: 416 CVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIH 475
C+T+ S+S L NG+ P GLRQGDPLSP+LFI+C E LS A GS +
Sbjct: 435 CITTVSYSFLLNGSAQGAVTPERGLRQGDPLSPFLFIICSEVLSGLCRKAQLDGSLLGLR 494
Query: 476 VSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVT 535
VS P+++HLLFADD + F +++ + + ++ + SG IN KS A S T
Sbjct: 495 VSKGNPRVNHLLFADDTIFFCRSDLKSCKTFLCILKKYEEASGQMINKSKS-AITFSRKT 553
Query: 536 QDKINT-LTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKP 594
D I T I GI+ L KYLG P + GR K+ F I+D++++R SW ++ L+
Sbjct: 554 PDHIKTEAQQILGIQLVGGLGKYLGLPKMFGRKKRDLFNQIVDRIRQRSLSWSSRFLSTA 613
Query: 595 GRLALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIWRDS 637
G+ + VL+S+ TY M L S+C I +F W S
Sbjct: 614 GKTTMLKSVLASMPTYTMSCFKLLVSLCKRIQSALTHFWWDSS 656
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 275 bits (702), Expect = 7e-74
Identities = 150/396 (37%), Positives = 214/396 (53%), Gaps = 1/396 (0%)
Query: 240 KVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIAL 299
+V AAL P KAPGPDG +FF++ I+ D+ +L S FD ++ T I L
Sbjct: 216 EVRAALFMIHPEKAPGPDGMTALFFQKSWAIIKSDLLSLVNSFLQEGVFDKRLNTTNICL 275
Query: 300 IPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAI 359
IPK + P + RPISLCN+ YK+I+K+L RL+ +L N+I QS+F+ GR +DN +
Sbjct: 276 IPKTERPTRMTELRPISLCNVGYKVISKILCQRLKTVLPNLISETQSAFVDGRLISDNIL 335
Query: 360 VLQEVIHYMRISKK-KKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVT 418
+ QE+ H +R + K ++A K D+ KA+D V W+F+ + L GF + I IM C+T
Sbjct: 336 IAQEMFHGLRTNSSCKDKFMAIKTDMSKAYDQVEWNFIEALLRKMGFCEKWISWIMWCIT 395
Query: 419 SSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSN 478
+ + +L NG P GLRQGDPLSPYLFILC E L + I A +Q I V+
Sbjct: 396 TVQYKVLINGQPKGLIIPERGLRQGDPLSPYLFILCTEVLIANIRKAERQNLITGIKVAT 455
Query: 479 PGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDK 538
P P +SHLLFADD L F KAN Q I ++ ++ SG +IN KS + V
Sbjct: 456 PSPAVSHLLFADDSLFFCKANKEQCGIILEILKQYESVSGQQINFSKSSIQFGHKVEDSI 515
Query: 539 INTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKPGRLA 598
+ I GI + YLG P G K F F+ D++Q R+ W K L+K G+
Sbjct: 516 KADIKLILGIHNLGGMGSYLGLPESLGGSKTKVFSFVRDRLQSRINGWSAKFLSKGGKEV 575
Query: 599 LASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
+ V +++ Y M LP+++ + F W
Sbjct: 576 MIKSVAATLPRYVMSCFRLPKAITSKLTSAVAKFWW 611
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 265 bits (676), Expect = 8e-71
Identities = 149/396 (37%), Positives = 212/396 (52%), Gaps = 1/396 (0%)
Query: 240 KVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIAL 299
+V A+ S APGPDGF +FF+Q+ +V + F T + T I L
Sbjct: 410 EVYNAVFSINKESAPGPDGFTALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICL 469
Query: 300 IPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAI 359
IPK+ P D RPISLC+++YKII+K+L RL+ L I+ QS+F+P R +DN +
Sbjct: 470 IPKITSPQRMSDLRPISLCSVLYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNIL 529
Query: 360 VLQEVIHYMRISKK-KKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVT 418
V E+IH +R + + K ++AFK D+ KA+D V W FL + + GF + I IM+CVT
Sbjct: 530 VAHEMIHSLRTNDRISKEHMAFKTDMSKAYDRVEWPFLETMMTALGFNNKWISWIMNCVT 589
Query: 419 SSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSN 478
S S+S+L NG PT G+RQGDPLSP LF+LC E L +N A Q G I +
Sbjct: 590 SVSYSVLINGQPYGHIIPTRGIRQGDPLSPALFVLCTEALIHILNKAEQAGKITGIQFQD 649
Query: 479 PGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDK 538
++HLLFADD LL KA + + ++ + SG INL+KS + V
Sbjct: 650 KKVSVNHLLFADDTLLMCKATKQECEELMQCLSQYGQLSGQMINLNKSAITFGKNVDIQI 709
Query: 539 INTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKPGRLA 598
+ + S SGI KYLG P K+ F FI +K+Q RL W K L++ G+
Sbjct: 710 KDWIKSRSGISLEGGTGKYLGLPECLSGSKRDLFGFIKEKLQSRLTGWYAKTLSQGGKEV 769
Query: 599 LASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
L + ++ Y M LP+++C + V +F W
Sbjct: 770 LLKSIALALPVYVMSCFKLPKNLCQKLTTVMMDFWW 805
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 264 bits (675), Expect = 1e-70
Identities = 135/404 (33%), Positives = 223/404 (54%), Gaps = 1/404 (0%)
Query: 236 LNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDT 295
+ K++V A S P+K PGPDG + ++Q+ +GD + + + F + + ++ T
Sbjct: 428 ITKEEVQRATFSINPHKCPGPDGMNGFLYQQFWETMGDQITEMVQAFFRSGSIEEGMNKT 487
Query: 296 LIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTT 355
I LIPK+ DFRPISLCN+IYK+I K++ +RL+ IL ++I Q++F+ GR +
Sbjct: 488 NICLIPKILKAEKMTDFRPISLCNVIYKVIGKLMANRLKKILPSLISETQAAFVKGRLIS 547
Query: 356 DNAIVLQEVIHYMRISKK-KKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIM 414
DN ++ E++H + + K + ++A K D+ KA+D V W FL + GF D I+LIM
Sbjct: 548 DNILIAHELLHALSSNNKCSEEFIAIKTDISKAYDRVEWPFLEKAMRGLGFADHWIRLIM 607
Query: 415 HCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPI 474
CV S + +L NG P+ GLRQGDPLSPYLF++C E L + +A Q+ +
Sbjct: 608 ECVKSVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLFVICTEMLVKMLQSAEQKNQITGL 667
Query: 475 HVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGV 534
V+ P +SHLLFADD + + K N L I + + +S SG ++N KS ++ +
Sbjct: 668 KVARGAPPISHLLFADDSMFYCKVNDEALGQIIRIIEEYSLASGQRVNYLKSSIYFGKHI 727
Query: 535 TQDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKP 594
++++ + GI YLG P K + ++ D++ K++ WQ+ L+
Sbjct: 728 SEERRCLVKRKLGIEREGGEGVYLGLPESFQGSKVATLSYLKDRLGKKVLGWQSNFLSPG 787
Query: 595 GRLALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIWRDSR 638
G+ L V ++ TY M +P+++C I+ V F W++ +
Sbjct: 788 GKEILLKAVAMALPTYTMSCFKIPKTICQQIESVMAEFWWKNKK 831
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 263 bits (671), Expect = 3e-70
Identities = 145/400 (36%), Positives = 218/400 (54%), Gaps = 1/400 (0%)
Query: 236 LNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDT 295
+N++++ A+ S APGPDGF +FF++ +V + + + F T + T
Sbjct: 157 VNEQEIYKAVFSINAESAPGPDGFTALFFQRQWPLVKNQIISDIELFFQTGILPEDWNHT 216
Query: 296 LIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTT 355
+ LIPK+ P D RPISLC+++YKII+K+L RL+ L I+ P QS+F+ R +
Sbjct: 217 HLCLIPKITKPARMADIRPISLCSVMYKIISKILSARLKKYLPVIVSPTQSAFVAERLVS 276
Query: 356 DNAIVLQEVIHYMRISKK-KKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIM 414
DN I+ E++H +R ++K K ++ FK D+ KA+D V W FL+ L GF T I +M
Sbjct: 277 DNIILAHEIVHNLRTNEKISKDFMVFKTDMSKAYDRVEWPFLKGILLALGFNSTWINWMM 336
Query: 415 HCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPI 474
CV+S S+S+L NG P GLRQGDPLSP+LF+LC E L +N A + G I
Sbjct: 337 ACVSSVSYSVLINGQPFGHITPHRGLRQGDPLSPFLFVLCTEALIHILNQAEKIGKISGI 396
Query: 475 HVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGV 534
+ GP ++HLLFADD LL KA+ + I ++ SG IN KS + + V
Sbjct: 397 QFNGTGPSVNHLLFADDTLLICKASQLECAEIMHCLSQYGHISGQMINSEKSAITFGAKV 456
Query: 535 TQDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKP 594
++ + + SGI+ KYLG P K+ F FI +K+Q RL+ W K L++
Sbjct: 457 NEETKQWIMNRSGIQTEGGTGKYLGLPECFQGSKQVLFGFIKEKLQSRLSGWYAKTLSQG 516
Query: 595 GRLALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
G+ L + + Y M L +++C + V +F W
Sbjct: 517 GKDILLKSIAMAFPVYAMTCFRLSKTLCTKLTSVMMDFWW 556
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 254 bits (650), Expect = 8e-68
Identities = 137/401 (34%), Positives = 218/401 (54%), Gaps = 1/401 (0%)
Query: 235 ALNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISD 294
++ ++V A+ + +APG DGF F+ ++G+DV + F + D I+
Sbjct: 789 SVTDQEVRDAVFAIGADRAPGFDGFTAAFYHHLWDLIGNDVCLMVRHFFESDVMDNQINQ 848
Query: 295 TLIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRST 354
T I LIPK+ P D+RPISLC YKII+K+L+ RL+ L ++I Q++F+PG++
Sbjct: 849 TQICLIPKIIDPKHMSDYRPISLCTASYKIISKILIKRLKQCLGDVISDSQAAFVPGQNI 908
Query: 355 TDNAIVLQEVIHYMRISKK-KKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLI 413
+DN +V E++H ++ ++ + GYVA K D+ KA+D V W+FL + GF +K I
Sbjct: 909 SDNVLVAHELLHSLKSRRECQSGYVAVKTDISKAYDRVEWNFLEKVMIQLGFAPRWVKWI 968
Query: 414 MHCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEP 473
M CVTS S+ +L NG+ P+ G+RQGDPLSPYLF+ C E LS+ + A
Sbjct: 969 MTCVTSVSYEVLINGSPYGKIFPSRGIRQGDPLSPYLFLFCAEVLSNMLRKAEVNKQIHG 1028
Query: 474 IHVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSG 533
+ ++ +SHLLFADD L F +A++ + + +F ++ + SG KIN KS +
Sbjct: 1029 MKITKDCLAISHLLFADDSLFFCRASNQNIEQLALIFKKYEEASGQKINYAKSSIIFGQK 1088
Query: 534 VTQDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNK 593
+ + L + GI KYLG P GR K F +I+ K+++R W L+
Sbjct: 1089 IPTMRRQRLHRLLGIDNVRGGGKYLGLPEQLGRRKVELFEYIVTKVKERTEGWAYNYLSP 1148
Query: 594 PGRLALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
G+ + + ++ Y M LP +C+ I+ + F W
Sbjct: 1149 AGKEIVIKAIAMALPVYSMNCFLLPTLICNEINSLITAFWW 1189
>At2g07730 putative non-LTR retroelement reverse transcriptase
Length = 970
Score = 252 bits (644), Expect = 4e-67
Identities = 128/334 (38%), Positives = 199/334 (59%), Gaps = 18/334 (5%)
Query: 293 SDTLIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGR 352
+D L+ LI KV P FRP+SLCN+++KIITK++V+RL+ ++ +IGP Q++F+PGR
Sbjct: 199 NDALLVLIAKVAKPERITQFRPMSLCNVLFKIITKMIVNRLKKVILKLIGPAQANFIPGR 258
Query: 353 STTDNAIVLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKL 412
+ DN +++QE +H MR K +KG++ K+DLEKA+D + W+FL+ L G +
Sbjct: 259 LSIDNIVLVQEAVHSMRRKKGRKGWMLLKLDLEKAYDRIRWNFLQETLVATGLSEVWTHR 318
Query: 413 IMHCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWE 472
IM VT S S+LWNG + F P GLRQGDPLSPYLF+LC+E+L I +V W+
Sbjct: 319 IMAGVTDPSMSVLWNGGKTDSFVPARGLRQGDPLSPYLFVLCLERLCHLIEASVGTWDWK 378
Query: 473 PIHVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSS 532
P+ +S +A+ Q+ IR + ++F + SG ++L KS+ +S+
Sbjct: 379 PMALS------------------CEASVAQILIIRRVLEQFCEASGQNVSLEKSKIVFSN 420
Query: 533 GVTQDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLN 592
V++D ++ SGI CT L KYLG PI++ R K +++ + LA W+++ L+
Sbjct: 421 NVSRDMERLISGESGIGCTRELGKYLGMPILQKRMNKETSGEVLEHVSSSLAGWKSRSLS 480
Query: 593 KPGRLALASFVLSSISTYYMQINWLPQSVCDNID 626
R+ L VLS+I + M LP + D++D
Sbjct: 481 LAWRITLTKAVLSTIPVHVMSAIILPVATLDSLD 514
>At1g24640 hypothetical protein
Length = 1270
Score = 244 bits (624), Expect = 8e-65
Identities = 136/400 (34%), Positives = 212/400 (53%), Gaps = 1/400 (0%)
Query: 236 LNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDT 295
++ +++ A+ S KP APGPDG +FF+ Y VG+ V + F + T
Sbjct: 391 VSAQEIKEAVFSIKPASAPGPDGMSALFFQHYWSTVGNQVTSEVKKFFADGIMPAEWNYT 450
Query: 296 LIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTT 355
+ LIPK P D RPISLC+++YKII+K++ RL+P L I+ QS+F+ R T
Sbjct: 451 HLCLIPKTQHPTEMVDLRPISLCSVLYKIISKIMAKRLQPWLPEIVSDTQSAFVSERLIT 510
Query: 356 DNAIVLQEVIHYMRISKK-KKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIM 414
DN +V E++H +++ + ++A K D+ KA+D V W +LRS L GF + IM
Sbjct: 511 DNILVAHELVHSLKVHPRISSEFMAVKSDMSKAYDRVEWSYLRSLLLSLGFHLKWVNWIM 570
Query: 415 HCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPI 474
CV+S ++S+L N GLRQGDPLSP+LF+LC E L+ +N A +G+ E I
Sbjct: 571 VCVSSVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGLTHLLNKAQWEGALEGI 630
Query: 475 HVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGV 534
S GP + HLLFADD L KA+ Q ++ + + +G INL+KS + V
Sbjct: 631 QFSENGPMVHHLLFADDSLFLCKASREQSLVLQKILKVYGNATGQTINLNKSSITFGEKV 690
Query: 535 TQDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKP 594
+ T+ + GI YLG P K ++ D+++++L W + L++
Sbjct: 691 DEQLKGTIRTCLGIFTEGGAGTYLGLPECFSGSKVDMLHYLKDRLKEKLDVWFTRCLSQG 750
Query: 595 GRLALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
G+ L V ++ + M LP + C+N++ +F W
Sbjct: 751 GKEVLLKSVALAMPVFAMSCFKLPITTCENLESAMASFWW 790
>At3g44650 putative protein
Length = 762
Score = 236 bits (603), Expect = 2e-62
Identities = 143/405 (35%), Positives = 229/405 (56%), Gaps = 15/405 (3%)
Query: 236 LNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPS-ISD 294
++ +++ L S K+PGPDGF FFK+ I+G + F LA +F F P ++
Sbjct: 6 VSAEEIKKVLFSMPNDKSPGPDGFTSEFFKESWEILGPE-FILAIQSFFALGFLPKGVNS 64
Query: 295 TLIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRST 354
T++ALIPK KD+RPIS CN++YK+I+K+L +RL+ +L I QSSF+ R
Sbjct: 65 TILALIPKKLESKEMKDYRPISCCNVMYKVISKILANRLKLLLPQFIAGNQSSFVKDRLL 124
Query: 355 TDNAIVLQEVI---HYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIK 411
+N ++ +++ H IS++ A KID+ KA D+V W FL + L FP+ I
Sbjct: 125 IENVLLATDLVKDYHKDSISER----CAIKIDISKASDSVQWSFLINTLTAMHFPEMFIH 180
Query: 412 LIMHCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQG-- 469
I C+T+ SFS+ NG F+ + GLRQG LSPYLF++CM+ LS ++ V G
Sbjct: 181 WIRLCITTPSFSVQVNGELAGFFQSSRGLRQGCALSPYLFVICMDVLSKLLDKVVGIGRI 240
Query: 470 SWEPIHVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAF 529
+ P H G L+HL FADD+++ T + I ++FD FSK+SGLKI++ KS F
Sbjct: 241 GYHP-HCKRMG--LTHLSFADDLMILTDGQCRSIEGIIEVFDLFSKWSGLKISMEKSTIF 297
Query: 530 YSSGVTQDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNK 589
S+G++ L + +YLG P++ R D+ +I++++KR+ SW ++
Sbjct: 298 -SAGLSSTSRAQLHTHFPFEVGELPIRYLGLPLVTKRLSSVDYAPLIEQIRKRIGSWSSR 356
Query: 590 LLNKPGRLALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
L+ GR L S ++ S +++ LP++ I+++ +F+W
Sbjct: 357 FLSFAGRFNLISSIIWSSCNFWLSAFQLPRACIQEIEKLCSSFLW 401
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 236 bits (602), Expect = 3e-62
Identities = 125/314 (39%), Positives = 182/314 (57%), Gaps = 1/314 (0%)
Query: 236 LNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDT 295
++ ++V AL S P KAPGPDG F++ Y + G D+ L + T FD +++T
Sbjct: 375 ISPEEVKRALFSLNPDKAPGPDGMTAFFYQHYWDLTGPDLIKLVQNFHSTGFFDERLNET 434
Query: 296 LIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTT 355
I LIPK + P +FRPISLCN+ YK+I+KVL RL+ +L +I QS+F+ R T
Sbjct: 435 NICLIPKTERPRKMAEFRPISLCNVSYKVISKVLSSRLKRLLPELISETQSAFVAERLIT 494
Query: 356 DNAIVLQEVIHYMRISKK-KKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIM 414
DN ++ QE H +R + KK Y+A K D+ KA+D V W FLR+ + GF + I+
Sbjct: 495 DNILIAQENFHALRTNPACKKKYMAIKTDMSKAYDRVEWSFLRALMLKMGFAQKWVDWII 554
Query: 415 HCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPI 474
C++S S+ IL NG+ +P+ G+RQGDP+SP+LFILC E L + + +A G + +
Sbjct: 555 FCISSVSYKILLNGSPKGFIKPSRGIRQGDPISPFLFILCTEALVAKLKDAEWHGRIQGL 614
Query: 475 HVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGV 534
+S P SHLLFADD L F KA+ Q + I D+ + + SG ++N KS + V
Sbjct: 615 QISRASPSTSHLLFADDSLFFCKADPLQGKEIIDILRLYGEASGQQLNPDKSSVMFGHEV 674
Query: 535 TQDKINTLTSISGI 548
NT+ GI
Sbjct: 675 DNSIRNTIKVSLGI 688
>At1g25430 hypothetical protein
Length = 1213
Score = 236 bits (601), Expect = 4e-62
Identities = 139/399 (34%), Positives = 219/399 (54%), Gaps = 6/399 (1%)
Query: 237 NKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTL 296
+ + + AAL S K+ GPDGF FF IVG +V + F + + T
Sbjct: 448 SNEDIRAALFSLPRNKSCGPDGFTAEFFIDSWSIVGAEVTDAIKEFFSSGCLLKQWNATT 507
Query: 297 IALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTD 356
I LIPK+ P DFRPIS N +YK+I ++L RL+ +L+ +I QS+FLPGRS +
Sbjct: 508 IVLIPKIVNPTCTSDFRPISCLNTLYKVIARLLTDRLQRLLSGVISSAQSAFLPGRSLAE 567
Query: 357 NAIVLQEVIH-YMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMH 415
N ++ +++H Y + +G + K+DL+KAFD+V W+F+ + L P+ I I
Sbjct: 568 NVLLATDLVHGYNWSNISPRGML--KVDLKKAFDSVRWEFVIAALRALAIPEKFINWISQ 625
Query: 416 CVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIH 475
C+++ +F++ NG F+ T GLRQGDPLSPYLF+L ME S+ +++ + G H
Sbjct: 626 CISTPTFTVSINGGNGGFFKSTKGLRQGDPLSPYLFVLAMEAFSNLLHSRYESGLIH-YH 684
Query: 476 VSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVT 535
+SHL+FADDV++F S L I + D F+ +SGLK+N KS Y +G+
Sbjct: 685 PKASNLSISHLMFADDVMIFFDGGSFSLHGICETLDDFASWSGLKVNKDKSH-LYLAGLN 743
Query: 536 QDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKPG 595
Q + N + G T +YLG P++ + + +++ +++K+ R SW NK L+ G
Sbjct: 744 QLESNA-NAAYGFPIGTLPIRYLGLPLMNRKLRIAEYEPLLEKITARFRSWVNKCLSFAG 802
Query: 596 RLALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
R+ L S V+ ++M LP+ I+ + F+W
Sbjct: 803 RIQLISSVIFGSINFWMSTFLLPKGCIKRIESLCSRFLW 841
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.350 0.156 0.550
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,411,796
Number of Sequences: 26719
Number of extensions: 546212
Number of successful extensions: 2453
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 61
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 2267
Number of HSP's gapped (non-prelim): 89
length of query: 638
length of database: 11,318,596
effective HSP length: 106
effective length of query: 532
effective length of database: 8,486,382
effective search space: 4514755224
effective search space used: 4514755224
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC136840.7


