

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136840.1 + phase: 0 /pseudo
(1308 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
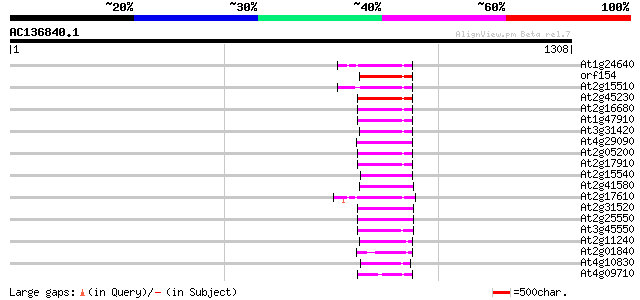
Score E
Sequences producing significant alignments: (bits) Value
At1g24640 hypothetical protein 118 2e-26
orf154 -mitochondrial genome- 111 2e-24
At2g15510 putative non-LTR retroelement reverse transcriptase 104 3e-22
At2g45230 putative non-LTR retroelement reverse transcriptase 103 6e-22
At2g16680 putative non-LTR retroelement reverse transcriptase 101 2e-21
At1g47910 reverse transcriptase, putative 101 2e-21
At3g31420 hypothetical protein 100 7e-21
At4g29090 putative protein 100 9e-21
At2g05200 putative non-LTR retroelement reverse transcriptase 99 1e-20
At2g17910 putative non-LTR retroelement reverse transcriptase 99 2e-20
At2g15540 putative non-LTR retroelement reverse transcriptase 99 2e-20
At2g41580 putative non-LTR retroelement reverse transcriptase 97 5e-20
At2g17610 putative non-LTR retroelement reverse transcriptase 97 8e-20
At2g31520 putative non-LTR retroelement reverse transcriptase 89 2e-17
At2g25550 putative non-LTR retroelement reverse transcriptase 89 2e-17
At3g45550 putative protein 86 1e-16
At2g11240 pseudogene 85 2e-16
At2g01840 putative non-LTR retroelement reverse transcriptase 85 3e-16
At4g10830 putative protein 81 3e-15
At4g09710 RNA-directed DNA polymerase -like protein 77 5e-14
>At1g24640 hypothetical protein
Length = 1270
Score = 118 bits (296), Expect = 2e-26
Identities = 68/176 (38%), Positives = 101/176 (56%), Gaps = 8/176 (4%)
Query: 765 EDKTSSVQNIGFGRSL-RGGKEILCLLQQEMY*LKLWSKKSLPM**VVPNG*CEHIESMI 823
+D+ ++ F R L +GGKE+L L+ + +++ + P CE++ES +
Sbjct: 732 KDRLKEKLDVWFTRCLSQGGKEVL--LKSVALAMPVFAMSCFKL----PITTCENLESAM 785
Query: 824 SRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQGWRILHNEELLLS 883
+ FWW S RKIHW + +C K GG+GFR + N A+L+KQ WR+LH + LLS
Sbjct: 786 ASFWWDSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCLLS 845
Query: 884 *CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGDGRNIRVWKD 939
LK RYF DFL A++ PS+ WRSI+ + E++ K RVGDG ++ VW D
Sbjct: 846 RLLKSRYFDATDFLDAALSQRPSFGWRSILFGR-ELLSKGLQKRVGDGASLFVWID 900
>orf154 -mitochondrial genome-
Length = 154
Score = 111 bits (278), Expect = 2e-24
Identities = 52/125 (41%), Positives = 80/125 (63%), Gaps = 2/125 (1%)
Query: 816 CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKV-GGMGFRTFKELNLAMLSKQGWRI 874
C+ + S ++ FWW S + +RKI W+ ++CK+K+ GG+GFR N A+L+KQ +RI
Sbjct: 18 CKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRDLGWFNQALLAKQSFRI 77
Query: 875 LHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGDGRNI 934
+H LLS L+ RYFP ++ SVG PSY WRSI+H + E++ + + +GDG +
Sbjct: 78 IHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGR-ELLSRGLLRTIGDGIHT 136
Query: 935 RVWKD 939
+VW D
Sbjct: 137 KVWLD 141
>At2g15510 putative non-LTR retroelement reverse transcriptase
Length = 1138
Score = 104 bits (260), Expect = 3e-22
Identities = 61/179 (34%), Positives = 97/179 (54%), Gaps = 14/179 (7%)
Query: 765 EDKTSSVQNIGFGRSLR-GGKEILC---LLQQEMY*LKLWSKKSLPM**VVPNG*CEHIE 820
+DK S + F R+L GGKEIL + ++Y + + C+++
Sbjct: 545 KDKLKSRLSGWFARTLSLGGKEILLKAVAMAMQVYAMSCFKLTKTT---------CKNLT 595
Query: 821 SMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQGWRILHNEEL 880
S +S FWW + + +RK HW+ +++C K+ GG+GFR + N A+L+KQ WRIL
Sbjct: 596 SAMSDFWWNALEHKRKTHWVSWEKLCLAKESGGLGFRDIESFNQALLAKQSWRILQYPSF 655
Query: 881 LLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGDGRNIRVWKD 939
L + K RYF +FL+A +G PSY SI+ + E++ K VG+G+++ VW D
Sbjct: 656 LFARFFKSRYFDDEEFLEADLGVRPSYACCSILFGR-ELLAKGLRKEVGNGKSLNVWMD 713
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 103 bits (257), Expect = 6e-22
Identities = 50/130 (38%), Positives = 79/130 (60%), Gaps = 3/130 (2%)
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQ 870
+P C+ IES+++ FWW +K+ R +HW + + K VGG+GF+ + N+A+L KQ
Sbjct: 810 IPKTICQQIESVMAEFWWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQ 869
Query: 871 GWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRV-G 929
WR++ ++ L++ K RYF + D L A +G PS+ W+SI A +V+ K G+ V G
Sbjct: 870 LWRMITEKDSLMAKVFKSRYFSKSDPLNAPLGSRPSFAWKSIYEA--QVLIKQGIRAVIG 927
Query: 930 DGRNIRVWKD 939
+G I VW D
Sbjct: 928 NGETINVWTD 937
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 101 bits (252), Expect = 2e-21
Identities = 49/129 (37%), Positives = 74/129 (56%), Gaps = 1/129 (0%)
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQ 870
+P G C + S++ FWW S + KIHWI ++ K +GG GF+ + N A+L+KQ
Sbjct: 762 LPKGLCTKLTSVMMDFWWNSMEFSNKIHWIGGKKLTLPKSLGGFGFKDLQCFNQALLAKQ 821
Query: 871 GWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGD 930
WR+ + + ++S K RYF DFL A G PSYTWRSI++ + E+++ +G+
Sbjct: 822 AWRLFSDSKSIVSQIFKSRYFMNTDFLNARQGTRPSYTWRSILYGR-ELLNGGLKRLIGN 880
Query: 931 GRNIRVWKD 939
G VW D
Sbjct: 881 GEQTNVWID 889
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 101 bits (252), Expect = 2e-21
Identities = 47/129 (36%), Positives = 77/129 (59%), Gaps = 1/129 (0%)
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQ 870
+P + S +++FWW S R +HW+ D++C +K GG+GFR + N A+L+KQ
Sbjct: 594 LPKAITSKLTSAVAKFWWSSNGDSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAKQ 653
Query: 871 GWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGD 930
WR++ + L + KGRYF + + L + ++PSY WRS++ A+ ++ K + RVG
Sbjct: 654 LWRLITAPDSLFAKVFKGRYFRKSNPLDSIKSYSPSYGWRSMISAR-SLVYKGLIKRVGS 712
Query: 931 GRNIRVWKD 939
G +I VW D
Sbjct: 713 GASISVWND 721
>At3g31420 hypothetical protein
Length = 1491
Score = 100 bits (248), Expect = 7e-21
Identities = 47/124 (37%), Positives = 71/124 (56%), Gaps = 1/124 (0%)
Query: 816 CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQGWRIL 875
CE I+S+++RFWW S + +HW + KK GG+GF+ + NLA+L KQ W +L
Sbjct: 973 CEEIDSLLARFWWSSGNETKGMHWFTWKRMSIPKKEGGLGFKELENFNLALLGKQTWHLL 1032
Query: 876 HNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGDGRNIR 935
+ L++ L+GRYFP + + A G S+ W+SI+H + ++ K VGDG I
Sbjct: 1033 QHPNCLMARVLRGRYFPETNVMNAVQGRRASFVWKSILHGR-NLLKKGLRCCVGDGSLIN 1091
Query: 936 VWKD 939
W D
Sbjct: 1092 AWLD 1095
>At4g29090 putative protein
Length = 575
Score = 99.8 bits (247), Expect = 9e-21
Identities = 49/129 (37%), Positives = 77/129 (58%), Gaps = 1/129 (0%)
Query: 810 VVPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSK 869
++P C+ I S+++ FWW +KQ + +HW D + K GG+GF+ + NLA+L K
Sbjct: 12 LLPKTVCKQIISVLADFWWRNKQEAKGMHWKAWDHLSCYKAEGGIGFKDIEAFNLALLGK 71
Query: 870 QGWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVG 929
Q WR+L E L++ K RYF + D L A +G PS+ W+SI HA E++ + VG
Sbjct: 72 QMWRMLSRPESLMAKVFKSRYFHKSDPLNAPLGSRPSFVWKSI-HASQEILRQGARAVVG 130
Query: 930 DGRNIRVWK 938
+G +I +W+
Sbjct: 131 NGEDIIIWR 139
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 99.4 bits (246), Expect = 1e-20
Identities = 44/129 (34%), Positives = 76/129 (58%), Gaps = 1/129 (0%)
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQ 870
+P+ C I+S+++RFWW +K RK W+ ++ K GG+GFR + N ++L+K
Sbjct: 705 LPSALCRKIQSLLTRFWWDTKPDVRKTSWVAWSKLTNPKNAGGLGFRDIERCNDSLLAKL 764
Query: 871 GWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGD 930
GWR+L++ E LLS L G+Y F++ + PS+ WRSI+ + E++ + W + +
Sbjct: 765 GWRLLNSPESLLSRILLGKYCHSSSFMECKLPSQPSHGWRSIIAGR-EILKEGLGWLITN 823
Query: 931 GRNIRVWKD 939
G + +W D
Sbjct: 824 GEKVSIWND 832
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 99.0 bits (245), Expect = 2e-20
Identities = 48/129 (37%), Positives = 73/129 (56%), Gaps = 1/129 (0%)
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQ 870
+P C+ + +++ FWW S Q +RKIHW+ + K GG GF+ + N A+L+KQ
Sbjct: 788 LPKNLCQKLTTVMMDFWWNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLAKQ 847
Query: 871 GWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGD 930
WR+L + L S + RYF DFL A+ G PSY WRSI+ + E++ + +G+
Sbjct: 848 AWRVLQEKGSLFSRVFQSRYFSNSDFLSATRGSRPSYAWRSILFGR-ELLMQGLRTVIGN 906
Query: 931 GRNIRVWKD 939
G+ VW D
Sbjct: 907 GQKTFVWTD 915
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 98.6 bits (244), Expect = 2e-20
Identities = 50/122 (40%), Positives = 74/122 (59%), Gaps = 3/122 (2%)
Query: 819 IESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQGWRILHNE 878
+ S+++ FWW +++ IHWI D++C GG+GFRT +E NL +L+KQ WR++
Sbjct: 698 LSSVVANFWWKTREESNGIHWIAWDKLCTPFSDGGLGFRTLEEFNLVLLAKQLWRLIRFP 757
Query: 879 ELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWR-VGDGRNIRVW 937
LLS L+GRYF D ++ PS+ WRSIM AK ++ G+ R +G G RVW
Sbjct: 758 NSLLSRVLRGRYFRYSDPIQIGKANRPSFGWRSIMAAKPLLLS--GLRRTIGSGMLTRVW 815
Query: 938 KD 939
+D
Sbjct: 816 ED 817
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 97.4 bits (241), Expect = 5e-20
Identities = 49/126 (38%), Positives = 76/126 (59%), Gaps = 3/126 (2%)
Query: 816 CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQGWRIL 875
C + S++ FWW S Q ++KIHWI ++ K +GG GF+ + N A+L+KQ R+
Sbjct: 544 CTKLTSVMMDFWWNSVQDKKKIHWIGAQKLMLPKFLGGFGFKDLQCFNQALLAKQASRLH 603
Query: 876 HNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWR-VGDGRNI 934
+ + LLS LK RY+ DFL A+ G PSY W+SI++ + ++ G+ + +G+G N
Sbjct: 604 TDSDSLLSQILKSRYYMNSDFLSATKGTRPSYAWQSILYGRELLVS--GLKKIIGNGENT 661
Query: 935 RVWKDN 940
VW DN
Sbjct: 662 YVWMDN 667
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 96.7 bits (239), Expect = 8e-20
Identities = 62/202 (30%), Positives = 102/202 (49%), Gaps = 18/202 (8%)
Query: 755 KILGLTSF*GEDKTSSVQNIG----------FGRSLR-GGKEILCLLQQEMY*LKLWSKK 803
+ LGL F G +KT++ I + + L GKE+L ++ + + +S
Sbjct: 204 RYLGLPEFVGRNKTNAFSFIAQTMDQKMDNWYNKLLSPAGKEVL--IKSIVTAIPTYSMS 261
Query: 804 SLPM**VVPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELN 863
++P I S + FWW + + + KI W+ ++ KK+GG+ R K+ N
Sbjct: 262 CF----LLPMRLIHQITSAMRWFWWSNTKVKHKIPWVAWSKLNDPKKMGGLAIRDLKDFN 317
Query: 864 LAMLSKQGWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKV 923
+A+L+KQ WRIL L++ K +YFP+ L A SY W+SI+H ++I +
Sbjct: 318 IALLAKQSWRILQQPFSLMARVFKAKYFPKERLLDAKATSQSSYAWKSILHGT-KLISRG 376
Query: 924 GVWRVGDGRNIRVWKDNCSLLN 945
+ G+G NI++WKDN LN
Sbjct: 377 LKYIAGNGNNIQLWKDNWLPLN 398
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 88.6 bits (218), Expect = 2e-17
Identities = 46/130 (35%), Positives = 68/130 (51%), Gaps = 1/130 (0%)
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQ 870
+P G IES++ FWW +R I W+ + +KK GG+GFR + N A+L+KQ
Sbjct: 966 LPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQ 1025
Query: 871 GWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGD 930
WR++ L + +K RYF L A V SY W S++ ++ K +GD
Sbjct: 1026 AWRLIQYPNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLLDG-IALLKKGTRHLIGD 1084
Query: 931 GRNIRVWKDN 940
G+NIR+ DN
Sbjct: 1085 GQNIRIGLDN 1094
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 88.6 bits (218), Expect = 2e-17
Identities = 46/130 (35%), Positives = 68/130 (51%), Gaps = 1/130 (0%)
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQ 870
+P G IES++ FWW +R I W+ + +KK GG+GFR + N A+L+KQ
Sbjct: 1192 LPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQ 1251
Query: 871 GWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGD 930
WR++ L + +K RYF L A V SY W S++ ++ K +GD
Sbjct: 1252 AWRLIQYPNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLLDG-IALLKKGTRHLIGD 1310
Query: 931 GRNIRVWKDN 940
G+NIR+ DN
Sbjct: 1311 GQNIRIGLDN 1320
>At3g45550 putative protein
Length = 851
Score = 85.9 bits (211), Expect = 1e-16
Identities = 44/130 (33%), Positives = 68/130 (51%), Gaps = 1/130 (0%)
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQ 870
+P G IES++ FWW +R I W+ + +KK GG+GFR + N A+L+KQ
Sbjct: 431 LPQGIVSEIESLLMNFWWEKASNKRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQ 490
Query: 871 GWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGD 930
WRI+ L + +K RYF + A SY W S++ + ++ K + +GD
Sbjct: 491 AWRIIQYPNSLFARVMKARYFKDNSIIDAKTRSQQSYGWSSLL-SGIALLRKGTRYVIGD 549
Query: 931 GRNIRVWKDN 940
G+ IR+ DN
Sbjct: 550 GKTIRLGIDN 559
>At2g11240 pseudogene
Length = 1044
Score = 85.1 bits (209), Expect = 2e-16
Identities = 43/124 (34%), Positives = 67/124 (53%), Gaps = 1/124 (0%)
Query: 816 CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQGWRIL 875
C+ I+S ++ FWW S ++K+ WI ++ KNKK GG+GF+ N A+L+K WRI+
Sbjct: 641 CKRIQSALTHFWWDSSADKKKMCWIAWSKMAKNKKEGGLGFKDITNFNDALLAKLSWRIV 700
Query: 876 HNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGDGRNIR 935
+ +L L G+Y FL SV S+ WR I K + ++G +G G +
Sbjct: 701 QSPSCVLVRILLGKYCRTSSFLDCSVTAASSHGWRGICTGKDLIKSQLGK-VIGSGLDTL 759
Query: 936 VWKD 939
VW +
Sbjct: 760 VWNE 763
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 84.7 bits (208), Expect = 3e-16
Identities = 46/130 (35%), Positives = 70/130 (53%), Gaps = 19/130 (14%)
Query: 810 VVPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSK 869
++P C I S+I+ FWWG K+ E G +GF+ + N A+L+K
Sbjct: 1171 LLPTLICNEINSLITAFWWG-KENE-----------------GDLGFKDLHQFNRALLAK 1212
Query: 870 QGWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVG 929
Q WRIL N + LL+ KG Y+P +L+A+ G + SY W SI K + + V R+G
Sbjct: 1213 QAWRILTNPQSLLARLYKGLYYPNTTYLRANKGGHASYGWNSIQEGKLLLQQGLRV-RLG 1271
Query: 930 DGRNIRVWKD 939
DG+ ++W+D
Sbjct: 1272 DGQTTKIWED 1281
>At4g10830 putative protein
Length = 1294
Score = 81.3 bits (199), Expect = 3e-15
Identities = 43/116 (37%), Positives = 65/116 (55%), Gaps = 1/116 (0%)
Query: 819 IESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQGWRILHNE 878
IE+++ FWW +R+I WI + +KK GG+GFR + N A+L+KQ WR+++N
Sbjct: 1180 IEALLMNFWWEKNAKKREIPWIAWKRLQYSKKEGGLGFRDLAKFNDALLAKQVWRMINNP 1239
Query: 879 ELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGDGRNI 934
L + +K RYF L A SY W S++ A +VI K + VGDG+ +
Sbjct: 1240 NSLFARIMKARYFREDSILDAKRQRYQSYGWTSML-AGLDVIKKGSRFIVGDGKTV 1294
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 77.4 bits (189), Expect = 5e-14
Identities = 43/130 (33%), Positives = 69/130 (53%), Gaps = 9/130 (6%)
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQ 870
+P C+ I+S+++RFWW SK +RK+ W+ D++ GG+GFR + +K
Sbjct: 751 LPASLCKQIQSVLTRFWWDSKPDKRKMAWVSWDKLTLPINEGGLGFREIE-------AKL 803
Query: 871 GWRILHNEELLLS*CLKGRYFPRGDFLKASVGFN-PSYTWRSIMHAKFEVIDKVGVWRVG 929
WRIL LLS L G+Y F+ S + S+ WR I+ + +++ K W +G
Sbjct: 804 SWRILKEPHSLLSRVLLGKYCNTSSFMDCSASPSFASHGWRGILAGR-DLLRKGLGWSIG 862
Query: 930 DGRNIRVWKD 939
G +I VW +
Sbjct: 863 QGDSINVWTE 872
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.358 0.160 0.572
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,120,776
Number of Sequences: 26719
Number of extensions: 893549
Number of successful extensions: 3303
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 3198
Number of HSP's gapped (non-prelim): 76
length of query: 1308
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1197
effective length of database: 8,352,787
effective search space: 9998286039
effective search space used: 9998286039
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC136840.1


