

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136507.9 + phase: 0 /pseudo
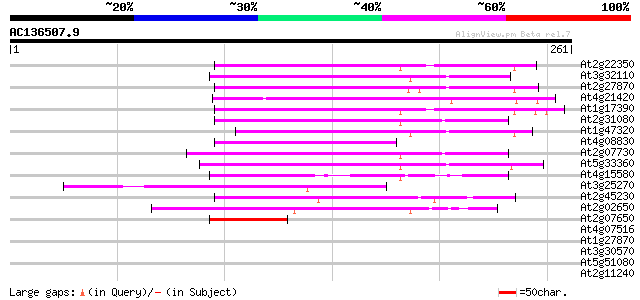
(261 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g22350 putative non-LTR retroelement reverse transcriptase 81 6e-16
At3g32110 non-LTR reverse transcriptase, putative 73 1e-13
At2g27870 putative non-LTR retroelement reverse transcriptase 73 2e-13
At4g21420 hypothetical protein 67 1e-11
At1g17390 hypothetical protein 66 2e-11
At2g31080 putative non-LTR retroelement reverse transcriptase 65 5e-11
At1g47320 hypothetical protein 59 2e-09
At4g08830 putative protein 58 4e-09
At2g07730 putative non-LTR retroelement reverse transcriptase 57 1e-08
At5g33360 putative protein 56 2e-08
At4g15580 splicing factor like protein 55 5e-08
At3g25270 hypothetical protein 52 4e-07
At2g45230 putative non-LTR retroelement reverse transcriptase 52 4e-07
At2g02650 putative reverse transcriptase 43 2e-04
At2g07650 putative non-LTR retrolelement reverse transcriptase 42 4e-04
At4g07516 putative protein 39 0.002
At1g27870 hypothetical protein 39 0.002
At3g30570 putative reverse transcriptase 37 0.008
At5g51080 unknown protein 37 0.011
At2g11240 pseudogene 37 0.011
>At2g22350 putative non-LTR retroelement reverse transcriptase
Length = 321
Score = 80.9 bits (198), Expect = 6e-16
Identities = 56/155 (36%), Positives = 77/155 (49%), Gaps = 8/155 (5%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
V W+ P W+K+NTDGA NP A+ GGV RDHN +IG F N+ SA A+L GV
Sbjct: 151 VSWVSPEDGWVKLNTDGASRGNPGFATAGGVLRDHNGAWIGGFAVNIGVCSAPLAELWGV 210
Query: 156 ITAINIAGENNWRNIWLETDLQLAV----LAFKNLNMVPWSLRNKWLNYFEHVRSMNFLI 211
+ IA R + LE D ++ V + + + + LR L Y + I
Sbjct: 211 YYGLFIAWGRGARRVELEVDSKMVVGFLTTGIADSHPLSFLLR---LCYDFLSKGWIVRI 267
Query: 212 THIYIEGNSCADCMANVGLTLS-SFVWLPSLPDCI 245
+H+Y E N AD +AN +LS L S PD +
Sbjct: 268 SHVYREANRLADGLANYAFSLSLGLHLLESRPDVV 302
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 73.2 bits (178), Expect = 1e-13
Identities = 49/142 (34%), Positives = 69/142 (48%), Gaps = 3/142 (2%)
Query: 94 KEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLL 153
K + W PP+ W K+NTDGA NP AS GGV R+ + G F N+ SA A+L
Sbjct: 1739 KPIRWTPPMEGWYKINTDGASRGNPGLASAGGVLRNSAGAWCGGFAVNIGRCSAPLAELW 1798
Query: 154 GVITAINIAGENNWRNIWLETDLQLAVLAFKN--LNMVPWSLRNKWLNYFEHVRSMNFLI 211
GV + +A ++ LE D ++ V K P S + + F + I
Sbjct: 1799 GVYYGLYMAWAKQLTHLELEVDSEVVVGFLKTGIGETHPLSFLVRLCHNFLS-KDWTVRI 1857
Query: 212 THIYIEGNSCADCMANVGLTLS 233
+H+Y E NS AD +AN +LS
Sbjct: 1858 SHVYREANSLADGLANHAFSLS 1879
>At2g27870 putative non-LTR retroelement reverse transcriptase
Length = 314
Score = 72.8 bits (177), Expect = 2e-13
Identities = 54/154 (35%), Positives = 74/154 (47%), Gaps = 4/154 (2%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
+ W P W K+NTDGA NP AS GGV RD + G F N+ SA A+L GV
Sbjct: 144 IAWSKPEEGWWKLNTDGASRGNPGLASAGGVLRDEEGAWRGGFALNIGVCSAPLAELWGV 203
Query: 156 ITAINIAGENNWRNIWLETDLQLAVLAFK-NLNMV-PWSLRNKWLNYFEHVRSMNFLITH 213
+ IA E + +E D ++ V K +N V P S + + F R I+H
Sbjct: 204 YYGLYIAWERRVTRLEIEVDSEIVVGFLKIGINEVHPLSFLVRLCHDFIS-RDWRVRISH 262
Query: 214 IYIEGNSCADCMANVGLTLS-SFVWLPSLPDCIK 246
+Y E N AD +AN +L F L +PD ++
Sbjct: 263 VYREANRLADGLANYAFSLPLGFHSLSLVPDSLR 296
>At4g21420 hypothetical protein
Length = 229
Score = 67.0 bits (162), Expect = 1e-11
Identities = 46/166 (27%), Positives = 82/166 (48%), Gaps = 7/166 (4%)
Query: 95 EVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLG 154
+V+W P IK+N DG+ + +AS GGVFR+H F+ + +++ ++ A+
Sbjct: 60 KVVWKKPENGRIKLNFDGSRGREG-QASIGGVFRNHKAEFLLGYSESIGEATSTMAEFAA 118
Query: 155 VITAINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRNKWLNYFEHV--RSMNFLIT 212
+ + +A EN ++WLE D ++ + + NK +NY + V N +++
Sbjct: 119 LKRGLELALENGLTDLWLEGDAKIIMDIISRRGRLRCEKTNKHVNYIKVVMPELNNCVLS 178
Query: 213 HIYIEGNSCADCMANVGLTLSS-FVWLPSLPDC---IKHDYGRNRL 254
H+Y EGN AD +A +G VW P+ I HD + ++
Sbjct: 179 HVYREGNRVADKLAKLGHQFQDPKVWRVQPPEIVLPIMHDDAKGKI 224
>At1g17390 hypothetical protein
Length = 322
Score = 65.9 bits (159), Expect = 2e-11
Identities = 55/177 (31%), Positives = 80/177 (45%), Gaps = 17/177 (9%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
+ W PP W K+NTDGA NP A+ GGV RD + + F N+ SA A+L G
Sbjct: 102 IAWSPPRVGWFKLNTDGASRGNPRLATAGGVVRDGDGNWCYGFSLNIGICSAPLAELWGA 161
Query: 156 ITAINIAGENNWRNIWLETDLQLAV----LAFKNLNMVPWSLRNKWLNYFEHVRSMNFLI 211
+NIA E + +E D ++ V + + + + +R L + + + I
Sbjct: 162 YYGLNIAWERGVTQLEMEIDSEMVVGFLRTGIDDSHPLSFLVR---LCHGLLSKDWSVRI 218
Query: 212 THIYIEGNSCADCMANVGLTLS-SFVWLPSLPD---CIKHD------YGRNRLGMHS 258
+H+Y E N AD +AN L F S PD I HD Y RN G ++
Sbjct: 219 SHVYREANRLADGLANYAFFLPLGFHLFNSTPDIVMSIVHDDVAGSAYPRNESGKNA 275
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 64.7 bits (156), Expect = 5e-11
Identities = 43/139 (30%), Positives = 66/139 (46%), Gaps = 3/139 (2%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
+ W P W+K+ TDGA N A+ GG R+ ++G F N+ + +A A+L G
Sbjct: 1061 IRWQVPSDGWVKITTDGASRGNHGLAAAGGAIRNGQGEWLGGFALNIGSCAAPLAELWGA 1120
Query: 156 ITAINIAGENNWRNIWLETDLQLAV--LAFKNLNMVPWSLRNKWLNYFEHVRSMNFLITH 213
+ IA + +R + L+ D +L V L+ N P S + F R ++H
Sbjct: 1121 YYGLLIAWDKGFRRVELDLDCKLVVGFLSTGVSNAHPLSFLVRLCQGF-FTRDWLVRVSH 1179
Query: 214 IYIEGNSCADCMANVGLTL 232
+Y E N AD +AN TL
Sbjct: 1180 VYREANRLADGLANYAFTL 1198
>At1g47320 hypothetical protein
Length = 259
Score = 59.3 bits (142), Expect = 2e-09
Identities = 45/141 (31%), Positives = 70/141 (48%), Gaps = 4/141 (2%)
Query: 106 IKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITAINIAGEN 165
+K+NTDGA NP A+ GGV +D+ + G F N+ +SA A+L G + +A E
Sbjct: 101 LKINTDGASRGNPGLATAGGVLQDNEGRWCGGFSLNIGRSSAPMAELWGAYYGLYLAWER 160
Query: 166 NWRNIWLETDLQLAVLAFKN--LNMVPWSLRNKWLNYFEHVRSMNFLITHIYIEGNSCAD 223
+I LE D ++ V K + P S + + F + I H+Y E N AD
Sbjct: 161 KSSHIELEVDSEIVVGFLKTGISDHHPLSFLVRLCHGFIS-KDWRVRIFHVYREANRFAD 219
Query: 224 CMANVGLTLS-SFVWLPSLPD 243
+AN +L F ++P++ D
Sbjct: 220 GLANYVFSLPLGFHFVPNVVD 240
>At4g08830 putative protein
Length = 947
Score = 58.2 bits (139), Expect = 4e-09
Identities = 33/85 (38%), Positives = 45/85 (52%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
V W PP W+K+NTDGA N A+ GGV RD + G F ++ SA A+L GV
Sbjct: 839 VAWKPPDGEWVKLNTDGASRGNLGLATTGGVLRDGIGHWCGGFALDIGVCSAPLAELWGV 898
Query: 156 ITAINIAGENNWRNIWLETDLQLAV 180
+ +A E + + LE D +L V
Sbjct: 899 YYGLYMAWERRFTRVELEVDSELVV 923
>At2g07730 putative non-LTR retroelement reverse transcriptase
Length = 970
Score = 57.0 bits (136), Expect = 1e-08
Identities = 42/152 (27%), Positives = 67/152 (43%), Gaps = 3/152 (1%)
Query: 83 INIHPPRAPLIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNL 142
+N H RA + + + W P W+K+ TDGA + A+ G + ++G F N+
Sbjct: 787 LNNHVKRARVERMIRWKAPSDRWVKLTTDGASRGHQGLAAASGAILNLQGEWLGGFALNI 846
Query: 143 NTNSAFNAKLLGVITAINIAGENNWRNIWLETDLQLAV--LAFKNLNMVPWSLRNKWLNY 200
+ A A+L G + IA + +R + L D +L V L+ P S +
Sbjct: 847 GSCDAPLAELWGAYYGLLIAWDKGFRRVELNLDSELVVGFLSTGISKAHPLSFLVRLCQG 906
Query: 201 FEHVRSMNFLITHIYIEGNSCADCMANVGLTL 232
F R ++H+Y E N AD +AN L
Sbjct: 907 F-FTRDWLVRVSHVYREANRLADGLANYAFFL 937
>At5g33360 putative protein
Length = 306
Score = 56.2 bits (134), Expect = 2e-08
Identities = 51/166 (30%), Positives = 71/166 (42%), Gaps = 7/166 (4%)
Query: 89 RAPLIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAF 148
R+ + ++V W P W K+NTDGA NP A GG R+ + F N+ SA
Sbjct: 129 RSRMERQVRWSKPSLGWCKLNTDGASHGNPGLAIAGGALRNEYGEWCFGFALNIGRCSAP 188
Query: 149 NAKLLGVITAINIAGENNWRNIWLETDLQLAV--LAFKNLNMVPWSLRNKWLNYFEHVRS 206
A+L GV + +A + + LE D ++ V L + P S + + F R
Sbjct: 189 LAELWGVYYGLFMAWDRGITRLELEVDSEMVVGFLRTGIGSSHPLSFLVRMCHGFLS-RD 247
Query: 207 MNFLITHIYIEGNSCADCMANVGLTL----SSFVWLPSLPDCIKHD 248
I H+Y E N AD +AN L F PS D I D
Sbjct: 248 WIVRIGHVYREANRLADELANYAFDLPLGYHGFASPPSSLDSILRD 293
>At4g15580 splicing factor like protein
Length = 559
Score = 54.7 bits (130), Expect = 5e-08
Identities = 44/151 (29%), Positives = 66/151 (43%), Gaps = 37/151 (24%)
Query: 94 KEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLL 153
+ + W P W+KV+TDGA NP A+ GGV RD + +++G F L AF
Sbjct: 24 RHIAWTKPPEGWVKVSTDGASRGNPGPAAAGGVIRDEDGLWVGGFALQL----AF----- 74
Query: 154 GVITAINIAGENNWRNIWLETDLQLAV------------LAFKNLNMVPWSLRNKWLNYF 201
+ + ++ + LE D +L V LAF + + L WL
Sbjct: 75 -----VRLRWQSCGGRVELEVDSELVVGFLQSGISEAHPLAFL-VRLCHGLLSKDWL--- 125
Query: 202 EHVRSMNFLITHIYIEGNSCADCMANVGLTL 232
VR ++H+Y E N AD +AN +L
Sbjct: 126 --VR-----VSHVYREANRLADGLANYAFSL 149
>At3g25270 hypothetical protein
Length = 343
Score = 51.6 bits (122), Expect = 4e-07
Identities = 34/151 (22%), Positives = 62/151 (40%), Gaps = 10/151 (6%)
Query: 26 ILSAIWFIRNQARFKDKKNHWKSAIDNIIVAVSLTGNRTKLTSFVNMDEFVFLKAFKINI 85
IL +W RNQ F+ K W++ + R + + + + +V +++
Sbjct: 123 ILWRLWKSRNQLVFQQKSISWQNTLQRA---------RNDVQEWEDTNTYVQSLNQQVHS 173
Query: 86 HPPRAPLIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGA-FVQNLNT 144
+ P + W P WIK N DGA A G + RD N +++G+ T
Sbjct: 174 SRHQQPTMARTKWQRPPSTWIKYNYDGAFNHQTRNAKAGWLMRDENGVYMGSGQAIGSTT 233
Query: 145 NSAFNAKLLGVITAINIAGENNWRNIWLETD 175
+ + ++ +I A+ A +R + E D
Sbjct: 234 SDSLESEFQALIIAMQHAWSQGYRKVIFEGD 264
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 51.6 bits (122), Expect = 4e-07
Identities = 41/144 (28%), Positives = 64/144 (43%), Gaps = 7/144 (4%)
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNL-NTNSAFNAKLLG 154
V W PP W+K NTDGA +K+ G V R+H + ++ L + S ++
Sbjct: 1213 VKWQPPSHGWVKCNTDGAWSKDLGNCGVGWVLRNHTGRLLWLGLRALPSQQSVLETEVEA 1272
Query: 155 VITAINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRNK---WLNYFEHVRSMNFLI 211
+ A+ N+R + E+D Q V +N +P SL + N H + F
Sbjct: 1273 LRWAVLSLSRFNYRRVIFESDSQYLVSLIQNEMDIP-SLAPRIQDIRNLLRHFEEVKFQF 1331
Query: 212 THIYIEGNSCADCMANVGLTLSSF 235
T EGN+ AD A L+L ++
Sbjct: 1332 TR--REGNNVADRTARESLSLMNY 1353
>At2g02650 putative reverse transcriptase
Length = 365
Score = 42.7 bits (99), Expect = 2e-04
Identities = 37/168 (22%), Positives = 71/168 (42%), Gaps = 13/168 (7%)
Query: 67 TSFVNMDEFVFLKAFKINIHPPRAPLIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGV 126
T ++N +E + +P + W PP W+K N D T+ G
Sbjct: 175 TEWLNANETTENTNVHVATNPIQTSRRDSSQWNPPPEGWVKCNFDSGYTQGSPYTRSGWT 234
Query: 127 FRDHN-NIFIGAFVQNLNTNSAFNAKLLGVITAINIAGENNWRNIWLETDLQLAVLAFKN 185
R+ N +I + + ++ + +A+ LG + A+ + + R +W E+D + V N
Sbjct: 235 IRECNGHIVLCGNAKLQSSTCSLHAEALGFLHALQVIWAHGLRYVWFESDSKSLVTLINN 294
Query: 186 ------LNMVPWSLRNKWLNYFEHVRSMNFLITHIYIEGNSCADCMAN 227
L + + +R+ W+ + S+ F + E NS AD +A+
Sbjct: 295 GEDHSLLGTLIYDIRH-WMLKLPYC-SLEF----VNRERNSAADALAS 336
>At2g07650 putative non-LTR retrolelement reverse transcriptase
Length = 732
Score = 41.6 bits (96), Expect = 4e-04
Identities = 17/36 (47%), Positives = 23/36 (63%)
Query: 94 KEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRD 129
++V W P W+K+NTDGA NP +A+ GG RD
Sbjct: 660 RQVSWRKPNEGWVKLNTDGASHGNPGEATAGGALRD 695
>At4g07516 putative protein
Length = 740
Score = 39.3 bits (90), Expect = 0.002
Identities = 26/73 (35%), Positives = 35/73 (47%)
Query: 111 DGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITAINIAGENNWRNI 170
DGA NP A+ GV RD + + G F N+ SA A+L GV + IA E +
Sbjct: 606 DGASPGNPGLATASGVLRDEHGNWRGDFALNIGICSAPLAELWGVYYKLYIAWEMRITRL 665
Query: 171 WLETDLQLAVLAF 183
LE D ++ F
Sbjct: 666 ELEVDSEIVSAFF 678
>At1g27870 hypothetical protein
Length = 213
Score = 39.3 bits (90), Expect = 0.002
Identities = 37/156 (23%), Positives = 68/156 (42%), Gaps = 19/156 (12%)
Query: 98 WIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIF-IGAFVQNLNTNSAFNAKLLGVI 156
W P WIK N DG+ K+ G V RD N + + ++A +++ +I
Sbjct: 52 WRRPERGWIKCNFDGSFVNGDVKSKAGWVVRDSNGSYLLAGQAIGRKVDNALESEIQALI 111
Query: 157 TAINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRN------KWLNYFEHVRSMNFL 210
++ + ++ + E D ++ + N + V + + N WL FE + N++
Sbjct: 112 ISMQHCWSHGYKRVCFEGDNKM-LFDLINGSKVYFGVHNWIRDIHWWLRKFESIH-FNWI 169
Query: 211 ITHIYIEGNSCADCMA-----NVGLTLSSFVWLPSL 241
H N+ AD +A N L L+ F W+P++
Sbjct: 170 RRH----HNTPADILAKAPLQNQSLFLNHF-WVPNV 200
>At3g30570 putative reverse transcriptase
Length = 1099
Score = 37.4 bits (85), Expect = 0.008
Identities = 16/38 (42%), Positives = 21/38 (55%)
Query: 94 KEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHN 131
+++ W P W K+N DGA NP A+ GG RD N
Sbjct: 1017 QQIGWRVPSAGWYKLNMDGASRGNPGLAAAGGALRDSN 1054
>At5g51080 unknown protein
Length = 322
Score = 37.0 bits (84), Expect = 0.011
Identities = 32/132 (24%), Positives = 55/132 (41%), Gaps = 10/132 (7%)
Query: 108 VNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITAINIAGENNW 167
+ DGA NP + V + + I Q L + A+ G+I + A E +
Sbjct: 188 IEFDGASKGNPGLSGAAAVLKTEDGSLIFKMRQGLGIATNNAAEYHGLILGLKHAIEKGY 247
Query: 168 RNIWLETDLQLAVLAFKNLNMVPWSLRNKWLNYFEHV------RSMNFLITHIYIEGNSC 221
I ++TD +L + K W + ++ L+ + ++F I+H+ NS
Sbjct: 248 TKIKVKTDSKLVCMQMKG----QWKVNHEVLSKLHKEAKQLSDKCLSFEISHVLRSLNSD 303
Query: 222 ADCMANVGLTLS 233
AD AN+ LS
Sbjct: 304 ADEQANMAARLS 315
>At2g11240 pseudogene
Length = 1044
Score = 37.0 bits (84), Expect = 0.011
Identities = 43/159 (27%), Positives = 68/159 (42%), Gaps = 18/159 (11%)
Query: 94 KEVMWIPPIPP------WIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAF-VQNLNTNS 146
K + IP I P I+ +TD + + +A G VF DH+N +N S
Sbjct: 891 KSKIVIPGISPSEIDLDTIQCSTDASWREETLQAGFGWVFVDHSNHLESHHKAAAMNIRS 950
Query: 147 AFNAKLLGVITAINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRNKWLNYFEHVRS 206
AK + AI A + ++ + + +D Q V K LN P + + + V S
Sbjct: 951 PLLAKASALSLAIQHAADLGFKKLVVASDSQQLV---KVLNGEPHPMELHGIVFDISVLS 1007
Query: 207 MNF---LITHIYIEGNSCADCMANVGLTLSSFVWLPSLP 242
+NF + + E NS AD +A L L +P++P
Sbjct: 1008 LNFEENSFSFVKRENNSKADALAKAALLL-----IPAVP 1041
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.139 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,222,775
Number of Sequences: 26719
Number of extensions: 254847
Number of successful extensions: 774
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 729
Number of HSP's gapped (non-prelim): 47
length of query: 261
length of database: 11,318,596
effective HSP length: 97
effective length of query: 164
effective length of database: 8,726,853
effective search space: 1431203892
effective search space used: 1431203892
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC136507.9


