

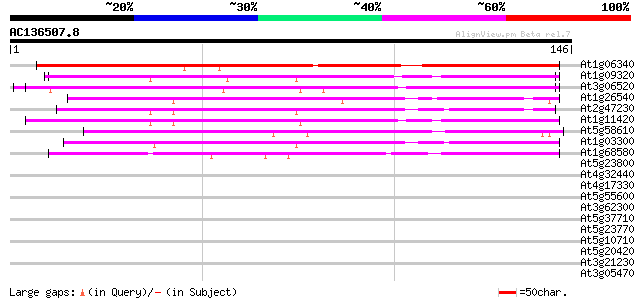
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136507.8 + phase: 0
(146 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g06340 unknown protein 113 3e-26
At1g09320 unknown protein 82 1e-16
At3g06520 hypothetical protein 80 4e-16
At1g26540 unknown protein 67 3e-12
At2g47230 unknown protein 67 4e-12
At1g11420 hypothetical protein 60 5e-10
At5g58610 putative protein 52 1e-07
At1g03300 unknown protein 42 2e-04
At1g68580 unknown protein 41 2e-04
At5g23800 putative protein 37 0.004
At4g32440 unknown protein 36 0.008
At4g17330 G2484-1 protein 35 0.014
At5g55600 unknown protein 35 0.018
At3g62300 putative protein 33 0.070
At5g37710 calmodulin-binding heat-shock protein -like 32 0.16
At5g23770 putative protein 30 0.59
At5g10710 unknown protein 30 0.59
At5g20420 putative protein 27 3.9
At3g21230 putative 4-coumarate:CoA ligase 2 27 3.9
At3g05470 unknown protein 27 5.0
>At1g06340 unknown protein
Length = 134
Score = 113 bits (283), Expect = 3e-26
Identities = 59/138 (42%), Positives = 86/138 (61%), Gaps = 8/138 (5%)
Query: 8 VDYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYV-IRYKNLLKD-DESELLMETLFP 65
+++ GD+VEVCS+++GF+GSYF AT+VS G Y I+YKNL+ D D+S+ L+E +
Sbjct: 1 MEFVKGDQVEVCSKEDGFLGSYFGATVVSKTPEGSYYKIKYKNLVSDTDQSKRLVEVISA 60
Query: 66 KDLRPIPPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIY 125
+LRP+PP + L + KVD FD DGWWVG++ + + YSVYF + +
Sbjct: 61 DELRPMPPKSLHVL-IRCGDKVDAFDKDGWWVGEVTA-----VRRNIYSVYFSTTDEELE 114
Query: 126 YPCDQIRVHQELVWGDWI 143
YP +R H E V G W+
Sbjct: 115 YPLYSLRKHHEWVNGSWV 132
>At1g09320 unknown protein
Length = 517
Score = 81.6 bits (200), Expect = 1e-16
Identities = 46/134 (34%), Positives = 70/134 (51%), Gaps = 4/134 (2%)
Query: 10 YKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+ +G +EV E+EGF S+F A ++ K ++ Y NL +D E L E + +R
Sbjct: 380 FSIGTPIEVSPEEEGFEDSWFLAKLIEYRGKDKCLVEYDNLKAEDGKEPLREEVNVSRIR 439
Query: 70 PIPPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPCD 129
P+P F+ + KV+ NDGWWVG I K+L + S Y V F + + +
Sbjct: 440 PLPLESVMVSPFERHDKVNALYNDGWWVGVI--RKVLAKSS--YLVLFKNTQELLKFHHS 495
Query: 130 QIRVHQELVWGDWI 143
Q+R+HQE + G WI
Sbjct: 496 QLRLHQEWIDGKWI 509
Score = 74.3 bits (181), Expect = 2e-14
Identities = 41/133 (30%), Positives = 68/133 (50%), Gaps = 5/133 (3%)
Query: 10 YKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+ G VEV S++EGF G +F A +V + K+++ Y++L + D E L E +R
Sbjct: 221 FSSGTVVEVSSDEEGFQGCWFAAKVVEPVGEDKFLVEYRDLREKDGIEPLKEETDFLHIR 280
Query: 70 PIPPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPCD 129
P PP + + F + K++ F NDGWWVG + K +YF + + +
Sbjct: 281 PPPPRDED-IDFAVGDKINAFYNDGWWVGVVIDGM----KHGTVGIYFRQSQEKMRFGRQ 335
Query: 130 QIRVHQELVWGDW 142
+R+H++ V G W
Sbjct: 336 GLRLHKDWVDGTW 348
Score = 64.3 bits (155), Expect = 2e-11
Identities = 44/141 (31%), Positives = 71/141 (50%), Gaps = 13/141 (9%)
Query: 11 KVGDKVEVCSEDEGFVGSYFEATIV-----SCLESGKYVIRYKNLLKDDE-SELLMETLF 64
K G VE+ S++ GF GS++ ++ S +S K + Y L D E ++ L E +
Sbjct: 40 KPGSAVEISSDEIGFRGSWYMGKVITIPSSSDKDSVKCQVEYTTLFFDKEGTKPLKEVVD 99
Query: 65 PKDLRPIPP---HVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCH 121
LRP P + K + ++VD F NDGWW G + ++L + +SV+F
Sbjct: 100 MSQLRPPAPPMSEIEKKKKIVVGEEVDAFYNDGWWEGDVT--EVLDDGK--FSVFFRSSK 155
Query: 122 QTIYYPCDQIRVHQELVWGDW 142
+ I + D++R H+E V G W
Sbjct: 156 EQIRFRKDELRFHREWVDGAW 176
>At3g06520 hypothetical protein
Length = 466
Score = 80.1 bits (196), Expect = 4e-16
Identities = 47/150 (31%), Positives = 76/150 (50%), Gaps = 12/150 (8%)
Query: 2 RPPEKRVD--------YKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKD 53
+PP K++ + G +VEV S++ G+ S+F A IVS L +Y + Y+ L D
Sbjct: 317 KPPLKKLKSCERAEKVFNNGMEVEVRSDEPGYEASWFSAKIVSYLGENRYTVEYQTLKTD 376
Query: 54 DESELLMETLFPKDLRPIPPH-VRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCY 112
DE ELL E D+RP PP + +++L + VD + N+GWW G++ +
Sbjct: 377 DERELLKEEARGSDIRPPPPPLIPKGYRYELYELVDAWYNEGWWSGRVYK---INNNKTR 433
Query: 113 YSVYFDYCHQTIYYPCDQIRVHQELVWGDW 142
Y VYF +++ + + +R Q G W
Sbjct: 434 YGVYFQTTDESLEFAYNDLRPCQVWRNGKW 463
Score = 68.6 bits (166), Expect = 1e-12
Identities = 42/141 (29%), Positives = 75/141 (52%), Gaps = 4/141 (2%)
Query: 5 EKRVDYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDD-ESELLMETL 63
++R Y+ G VEV SE++ + GS++ A I+ L KY++ + +DD ES L + +
Sbjct: 164 KRRDQYEKGALVEVRSEEKAYKGSWYCARILCLLGDDKYIVEHLKFSRDDGESIPLRDVV 223
Query: 64 FPKDLRPIPPHVRNPLK-FKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQ 122
KD+RP+PP +P+ ++ VD + N WW +++ K+L S YSV+ +
Sbjct: 224 EAKDIRPVPPSELSPVVCYEPGVIVDAWFNKRWWTSRVS--KVLGGGSNKYSVFIISTGE 281
Query: 123 TIYYPCDQIRVHQELVWGDWI 143
+R H++ + G W+
Sbjct: 282 ETTILNFNLRPHKDWINGQWV 302
>At1g26540 unknown protein
Length = 695
Score = 67.0 bits (162), Expect = 3e-12
Identities = 48/135 (35%), Positives = 65/135 (47%), Gaps = 13/135 (9%)
Query: 16 VEVCSEDEGFVGSYFEATIVSCLESG---KYVIRYKNLLKDDESELLMETLFPKDLRPIP 72
VEV SE+EGF G++F A + + K +RY LL D S L+E + + +RP+P
Sbjct: 14 VEVSSEEEGFEGAWFRAVLEENPGNSSRRKLRVRYSTLLDMDGSSPLIEHIEQRFIRPVP 73
Query: 73 PHVRNPLKFKLNQ--KVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPCDQ 130
P L + VD DGWW G + + ME Y VYFD I + Q
Sbjct: 74 PEENQQKDVVLEEGLLVDADHKDGWWTGVVVKK---MEDD-NYLVYFDLPPDIIQFERKQ 129
Query: 131 IRVHQELVW--GDWI 143
+R H L+W G WI
Sbjct: 130 LRTH--LIWTGGTWI 142
>At2g47230 unknown protein
Length = 701
Score = 66.6 bits (161), Expect = 4e-12
Identities = 45/134 (33%), Positives = 69/134 (50%), Gaps = 10/134 (7%)
Query: 13 GDKVEVCSEDEGFVGSYFEATIV-SCLESG--KYVIRYKNLLKDDESELLMETLFPKDLR 69
G +VEV S +EGF ++F + + +SG K +RY LL DD L+E + P+ +R
Sbjct: 8 GSEVEVSSTEEGFADAWFRGILQENPTKSGRKKLRVRYLTLLNDDALSPLIENIEPRFIR 67
Query: 70 PIPP-HVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPC 128
P+PP + N + + VD DGWW G I + +E ++ VY+D I +
Sbjct: 68 PVPPENEYNGIVLEEGTVVDADHKDGWWTGVIIKK---LENGKFW-VYYDSPPDIIEFER 123
Query: 129 DQIRVHQELVWGDW 142
+Q+R H L W W
Sbjct: 124 NQLRPH--LRWSGW 135
>At1g11420 hypothetical protein
Length = 604
Score = 59.7 bits (143), Expect = 5e-10
Identities = 44/143 (30%), Positives = 70/143 (48%), Gaps = 8/143 (5%)
Query: 5 EKRVDYKVGDKVEVCSEDEGFVGSYFEATIV-SCLESG--KYVIRYKNLLKDDESELLME 61
E+ + + DKVEV SE+E GSY+ A + + +SG K +RY L + L E
Sbjct: 2 EQMIFIRKDDKVEVFSEEEELKGSYYRAILEDNPTKSGHNKLKVRYLTQLNEHRLAPLTE 61
Query: 62 TLFPKDLRPIPPH-VRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYC 120
+ + +RP+P V + + F VD + DGWW G + K + ++ + VYFD
Sbjct: 62 FVDQRFIRPVPSEDVNDGVVFVEGLMVDAYLKDGWWTGVVV--KTMEDEK--FLVYFDCP 117
Query: 121 HQTIYYPCDQIRVHQELVWGDWI 143
I + ++RVH + WI
Sbjct: 118 PDIIQFEKKKLRVHLDWTGFKWI 140
>At5g58610 putative protein
Length = 1065
Score = 52.0 bits (123), Expect = 1e-07
Identities = 43/152 (28%), Positives = 64/152 (41%), Gaps = 30/152 (19%)
Query: 20 SEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKD------------ 67
S +EG +GS++ T+ S + + IRY N+L DD S L+ET+ D
Sbjct: 27 SLEEGSLGSWYLGTVTSAKKRRRRCIRYDNILSDDGSGNLVETVDVSDIVEGLDDCTDVS 86
Query: 68 ------LRPIPPHVR-NPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYC 120
LRP+PP + L VDVF +D WW G + + EK V+F
Sbjct: 87 DTFRGRLRPVPPKLDVAKLNLAYGLCVDVFFSDAWWEGVLFDHENGSEKR---RVFFPDL 143
Query: 121 HQTIYYPCDQIRVHQEL-----VW---GDWIF 144
+ +R+ Q+ W G W+F
Sbjct: 144 GDELDADLQSLRITQDWNEATETWECRGSWLF 175
>At1g03300 unknown protein
Length = 670
Score = 41.6 bits (96), Expect = 2e-04
Identities = 33/135 (24%), Positives = 58/135 (42%), Gaps = 10/135 (7%)
Query: 15 KVEVCSEDEGFVGSYFEATIVS-----CLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+VE+ SE++GF +++ A + ES K Y + E T+ + +R
Sbjct: 8 EVEIFSEEDGFRNAWYRAILEETPTNPTSESKKLRFSYMTKSLNKEGSSSPPTVEQRFIR 67
Query: 70 PIPP-HVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYYSVYFDYCHQTIYYPC 128
P+PP ++ N + F+ VD W G + ++ ME Y V FD I +
Sbjct: 68 PVPPENLYNGVVFEEGTMVDADYKHRWRTGVVINK---MENDSYL-VLFDCPPDIIQFET 123
Query: 129 DQIRVHQELVWGDWI 143
+R H + +W+
Sbjct: 124 KHLRAHLDWTGSEWV 138
>At1g68580 unknown protein
Length = 648
Score = 41.2 bits (95), Expect = 2e-04
Identities = 39/153 (25%), Positives = 68/153 (43%), Gaps = 25/153 (16%)
Query: 11 KVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLL-KDDESELLMETLFP---- 65
K G +EV SED G G +F+A ++ K ++Y+++ DDES+ L E +
Sbjct: 364 KKGSLIEVLSEDSGIRGCWFKALVLK-KHKDKVKVQYQDIQDADDESKKLEEWILTSRVA 422
Query: 66 -------------KDLRPI--PPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKS 110
K +RP+ P + + VDV+ DGWW G I +++ EK
Sbjct: 423 AGDHLGDLRIKGRKVVRPMLKPSKENDVCVIGVGMPVDVWWCDGWWEG-IVVQEVSEEK- 480
Query: 111 CYYSVYFDYCHQTIYYPCDQIRVHQELVWGDWI 143
+ VY + + + +R +E + +W+
Sbjct: 481 --FEVYLPGEKKMSAFHRNDLRQSREWLDDEWL 511
>At5g23800 putative protein
Length = 552
Score = 37.0 bits (84), Expect = 0.004
Identities = 27/96 (28%), Positives = 44/96 (45%), Gaps = 1/96 (1%)
Query: 4 PEKRVDYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETL 63
P + + G +VE+ ++ G +++A + + S LLKDD S L E
Sbjct: 6 PSESLSLSEGCEVEISYKNNGNESVWYKAILEAKPNSIFKEELSVRLLKDDFSTPLNELR 65
Query: 64 FPKDLRPIPP-HVRNPLKFKLNQKVDVFDNDGWWVG 98
+RPIPP +V+ + ++ VD D WW G
Sbjct: 66 HKVLIRPIPPTNVQACIDIEIGTFVDADYKDAWWAG 101
Score = 36.6 bits (83), Expect = 0.005
Identities = 27/100 (27%), Positives = 53/100 (53%), Gaps = 7/100 (7%)
Query: 10 YKVGDKVEVCSE-DEGFVGSYFEATIVSCLESG---KYVIRYKNLLKDDESELLMETLFP 65
+ G VE+CS+ DEG V + A + + +Y+++ K L+ +T+
Sbjct: 165 FSPGTIVELCSKRDEGEV-VWVPALVYKEFKENDEYRYIVKDKPLIGRSYKSRPSKTVDL 223
Query: 66 KDLRPIPPHVRNPLKFKLNQKVDVF-DNDGWWVGKIASEK 104
+ LRPIPP +R +++L++ ++V+ D GW G++ +
Sbjct: 224 RSLRPIPPPIR-VKEYRLDEYIEVYHDGIGWRQGRVVKSE 262
>At4g32440 unknown protein
Length = 393
Score = 35.8 bits (81), Expect = 0.008
Identities = 29/91 (31%), Positives = 45/91 (48%), Gaps = 3/91 (3%)
Query: 13 GDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLRPIP 72
G +VEV S E G++ A IVS Y +R+ + + E E +ME + K +RP P
Sbjct: 6 GSRVEVFSNKEAPYGAWRCAEIVSG-NGHTYNVRFYSFQIEHE-EAVMEKVPRKIIRPCP 63
Query: 73 PHVRNPLKFKLNQKVDVFDNDGWWVGKIASE 103
P V + ++ + V+V DN W + E
Sbjct: 64 PLV-DVERWDTGELVEVLDNFSWKAATVREE 93
>At4g17330 G2484-1 protein
Length = 2037
Score = 35.0 bits (79), Expect = 0.014
Identities = 39/162 (24%), Positives = 66/162 (40%), Gaps = 30/162 (18%)
Query: 6 KRVDYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFP 65
K D K G VEV E+ G +++ A ++S LE K + + +L + ++ L E +
Sbjct: 1569 KNEDIKEGSNVEVFKEEPGLRTAWYSANVLS-LEDDKAYVLFSDLSVEQGTDKLKEWVAL 1627
Query: 66 KDLRPIPPHVRNPLK----------------------FKLNQKVDVFDNDGWWVGKIASE 103
K P +R P + +K+ +VD + +D W G I +
Sbjct: 1628 KGEGDQAPKIR-PARSVTALPYEGTRKRRRAALGDHIWKIGDRVDSWVHDSWLEGVITEK 1686
Query: 104 KILMEKSCYYSVYFDYCHQTIYYPCDQIRVHQELVW--GDWI 143
E + +V+F +T+ +R LVW G WI
Sbjct: 1687 NKKDENT--VTVHFPAEEETLTIKAWNLR--PSLVWKDGKWI 1724
>At5g55600 unknown protein
Length = 663
Score = 34.7 bits (78), Expect = 0.018
Identities = 36/152 (23%), Positives = 59/152 (38%), Gaps = 24/152 (15%)
Query: 11 KVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPK---- 66
K K+E +D G G +F T++ S K V + ++D++ +E P
Sbjct: 382 KTDAKIEFLCQDSGIRGCWFRCTVLDV--SRKQVKLQYDDIEDEDGYGNLEEWVPAFKSA 439
Query: 67 -------------DLRPIPPHVRNP-LKFKLNQKVDVFDNDGWWVG-KIASEKILMEKSC 111
+RP P V+ + + VD + NDGWW G IA+ K E
Sbjct: 440 MPDKLGIRLSNRPTIRPAPRDVKTAYFDLTIGEAVDAWWNDGWWEGVVIATGKPDTEDLK 499
Query: 112 YYSVYFDYCHQTIYYPCDQIRVHQELVWGDWI 143
Y + C + IR+ ++ V W+
Sbjct: 500 IYIPGENLCLTVLR---KDIRISRDWVGDSWV 528
>At3g62300 putative protein
Length = 708
Score = 32.7 bits (73), Expect = 0.070
Identities = 25/101 (24%), Positives = 47/101 (45%), Gaps = 4/101 (3%)
Query: 9 DYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDL 68
D+ G VEV ++ + + A ++ E G +++ K LK++E ++ ++
Sbjct: 160 DFSAGKSVEVRTKVDKLGDVWAPAMVIKEDEDGTMLVKLKT-LKEEEVNCTKISVSYSEI 218
Query: 69 RPIPPHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEK 109
RP P + +KL + VD GW G ++ K+L K
Sbjct: 219 RPSPLPI-GLRDYKLMENVDALVESGWCPGVVS--KVLAGK 256
Score = 31.2 bits (69), Expect = 0.20
Identities = 28/93 (30%), Positives = 41/93 (43%), Gaps = 7/93 (7%)
Query: 50 LLKDDESELLMETLFPKDLRPIPPHVRNP-LKFKLNQKVDVFDNDGWWVGKIASEKILME 108
LLK D S L++T + +RP+PP P + F+ VD GW G + K+L
Sbjct: 59 LLKYDYSPPLIKTTVHRFMRPVPPPDPFPEVDFEEGDVVDAAYKGGWCSGSVV--KVLGN 116
Query: 109 KSCYYSVYFDYCHQTIYYPCDQIRVHQELVWGD 141
+ + VY + I +R H VW D
Sbjct: 117 RR--FLVYLRFQPDVIELLRKDLRPH--FVWKD 145
>At5g37710 calmodulin-binding heat-shock protein -like
Length = 439
Score = 31.6 bits (70), Expect = 0.16
Identities = 20/71 (28%), Positives = 33/71 (46%), Gaps = 14/71 (19%)
Query: 65 PKDLRPIP-----------PHVRNPLKFKLNQKVDVFDNDGWWVGKIASEKILMEKSCYY 113
P++ PIP P +RNP K++ + FD + WV K + + +S Y
Sbjct: 39 PEEFEPIPRISRVILAVYEPDLRNP---KISPSLGTFDLNPEWVIKRVTHEKTQGRSPPY 95
Query: 114 SVYFDYCHQTI 124
+Y D+ H+ I
Sbjct: 96 IIYIDHDHREI 106
>At5g23770 putative protein
Length = 438
Score = 29.6 bits (65), Expect = 0.59
Identities = 18/60 (30%), Positives = 33/60 (55%), Gaps = 4/60 (6%)
Query: 42 KYVIRYKNLLKDDESELLMETLFPKDLRPIPPHVRNPLKFKLNQKVDVF-DNDGWWVGKI 100
KY+++ K+ + + +T+ LRPIP V +++L + V+VF D GW G++
Sbjct: 54 KYIVKDKSFSCEGKKARPNKTVDLSSLRPIPVSVD---EYQLEENVEVFLDGMGWRHGRV 110
>At5g10710 unknown protein
Length = 312
Score = 29.6 bits (65), Expect = 0.59
Identities = 21/63 (33%), Positives = 33/63 (52%), Gaps = 5/63 (7%)
Query: 4 PEKRVDYKVGDKVEVCSEDEGFVGSYFE-ATIVSCLESGKYVIRYKNLLKDDESELLMET 62
PE+R+ Y+VG+KV +C D +G FE +T E V+ K+ L E +++E
Sbjct: 102 PEERITYRVGNKV-ICCLDGSRIGIQFETSTAGETYEVYHCVLESKSFL---EKMIVLEH 157
Query: 63 LFP 65
P
Sbjct: 158 TIP 160
>At5g20420 putative protein
Length = 1261
Score = 26.9 bits (58), Expect = 3.9
Identities = 13/46 (28%), Positives = 24/46 (51%)
Query: 9 DYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDD 54
DY++ +++ +C G VGS + E K+ I K++ +DD
Sbjct: 590 DYRLEEEIGMCCRLCGHVGSEIKDVSAPFAEHKKWTIETKHIEEDD 635
>At3g21230 putative 4-coumarate:CoA ligase 2
Length = 570
Score = 26.9 bits (58), Expect = 3.9
Identities = 19/68 (27%), Positives = 33/68 (47%), Gaps = 8/68 (11%)
Query: 5 EKRVDYKVGDKVEVCSEDEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLF 64
+K + K + VC +D+G G +VS + G + + L + DE+ELL +
Sbjct: 157 DKLTNLKNDGVLIVCLDDDGDNG------VVSSSDDG--CVSFTELTQADETELLKPKIS 208
Query: 65 PKDLRPIP 72
P+D +P
Sbjct: 209 PEDTVAMP 216
>At3g05470 unknown protein
Length = 884
Score = 26.6 bits (57), Expect = 5.0
Identities = 10/15 (66%), Positives = 11/15 (72%)
Query: 65 PKDLRPIPPHVRNPL 79
P RPIPPH+R PL
Sbjct: 118 PAPPRPIPPHLRRPL 132
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.142 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,773,648
Number of Sequences: 26719
Number of extensions: 153809
Number of successful extensions: 419
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 375
Number of HSP's gapped (non-prelim): 29
length of query: 146
length of database: 11,318,596
effective HSP length: 90
effective length of query: 56
effective length of database: 8,913,886
effective search space: 499177616
effective search space used: 499177616
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC136507.8


