

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136507.13 - phase: 0
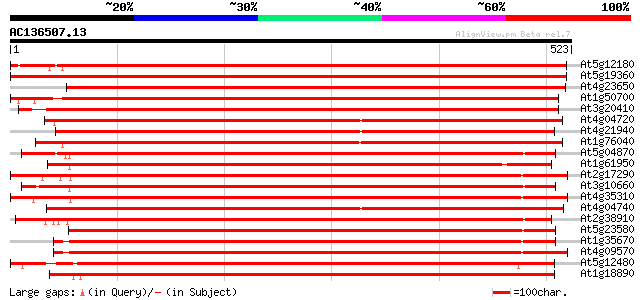
(523 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g12180 calcium-dependent protein kinase 868 0.0
At5g19360 calcium-dependent protein kinase - like 862 0.0
At4g23650 calcium-dependent protein kinase (CDPK6) 704 0.0
At1g50700 hypothetical protein 679 0.0
At3g20410 calmodulin-domain protein kinase CDPK isoform 9 675 0.0
At4g04720 putative calcium dependent protein kinase 666 0.0
At4g21940 calcium-dependent protein kinase like protein 657 0.0
At1g76040 calcium-dependent protein kinase (CPK29) 637 0.0
At5g04870 calcium-dependent protein kinase 628 e-180
At1g61950 hypothetical protein 624 e-179
At2g17290 calmodulin-domain protein kinase CDPK isoform 6 (CPK6) 619 e-177
At3g10660 calmodulin-domain protein kinase CDPK isoform 2 617 e-177
At4g35310 calmodulin-domain protein kinase CDPK isoform 5 (CPK5) 615 e-176
At4g04740 putative calcium dependent protein kinase 602 e-172
At2g38910 putative calcium-dependent protein kinase 600 e-172
At5g23580 calcium-dependent protein kinase (pir||S71196) 582 e-166
At1g35670 calcium-dependent protein kinase 575 e-164
At4g09570 calmodulin-domain protein kinase CDPK isoform 4 (CPK4) 574 e-164
At5g12480 calcium-dependent protein kinase - like protein 548 e-156
At1g18890 calcium-dependent protein kinase (ATCDPK1) 547 e-156
>At5g12180 calcium-dependent protein kinase
Length = 528
Score = 868 bits (2242), Expect = 0.0
Identities = 432/526 (82%), Positives = 470/526 (89%), Gaps = 9/526 (1%)
Query: 1 MGNCCSGGTEDPEDKGENNQQNNDANGDSSATTPP-----PWSKPSPQPSKP--SKPSAI 53
MGNCCS G D D G+ + A+ +++T P P SK +P PS P +K I
Sbjct: 1 MGNCCSHG-RDSADNGDALENGASASNAANSTGPTAEASVPQSKHAP-PSPPPATKQGPI 58
Query: 54 GPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDV 113
GPVLGRPMEDVKA+YS+GKELGRGQFGVTHLCT K TG Q+ACKTIAKRKL NKEDIEDV
Sbjct: 59 GPVLGRPMEDVKASYSLGKELGRGQFGVTHLCTQKATGHQFACKTIAKRKLVNKEDIEDV 118
Query: 114 RREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLL 173
RREVQIMHHLTGQPNIVEL GA+EDK SVHLVMELCAGGELFDRIIAKGHY+ERAAASLL
Sbjct: 119 RREVQIMHHLTGQPNIVELKGAYEDKHSVHLVMELCAGGELFDRIIAKGHYSERAAASLL 178
Query: 174 RTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSA 233
RTIVQIVHTCHSMGVIHRDLKPENFLLL+KDENSPLKATDFGLSVFYK G+ FKDIVGSA
Sbjct: 179 RTIVQIVHTCHSMGVIHRDLKPENFLLLNKDENSPLKATDFGLSVFYKPGEVFKDIVGSA 238
Query: 234 YYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWP 293
YYIAP+VLKRKYGPE DIWS+GVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWP
Sbjct: 239 YYIAPEVLKRKYGPEADIWSIGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWP 298
Query: 294 SISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQ 353
SISP AKDLV+KMLNSDPKQR+TA +VLNHPWIKEDGEAPD PLDNAV++RLKQF+AMN
Sbjct: 299 SISPQAKDLVKKMLNSDPKQRLTAAQVLNHPWIKEDGEAPDVPLDNAVMSRLKQFKAMNN 358
Query: 354 FKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQGTRLSETEVKQL 413
FKKVAL+VIA CLSEEEIMGLK+MFKGMDTD+SGTIT+EEL+QGLAKQGTRLSE EV+QL
Sbjct: 359 FKKVALRVIAGCLSEEEIMGLKEMFKGMDTDSSGTITLEELRQGLAKQGTRLSEYEVQQL 418
Query: 414 MEAADADGNGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYITIEELEQALHEY 473
MEAADADGNG IDY EFI ATMH+NRL+REEH+Y+AFQ FDKDNSGYIT+EELEQAL E+
Sbjct: 419 MEAADADGNGTIDYGEFIAATMHINRLDREEHLYSAFQHFDKDNSGYITMEELEQALREF 478
Query: 474 NMHDGRDIKEIISEVDADNDGRINYDEFVAMMGKGNPEANTKKRRD 519
M+DGRDIKEIISEVD DNDGRINYDEFVAMM KGNP+ KKRR+
Sbjct: 479 GMNDGRDIKEIISEVDGDNDGRINYDEFVAMMRKGNPDPIPKKRRE 524
>At5g19360 calcium-dependent protein kinase - like
Length = 523
Score = 862 bits (2226), Expect = 0.0
Identities = 420/519 (80%), Positives = 459/519 (87%)
Query: 1 MGNCCSGGTEDPEDKGENNQQNNDANGDSSATTPPPWSKPSPQPSKPSKPSAIGPVLGRP 60
MGNCCS G + ++K E +N ++ + P P +K IGPVLGRP
Sbjct: 1 MGNCCSHGRDSDDNKEEPRPENGGGGVGAAEASVRASKHPPASPPPATKQGPIGPVLGRP 60
Query: 61 MEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIM 120
MEDVK++Y++GKELGRGQFGVTHLCT K TG Q+ACKTIAKRKL NKEDIEDVRREVQIM
Sbjct: 61 MEDVKSSYTLGKELGRGQFGVTHLCTQKATGLQFACKTIAKRKLVNKEDIEDVRREVQIM 120
Query: 121 HHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIV 180
HHLTGQPNIVEL GA+EDK SVHLVMELCAGGELFDRIIAKGHY+ERAAASLLRTIVQI+
Sbjct: 121 HHLTGQPNIVELKGAYEDKHSVHLVMELCAGGELFDRIIAKGHYSERAAASLLRTIVQII 180
Query: 181 HTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSAYYIAPDV 240
HTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYK G+ FKDIVGSAYYIAP+V
Sbjct: 181 HTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKPGEVFKDIVGSAYYIAPEV 240
Query: 241 LKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPSAK 300
L+RKYGPE DIWS+GVMLYILLCGVPPFWAESENGIFNAIL G VDFSSDPWP ISP AK
Sbjct: 241 LRRKYGPEADIWSIGVMLYILLCGVPPFWAESENGIFNAILSGQVDFSSDPWPVISPQAK 300
Query: 301 DLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQFKKVALK 360
DLVRKMLNSDPKQR+TA +VLNHPWIKEDGEAPD PLDNAV++RLKQF+AMN FKKVAL+
Sbjct: 301 DLVRKMLNSDPKQRLTAAQVLNHPWIKEDGEAPDVPLDNAVMSRLKQFKAMNNFKKVALR 360
Query: 361 VIASCLSEEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQGTRLSETEVKQLMEAADAD 420
VIA CLSEEEIMGLK+MFKGMDTDNSGTIT+EEL+QGLAKQGTRLSE EV+QLMEAADAD
Sbjct: 361 VIAGCLSEEEIMGLKEMFKGMDTDNSGTITLEELRQGLAKQGTRLSEYEVQQLMEAADAD 420
Query: 421 GNGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYITIEELEQALHEYNMHDGRD 480
GNG IDY EFI ATMH+NRL+REEH+Y+AFQ FDKDNSGYIT EELEQAL E+ M+DGRD
Sbjct: 421 GNGTIDYGEFIAATMHINRLDREEHLYSAFQHFDKDNSGYITTEELEQALREFGMNDGRD 480
Query: 481 IKEIISEVDADNDGRINYDEFVAMMGKGNPEANTKKRRD 519
IKEIISEVD DNDGRINY+EFVAMM KGNP+ N KKRR+
Sbjct: 481 IKEIISEVDGDNDGRINYEEFVAMMRKGNPDPNPKKRRE 519
>At4g23650 calcium-dependent protein kinase (CDPK6)
Length = 529
Score = 704 bits (1818), Expect = 0.0
Identities = 335/465 (72%), Positives = 401/465 (86%)
Query: 54 GPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDV 113
G +LGRPME+V+ TY G+ELGRGQFGVT+L THK T +Q ACK+I R+L +K+DIEDV
Sbjct: 64 GRILGRPMEEVRRTYEFGRELGRGQFGVTYLVTHKETKQQVACKSIPTRRLVHKDDIEDV 123
Query: 114 RREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLL 173
RREVQIMHHL+G NIV+L GA+ED+ SV+L+MELC GGELFDRII+KG Y+ERAAA L
Sbjct: 124 RREVQIMHHLSGHRNIVDLKGAYEDRHSVNLIMELCEGGELFDRIISKGLYSERAAADLC 183
Query: 174 RTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSA 233
R +V +VH+CHSMGV+HRDLKPENFL LSKDENSPLKATDFGLSVF+K GD+FKD+VGSA
Sbjct: 184 RQMVMVVHSCHSMGVMHRDLKPENFLFLSKDENSPLKATDFGLSVFFKPGDKFKDLVGSA 243
Query: 234 YYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWP 293
YY+AP+VLKR YGPE DIWS GV+LYILL GVPPFW E+E GIF+AIL+G +DFS+DPWP
Sbjct: 244 YYVAPEVLKRNYGPEADIWSAGVILYILLSGVPPFWGENETGIFDAILQGQLDFSADPWP 303
Query: 294 SISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQ 353
++S AKDLVRKML DPK R+TA EVLNHPWI+EDGEA D PLDNAVL+R+KQFRAMN+
Sbjct: 304 ALSDGAKDLVRKMLKYDPKDRLTAAEVLNHPWIREDGEASDKPLDNAVLSRMKQFRAMNK 363
Query: 354 FKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQGTRLSETEVKQL 413
KK+ALKVIA LSEEEI+GLK+MFK +DTDN+G +T+EEL+ GL K G+++SE E++QL
Sbjct: 364 LKKMALKVIAENLSEEEIIGLKEMFKSLDTDNNGIVTLEELRTGLPKLGSKISEAEIRQL 423
Query: 414 MEAADADGNGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYITIEELEQALHEY 473
MEAAD DG+G IDY EFI+ATMHMNR+ RE+H+YTAFQFFD DNSGYIT+EELE A+ +Y
Sbjct: 424 MEAADMDGDGSIDYLEFISATMHMNRIEREDHLYTAFQFFDNDNSGYITMEELELAMKKY 483
Query: 474 NMHDGRDIKEIISEVDADNDGRINYDEFVAMMGKGNPEANTKKRR 518
NM D + IKEII+EVD D DG+INY+EFVAMM KGNPE +RR
Sbjct: 484 NMGDDKSIKEIIAEVDTDRDGKINYEEFVAMMKKGNPELVPNRRR 528
>At1g50700 hypothetical protein
Length = 521
Score = 679 bits (1752), Expect = 0.0
Identities = 339/523 (64%), Positives = 409/523 (77%), Gaps = 19/523 (3%)
Query: 1 MGNCCS---GGTEDPEDKGENNQQ--------NNDANGDSSATT-PPPWSKPSPQPSKPS 48
MGNC + G P+ GE + + + D + S+ T PPPW P+
Sbjct: 1 MGNCLAKKYGLVMKPQQNGERSVEIENRRRSTHQDPSKISTGTNQPPPWRNPA------- 53
Query: 49 KPSAIGPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKE 108
K S +L +P EDVK Y++ KELGRGQFGVT+LCT K+TGK++ACK+I+K+KL K
Sbjct: 54 KHSGAAAILEKPYEDVKLFYTLSKELGRGQFGVTYLCTEKSTGKRFACKSISKKKLVTKG 113
Query: 109 DIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERA 168
D ED+RRE+QIM HL+GQPNIVE GA+ED+++V+LVMELCAGGELFDRI+AKGHY+ERA
Sbjct: 114 DKEDMRREIQIMQHLSGQPNIVEFKGAYEDEKAVNLVMELCAGGELFDRILAKGHYSERA 173
Query: 169 AASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKD 228
AAS+ R IV +V+ CH MGV+HRDLKPENFLL SKDE + +KATDFGLSVF ++G +KD
Sbjct: 174 AASVCRQIVNVVNICHFMGVMHRDLKPENFLLSSKDEKALIKATDFGLSVFIEEGRVYKD 233
Query: 229 IVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHVDFS 288
IVGSAYY+AP+VLKR+YG E+DIWS G++LYILL GVPPFWAE+E GIF+AIL G +DF
Sbjct: 234 IVGSAYYVAPEVLKRRYGKEIDIWSAGIILYILLSGVPPFWAETEKGIFDAILEGEIDFE 293
Query: 289 SDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLNRLKQF 348
S PWPSIS SAKDLVR+ML DPK+RI+A EVL HPW++E GEA D P+D+AVL+R+KQF
Sbjct: 294 SQPWPSISNSAKDLVRRMLTQDPKRRISAAEVLKHPWLREGGEASDKPIDSAVLSRMKQF 353
Query: 349 RAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQGTRLSET 408
RAMN+ KK+ALKVIA + EEI GLK MF +DTDNSGTIT EELK+GLAK G+RL+E
Sbjct: 354 RAMNKLKKLALKVIAENIDTEEIQGLKAMFANIDTDNSGTITYEELKEGLAKLGSRLTEA 413
Query: 409 EVKQLMEAADADGNGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYITIEELEQ 468
EVKQLM+AAD DGNG IDY EFITATMH +RL E+VY AFQ FDKD SGYIT +ELE
Sbjct: 414 EVKQLMDAADVDGNGSIDYIEFITATMHRHRLESNENVYKAFQHFDKDGSGYITTDELEA 473
Query: 469 ALHEYNMHDGRDIKEIISEVDADNDGRINYDEFVAMMGKGNPE 511
AL EY M D IKEI+S+VDADNDGRINYDEF AMM GNP+
Sbjct: 474 ALKEYGMGDDATIKEILSDVDADNDGRINYDEFCAMMRSGNPQ 516
>At3g20410 calmodulin-domain protein kinase CDPK isoform 9
Length = 541
Score = 675 bits (1741), Expect = 0.0
Identities = 329/504 (65%), Positives = 399/504 (78%), Gaps = 13/504 (2%)
Query: 9 TEDPEDKGENNQQNNDANGDSSATTPPPWSKPSPQPS-KPSKPSAIGPVLGRPMEDVKAT 67
T+ PE G N Q PPPW + P P + +L EDVK
Sbjct: 43 TQQPEKPGSVNSQ------------PPPWRAAAAAPGLSPKTTTKSNSILENAFEDVKLF 90
Query: 68 YSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQP 127
Y++GKELGRGQFGVT+LCT +TGK+YACK+I+K+KL K D +D+RRE+QIM HL+GQP
Sbjct: 91 YTLGKELGRGQFGVTYLCTENSTGKKYACKSISKKKLVTKADKDDMRREIQIMQHLSGQP 150
Query: 128 NIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMG 187
NIVE GA+ED+++V+LVMELCAGGELFDRIIAKGHYTERAAAS+ R IV +V CH MG
Sbjct: 151 NIVEFKGAYEDEKAVNLVMELCAGGELFDRIIAKGHYTERAAASVCRQIVNVVKICHFMG 210
Query: 188 VIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSAYYIAPDVLKRKYGP 247
V+HRDLKPENFLL SKDE + +KATDFGLSVF ++G ++DIVGSAYY+AP+VL+R+YG
Sbjct: 211 VLHRDLKPENFLLSSKDEKALIKATDFGLSVFIEEGKVYRDIVGSAYYVAPEVLRRRYGK 270
Query: 248 EVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPSAKDLVRKML 307
EVDIWS G++LYILL GVPPFWAE+E GIF+AIL GH+DF S PWPSIS SAKDLVR+ML
Sbjct: 271 EVDIWSAGIILYILLSGVPPFWAETEKGIFDAILEGHIDFESQPWPSISSSAKDLVRRML 330
Query: 308 NSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQFKKVALKVIASCLS 367
+DPK+RI+A +VL HPW++E GEA D P+D+AVL+R+KQFRAMN+ KK+ALKVIA +
Sbjct: 331 TADPKRRISAADVLQHPWLREGGEASDKPIDSAVLSRMKQFRAMNKLKKLALKVIAENID 390
Query: 368 EEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQGTRLSETEVKQLMEAADADGNGIIDY 427
EEI GLK MF +DTDNSGTIT EELK+GLAK G++L+E EVKQLM+AAD DGNG IDY
Sbjct: 391 TEEIQGLKAMFANIDTDNSGTITYEELKEGLAKLGSKLTEAEVKQLMDAADVDGNGSIDY 450
Query: 428 DEFITATMHMNRLNREEHVYTAFQFFDKDNSGYITIEELEQALHEYNMHDGRDIKEIISE 487
EFITATMH +RL E++Y AFQ FDKD+SGYITI+ELE AL EY M D IKE++S+
Sbjct: 451 IEFITATMHRHRLESNENLYKAFQHFDKDSSGYITIDELESALKEYGMGDDATIKEVLSD 510
Query: 488 VDADNDGRINYDEFVAMMGKGNPE 511
VD+DNDGRINY+EF AMM GNP+
Sbjct: 511 VDSDNDGRINYEEFCAMMRSGNPQ 534
>At4g04720 putative calcium dependent protein kinase
Length = 531
Score = 666 bits (1718), Expect = 0.0
Identities = 325/488 (66%), Positives = 394/488 (80%), Gaps = 6/488 (1%)
Query: 33 TPPPWSKP-----SPQPSKPSKPSAIGPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTH 87
TP P ++P S S P +LG+P ED++ YS+GKELGRGQFG+T++C
Sbjct: 40 TPKPMTQPIHQQISTPSSNPVSVRDPDTILGKPFEDIRKFYSLGKELGRGQFGITYMCKE 99
Query: 88 KTTGKQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVME 147
TG YACK+I KRKL +K+D EDV+RE+QIM +L+GQPNIVE+ GA+ED+QS+HLVME
Sbjct: 100 IGTGNTYACKSILKRKLISKQDKEDVKREIQIMQYLSGQPNIVEIKGAYEDRQSIHLVME 159
Query: 148 LCAGGELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENS 207
LCAGGELFDRIIA+GHY+ERAAA ++R+IV +V CH MGV+HRDLKPENFLL SK+EN+
Sbjct: 160 LCAGGELFDRIIAQGHYSERAAAGIIRSIVNVVQICHFMGVVHRDLKPENFLLSSKEENA 219
Query: 208 PLKATDFGLSVFYKQGDQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPP 267
LKATDFGLSVF ++G ++DIVGSAYY+AP+VL+R YG E+DIWS GV+LYILL GVPP
Sbjct: 220 MLKATDFGLSVFIEEGKVYRDIVGSAYYVAPEVLRRSYGKEIDIWSAGVILYILLSGVPP 279
Query: 268 FWAESENGIFNAILRGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIK 327
FWAE+E GIF+ +++G +DF S+PWPSIS SAKDLVRKML DPK+RITA +VL HPWIK
Sbjct: 280 FWAENEKGIFDEVIKGEIDFVSEPWPSISESAKDLVRKMLTKDPKRRITAAQVLEHPWIK 339
Query: 328 EDGEAPDTPLDNAVLNRLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSG 387
GEAPD P+D+AVL+R+KQFRAMN+ KK+ALKVIA LSEEEI GLK MF +DTD SG
Sbjct: 340 -GGEAPDKPIDSAVLSRMKQFRAMNKLKKLALKVIAESLSEEEIKGLKTMFANIDTDKSG 398
Query: 388 TITIEELKQGLAKQGTRLSETEVKQLMEAADADGNGIIDYDEFITATMHMNRLNREEHVY 447
TIT EELK GL + G+RLSETEVKQLMEAAD DGNG IDY EFI+ATMH +L+R+EHVY
Sbjct: 399 TITYEELKTGLTRLGSRLSETEVKQLMEAADVDGNGTIDYYEFISATMHRYKLDRDEHVY 458
Query: 448 TAFQFFDKDNSGYITIEELEQALHEYNMHDGRDIKEIISEVDADNDGRINYDEFVAMMGK 507
AFQ FDKDNSG+IT +ELE A+ EY M D IKE+ISEVD DNDGRIN++EF AMM
Sbjct: 459 KAFQHFDKDNSGHITRDELESAMKEYGMGDEASIKEVISEVDTDNDGRINFEEFCAMMRS 518
Query: 508 GNPEANTK 515
G+ + K
Sbjct: 519 GSTQPQGK 526
>At4g21940 calcium-dependent protein kinase like protein
Length = 554
Score = 657 bits (1695), Expect = 0.0
Identities = 311/466 (66%), Positives = 382/466 (81%), Gaps = 1/466 (0%)
Query: 43 QPSKPSKPSAIGPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKR 102
QP KP +LG+P E+++ Y++GKELGRGQFG+T+ C +TG YACK+I KR
Sbjct: 77 QPLKPIVFRETETILGKPFEEIRKLYTLGKELGRGQFGITYTCKENSTGNTYACKSILKR 136
Query: 103 KLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKG 162
KL K+DI+DV+RE+QIM +L+GQ NIVE+ GA+ED+QS+HLVMELC G ELFDRIIA+G
Sbjct: 137 KLTRKQDIDDVKREIQIMQYLSGQENIVEIKGAYEDRQSIHLVMELCGGSELFDRIIAQG 196
Query: 163 HYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQ 222
HY+E+AAA ++R+++ +V CH MGVIHRDLKPENFLL S DEN+ LKATDFGLSVF ++
Sbjct: 197 HYSEKAAAGVIRSVLNVVQICHFMGVIHRDLKPENFLLASTDENAMLKATDFGLSVFIEE 256
Query: 223 GDQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILR 282
G ++DIVGSAYY+AP+VL+R YG E+DIWS G++LYILLCGVPPFW+E+E GIFN I++
Sbjct: 257 GKVYRDIVGSAYYVAPEVLRRSYGKEIDIWSAGIILYILLCGVPPFWSETEKGIFNEIIK 316
Query: 283 GHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVL 342
G +DF S PWPSIS SAKDLVRK+L DPKQRI+A + L HPWI+ GEAPD P+D+AVL
Sbjct: 317 GEIDFDSQPWPSISESAKDLVRKLLTKDPKQRISAAQALEHPWIR-GGEAPDKPIDSAVL 375
Query: 343 NRLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQG 402
+R+KQFRAMN+ KK+ALKVIA LSEEEI GLK MF MDTD SGTIT EELK GLAK G
Sbjct: 376 SRMKQFRAMNKLKKLALKVIAESLSEEEIKGLKTMFANMDTDKSGTITYEELKNGLAKLG 435
Query: 403 TRLSETEVKQLMEAADADGNGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYIT 462
++L+E EVKQLMEAAD DGNG IDY EFI+ATMH R +R+EHV+ AFQ+FDKDNSG+IT
Sbjct: 436 SKLTEAEVKQLMEAADVDGNGTIDYIEFISATMHRYRFDRDEHVFKAFQYFDKDNSGFIT 495
Query: 463 IEELEQALHEYNMHDGRDIKEIISEVDADNDGRINYDEFVAMMGKG 508
++ELE A+ EY M D IKE+I+EVD DNDGRINY+EF AMM G
Sbjct: 496 MDELESAMKEYGMGDEASIKEVIAEVDTDNDGRINYEEFCAMMRSG 541
>At1g76040 calcium-dependent protein kinase (CPK29)
Length = 561
Score = 637 bits (1642), Expect = 0.0
Identities = 308/494 (62%), Positives = 387/494 (77%), Gaps = 4/494 (0%)
Query: 25 ANGDSSATTPPPWSKPSPQPSKP---SKPSAIGPVLGRPMEDVKATYSMGKELGRGQFGV 81
+ G +S T P KP P P P S S IGP+L RPM D+ A Y + KELGRGQFG+
Sbjct: 66 SQGQTSNPTSNPQPKPKPAPPPPPSTSSGSQIGPILNRPMIDLSALYDLHKELGRGQFGI 125
Query: 82 THLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQS 141
T+ CT K+ G++YACK+I+KRKL ++DIEDVRREV I+ HLTGQPNIVE GA+EDK +
Sbjct: 126 TYKCTDKSNGREYACKSISKRKLIRRKDIEDVRREVMILQHLTGQPNIVEFRGAYEDKDN 185
Query: 142 VHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLL 201
+HLVMELC+GGELFDRII KG Y+E+ AA++ R IV +VH CH MGV+HRDLKPENFLL+
Sbjct: 186 LHLVMELCSGGELFDRIIKKGSYSEKEAANIFRQIVNVVHVCHFMGVVHRDLKPENFLLV 245
Query: 202 SKDENSPLKATDFGLSVFYKQGDQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYIL 261
S +E+SP+KATDFGLSVF ++G ++DIVGSAYY+AP+VL R YG E+D+WS GVMLYIL
Sbjct: 246 SNEEDSPIKATDFGLSVFIEEGKVYRDIVGSAYYVAPEVLHRNYGKEIDVWSAGVMLYIL 305
Query: 262 LCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVL 321
L GVPPFW E+E IF AIL G +D + PWP+IS SAKDL+RKML DPK+RITA E L
Sbjct: 306 LSGVPPFWGETEKTIFEAILEGKLDLETSPWPTISESAKDLIRKMLIRDPKKRITAAEAL 365
Query: 322 NHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGM 381
HPW+ D + D P+++AVL R+KQFRAMN+ KK+ALKVIA LSEEEI GLKQ FK M
Sbjct: 366 EHPWM-TDTKISDKPINSAVLVRMKQFRAMNKLKKLALKVIAENLSEEEIKGLKQTFKNM 424
Query: 382 DTDNSGTITIEELKQGLAKQGTRLSETEVKQLMEAADADGNGIIDYDEFITATMHMNRLN 441
DTD SGTIT +EL+ GL + G++L+E+E+KQLMEAAD D +G IDY EF+TATMH +RL
Sbjct: 425 DTDESGTITFDELRNGLHRLGSKLTESEIKQLMEAADVDKSGTIDYIEFVTATMHRHRLE 484
Query: 442 REEHVYTAFQFFDKDNSGYITIEELEQALHEYNMHDGRDIKEIISEVDADNDGRINYDEF 501
+EE++ AF++FDKD SG+IT +EL+ ++ EY M D I E+I++VD DNDGRINY+EF
Sbjct: 485 KEENLIEAFKYFDKDRSGFITRDELKHSMTEYGMGDDATIDEVINDVDTDNDGRINYEEF 544
Query: 502 VAMMGKGNPEANTK 515
VAMM KG +++ K
Sbjct: 545 VAMMRKGTTDSDPK 558
>At5g04870 calcium-dependent protein kinase
Length = 610
Score = 628 bits (1619), Expect = e-180
Identities = 307/508 (60%), Positives = 387/508 (75%), Gaps = 12/508 (2%)
Query: 12 PEDKGENNQQNNDANGDSSATTPPPWSKPSPQPSKPSKP------SAIG----PVLGRPM 61
PE E + ++ + + P SKP P P+KP KP S+ G VL R
Sbjct: 85 PETLEEISLESKPETKQETKSETKPESKPDP-PAKPKKPKHMKRVSSAGLRTESVLQRKT 143
Query: 62 EDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMH 121
E+ K YS+G++LG+GQFG T LC KTTGK++ACK+IAKRKL ED+EDVRRE+QIMH
Sbjct: 144 ENFKEFYSLGRKLGQGQFGTTFLCVEKTTGKEFACKSIAKRKLLTDEDVEDVRREIQIMH 203
Query: 122 HLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIVH 181
HL G PN++ + GA+ED +VHLVME CAGGELFDRII +GHYTER AA L RTIV +V
Sbjct: 204 HLAGHPNVISIKGAYEDVVAVHLVMECCAGGELFDRIIQRGHYTERKAAELTRTIVGVVE 263
Query: 182 TCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSAYYIAPDVL 241
CHS+GV+HRDLKPENFL +SK E+S LK DFGLS+F+K D F D+VGS YY+AP+VL
Sbjct: 264 ACHSLGVMHRDLKPENFLFVSKHEDSLLKTIDFGLSMFFKPDDVFTDVVGSPYYVAPEVL 323
Query: 242 KRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPSAKD 301
+++YGPE D+WS GV++YILL GVPPFWAE+E GIF +L G +DFSSDPWPSIS SAKD
Sbjct: 324 RKRYGPEADVWSAGVIVYILLSGVPPFWAETEQGIFEQVLHGDLDFSSDPWPSISESAKD 383
Query: 302 LVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQFKKVALKV 361
LVRKML DPK+R+TA++VL HPW++ DG APD PLD+AVL+R+KQF AMN+FKK+AL+V
Sbjct: 384 LVRKMLVRDPKKRLTAHQVLCHPWVQVDGVAPDKPLDSAVLSRMKQFSAMNKFKKMALRV 443
Query: 362 IASCLSEEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQGTRLSETEVKQLMEAADADG 421
IA LSEEEI GLK+MF +D D SG IT EELK GL + G L E+E+ LM+AAD D
Sbjct: 444 IAESLSEEEIAGLKEMFNMIDADKSGQITFEELKAGLKRVGANLKESEILDLMQAADVDN 503
Query: 422 NGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYITIEELEQALHEYNMHDGRDI 481
+G IDY EFI AT+H+N++ RE+H++ AF +FDKD SGYIT +EL+QA E+ + D R I
Sbjct: 504 SGTIDYKEFIAATLHLNKIEREDHLFAAFTYFDKDGSGYITPDELQQACEEFGVEDVR-I 562
Query: 482 KEIISEVDADNDGRINYDEFVAMMGKGN 509
+E++ +VD DNDGRI+Y+EFVAMM KG+
Sbjct: 563 EELMRDVDQDNDGRIDYNEFVAMMQKGS 590
>At1g61950 hypothetical protein
Length = 547
Score = 624 bits (1610), Expect = e-179
Identities = 309/482 (64%), Positives = 374/482 (77%), Gaps = 16/482 (3%)
Query: 36 PWSKPSPQPSKPSKPSAIG-----------PVLGRPMEDVKATYSMGKELGRGQFGVTHL 84
P P P P K I P+LGRP ED+K YS+G+ELGRGQFG+T++
Sbjct: 55 PHKLPLPLPQPQEKQKLINHQKQSTLQQPEPILGRPFEDIKEKYSLGRELGRGQFGITYI 114
Query: 85 CTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHL 144
CT ++GK +ACK+I KRKL +D EDVRRE+QIMH+L+GQPNIVE+ GA+ED+QSVHL
Sbjct: 115 CTEISSGKNFACKSILKRKLIRTKDREDVRREIQIMHYLSGQPNIVEIKGAYEDRQSVHL 174
Query: 145 VMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKD 204
VMELC GGELFD+I +GHY+E+AAA ++R++V++V CH MGVIHRDLKPENFLL SKD
Sbjct: 175 VMELCEGGELFDKITKRGHYSEKAAAEIIRSVVKVVQICHFMGVIHRDLKPENFLLSSKD 234
Query: 205 E-NSPLKATDFGLSVFYKQGDQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLC 263
E +S LKATDFG+SVF ++G ++DIVGSAYY+AP+VLKR YG +DIWS GV+LYILLC
Sbjct: 235 EASSMLKATDFGVSVFIEEGKVYEDIVGSAYYVAPEVLKRNYGKAIDIWSAGVILYILLC 294
Query: 264 GVPPFWAESENGIFNAILRGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNH 323
G PPFWAE++ GIF ILRG +DF S+PWPSIS SAKDLVR ML DPK+R TA +VL H
Sbjct: 295 GNPPFWAETDKGIFEEILRGEIDFESEPWPSISESAKDLVRNMLKYDPKKRFTAAQVLEH 354
Query: 324 PWIKEDGEAPDTPLDNAVLNRLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDT 383
PWI+E GEA D P+D+AVL+R+KQ RAMN+ KK+A K IA L EEE+ GLK MF MDT
Sbjct: 355 PWIREGGEASDKPIDSAVLSRMKQLRAMNKLKKLAFKFIAQNLKEEELKGLKTMFANMDT 414
Query: 384 DNSGTITIEELKQGLAKQGTRLSETEVKQLMEAADADGNGIIDYDEFITATMHMNRLNRE 443
D SGTIT +ELK GL K G+RL+ETEVKQL+E AD DGNG IDY EFI+ATM+ R+ RE
Sbjct: 415 DKSGTITYDELKSGLEKLGSRLTETEVKQLLEDADVDGNGTIDYIEFISATMNRFRVERE 474
Query: 444 EHVYTAFQFFDKDNSGYITIEELEQALHEYNMHDGRDIKEIISEVDADNDGRINYDEFVA 503
++++ AFQ FDKDNSG +ELE A+ EYNM D IKEIISEVDADNDG INY EF
Sbjct: 475 DNLFKAFQHFDKDNSG----QELETAMKEYNMGDDIMIKEIISEVDADNDGSINYQEFCN 530
Query: 504 MM 505
MM
Sbjct: 531 MM 532
>At2g17290 calmodulin-domain protein kinase CDPK isoform 6 (CPK6)
Length = 544
Score = 619 bits (1596), Expect = e-177
Identities = 307/537 (57%), Positives = 397/537 (73%), Gaps = 18/537 (3%)
Query: 1 MGNCCSGGTEDPEDKGENNQQNNDANGDS-------SATTPPPWSKPSPQPSK--PSKPS 51
MGN C G +D +G +++ ++ + S TT +SK + P+ P K
Sbjct: 1 MGNSCRGSFKDKIYEGNHSRPEENSKSTTTTVSSVHSPTTDQDFSKQNTNPALVIPVKEP 60
Query: 52 AIGP--------VLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRK 103
+ VLG +++ Y++ ++LG+GQFG T+LCT TG YACK+I+KRK
Sbjct: 61 IMRRNVDNQSYYVLGHKTPNIRDLYTLSRKLGQGQFGTTYLCTDIATGVDYACKSISKRK 120
Query: 104 LANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGH 163
L +KED+EDVRRE+QIMHHL G NIV + GA+ED VH+VMELCAGGELFDRII +GH
Sbjct: 121 LISKEDVEDVRREIQIMHHLAGHKNIVTIKGAYEDPLYVHIVMELCAGGELFDRIIHRGH 180
Query: 164 YTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQG 223
Y+ER AA L + IV +V CHS+GV+HRDLKPENFLL++KD++ LKA DFGLSVF+K G
Sbjct: 181 YSERKAAELTKIIVGVVEACHSLGVMHRDLKPENFLLVNKDDDFSLKAIDFGLSVFFKPG 240
Query: 224 DQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRG 283
FKD+VGS YY+AP+VL + YGPE D+W+ GV+LYILL GVPPFWAE++ GIF+A+L+G
Sbjct: 241 QIFKDVVGSPYYVAPEVLLKHYGPEADVWTAGVILYILLSGVPPFWAETQQGIFDAVLKG 300
Query: 284 HVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLN 343
++DF +DPWP IS SAKDL+RKML S P +R+TA+EVL HPWI E+G APD LD AVL+
Sbjct: 301 YIDFDTDPWPVISDSAKDLIRKMLCSSPSERLTAHEVLRHPWICENGVAPDRALDPAVLS 360
Query: 344 RLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQGT 403
RLKQF AMN+ KK+ALKVIA LSEEEI GL+ MF+ MDTDNSG IT +ELK GL + G+
Sbjct: 361 RLKQFSAMNKLKKMALKVIAESLSEEEIAGLRAMFEAMDTDNSGAITFDELKAGLRRYGS 420
Query: 404 RLSETEVKQLMEAADADGNGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYITI 463
L +TE++ LMEAAD D +G IDY EFI AT+H+N+L REEH+ +AFQ+FDKD SGYITI
Sbjct: 421 TLKDTEIRDLMEAADVDNSGTIDYSEFIAATIHLNKLEREEHLVSAFQYFDKDGSGYITI 480
Query: 464 EELEQALHEYNMHDGRDIKEIISEVDADNDGRINYDEFVAMMGKGNPEANTKKRRDS 520
+EL+Q+ E+ M D +++II EVD DNDGRI+Y+EFVAMM KGN + ++S
Sbjct: 481 DELQQSCIEHGMTD-VFLEDIIKEVDQDNDGRIDYEEFVAMMQKGNAGVGRRTMKNS 536
>At3g10660 calmodulin-domain protein kinase CDPK isoform 2
Length = 646
Score = 617 bits (1591), Expect = e-177
Identities = 297/502 (59%), Positives = 384/502 (76%), Gaps = 7/502 (1%)
Query: 12 PEDKGENNQQNNDANGDSSATTPPPWSKPSPQPSKPSKPSAIG----PVLGRPMEDVKAT 67
PE K E+ + ++++ T P +P + S+ G VL R E+ K
Sbjct: 128 PETKSESKPETTKP--ETTSETKPETKAEPQKPKHMRRVSSAGLRTESVLQRKTENFKEF 185
Query: 68 YSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQP 127
YS+G++LG+GQFG T LC K TG +YACK+I+KRKL ED+EDVRRE+QIMHHL G P
Sbjct: 186 YSLGRKLGQGQFGTTFLCLEKGTGNEYACKSISKRKLLTDEDVEDVRREIQIMHHLAGHP 245
Query: 128 NIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMG 187
N++ + GA+ED +VHLVMELC+GGELFDRII +GHYTER AA L RTIV ++ CHS+G
Sbjct: 246 NVISIKGAYEDVVAVHLVMELCSGGELFDRIIQRGHYTERKAAELARTIVGVLEACHSLG 305
Query: 188 VIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSAYYIAPDVLKRKYGP 247
V+HRDLKPENFL +S++E+S LK DFGLS+F+K + F D+VGS YY+AP+VL+++YGP
Sbjct: 306 VMHRDLKPENFLFVSREEDSLLKTIDFGLSMFFKPDEVFTDVVGSPYYVAPEVLRKRYGP 365
Query: 248 EVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPSAKDLVRKML 307
E D+WS GV++YILL GVPPFWAE+E GIF +L G +DFSSDPWPSIS SAKDLVRKML
Sbjct: 366 ESDVWSAGVIVYILLSGVPPFWAETEQGIFEQVLHGDLDFSSDPWPSISESAKDLVRKML 425
Query: 308 NSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQFKKVALKVIASCLS 367
DPK+R+TA++VL HPW++ DG APD PLD+AVL+R+KQF AMN+FKK+AL+VIA LS
Sbjct: 426 VRDPKRRLTAHQVLCHPWVQIDGVAPDKPLDSAVLSRMKQFSAMNKFKKMALRVIAESLS 485
Query: 368 EEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQGTRLSETEVKQLMEAADADGNGIIDY 427
EEEI GLKQMFK +D DNSG IT EELK GL + G L E+E+ LM+AAD D +G IDY
Sbjct: 486 EEEIAGLKQMFKMIDADNSGQITFEELKAGLKRVGANLKESEILDLMQAADVDNSGTIDY 545
Query: 428 DEFITATMHMNRLNREEHVYTAFQFFDKDNSGYITIEELEQALHEYNMHDGRDIKEIISE 487
EFI AT+H+N++ RE+H++ AF +FDKD SG+IT +EL+QA E+ + D R I+E++ +
Sbjct: 546 KEFIAATLHLNKIEREDHLFAAFSYFDKDESGFITPDELQQACEEFGVEDAR-IEEMMRD 604
Query: 488 VDADNDGRINYDEFVAMMGKGN 509
VD D DGRI+Y+EFVAMM KG+
Sbjct: 605 VDQDKDGRIDYNEFVAMMQKGS 626
>At4g35310 calmodulin-domain protein kinase CDPK isoform 5 (CPK5)
Length = 556
Score = 615 bits (1587), Expect = e-176
Identities = 303/549 (55%), Positives = 398/549 (72%), Gaps = 30/549 (5%)
Query: 1 MGNCCSGGTEDPEDKGENNQ-------------QNNDANGDSSATTPPPWSKPSPQPSKP 47
MGN C G +D D+G+NN+ N+D + +++ +SK + +
Sbjct: 1 MGNSCRGSFKDKLDEGDNNKPEDYSKTSTTNLSSNSDHSPNAADIIAQEFSKDNNSNNNS 60
Query: 48 SKPSAIGP----------------VLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTG 91
P+ + P VLG +++ Y++ ++LG+GQFG T+LCT +G
Sbjct: 61 KDPALVIPLREPIMRRNPDNQAYYVLGHKTPNIRDIYTLSRKLGQGQFGTTYLCTEIASG 120
Query: 92 KQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAG 151
YACK+I+KRKL +KED+EDVRRE+QIMHHL G +IV + GA+ED VH+VMELCAG
Sbjct: 121 VDYACKSISKRKLISKEDVEDVRREIQIMHHLAGHGSIVTIKGAYEDSLYVHIVMELCAG 180
Query: 152 GELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKA 211
GELFDRII +GHY+ER AA L + IV +V CHS+GV+HRDLKPENFLL++KD++ LKA
Sbjct: 181 GELFDRIIQRGHYSERKAAELTKIIVGVVEACHSLGVMHRDLKPENFLLVNKDDDFSLKA 240
Query: 212 TDFGLSVFYKQGDQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAE 271
DFGLSVF+K G F D+VGS YY+AP+VL ++YGPE D+W+ GV+LYILL GVPPFWAE
Sbjct: 241 IDFGLSVFFKPGQIFTDVVGSPYYVAPEVLLKRYGPEADVWTAGVILYILLSGVPPFWAE 300
Query: 272 SENGIFNAILRGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGE 331
++ GIF+A+L+G++DF SDPWP IS SAKDL+R+ML+S P +R+TA+EVL HPWI E+G
Sbjct: 301 TQQGIFDAVLKGYIDFESDPWPVISDSAKDLIRRMLSSKPAERLTAHEVLRHPWICENGV 360
Query: 332 APDTPLDNAVLNRLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSGTITI 391
APD LD AVL+RLKQF AMN+ KK+ALKVIA LSEEEI GL++MF+ MDTDNSG IT
Sbjct: 361 APDRALDPAVLSRLKQFSAMNKLKKMALKVIAESLSEEEIAGLREMFQAMDTDNSGAITF 420
Query: 392 EELKQGLAKQGTRLSETEVKQLMEAADADGNGIIDYDEFITATMHMNRLNREEHVYTAFQ 451
+ELK GL K G+ L +TE+ LM+AAD D +G IDY EFI AT+H+N+L REEH+ AFQ
Sbjct: 421 DELKAGLRKYGSTLKDTEIHDLMDAADVDNSGTIDYSEFIAATIHLNKLEREEHLVAAFQ 480
Query: 452 FFDKDNSGYITIEELEQALHEYNMHDGRDIKEIISEVDADNDGRINYDEFVAMMGKGNPE 511
+FDKD SG+ITI+EL+QA E+ M D +++II EVD +NDG+I+Y EFV MM KGN
Sbjct: 481 YFDKDGSGFITIDELQQACVEHGMAD-VFLEDIIKEVDQNNDGKIDYGEFVEMMQKGNAG 539
Query: 512 ANTKKRRDS 520
+ R+S
Sbjct: 540 VGRRTMRNS 548
>At4g04740 putative calcium dependent protein kinase
Length = 520
Score = 602 bits (1553), Expect = e-172
Identities = 294/483 (60%), Positives = 372/483 (76%), Gaps = 2/483 (0%)
Query: 35 PPWSKPS-PQPSKPSKPSAIGPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQ 93
P KP P PS P +LG+P ED++ YS+G+ELGRG G+T++C TG
Sbjct: 35 PDHRKPQIPSPSIPISVRDPETILGKPFEDIRKFYSLGRELGRGGLGITYMCKEIGTGNI 94
Query: 94 YACKTIAKRKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGE 153
YACK+I KRKL ++ EDV+ E+QIM HL+GQPN+VE+ G++ED+ SVHLVMELCAGGE
Sbjct: 95 YACKSILKRKLISELGREDVKTEIQIMQHLSGQPNVVEIKGSYEDRHSVHLVMELCAGGE 154
Query: 154 LFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATD 213
LFDRIIA+GHY+ERAAA +++IV +V CH GVIHRDLKPENFL SK+EN+ LK TD
Sbjct: 155 LFDRIIAQGHYSERAAAGTIKSIVDVVQICHLNGVIHRDLKPENFLFSSKEENAMLKVTD 214
Query: 214 FGLSVFYKQGDQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESE 273
FGLS F ++G +KD+VGS YY+AP+VL++ YG E+DIWS GV+LYILLCGVPPFWA++E
Sbjct: 215 FGLSAFIEEGKIYKDVVGSPYYVAPEVLRQSYGKEIDIWSAGVILYILLCGVPPFWADNE 274
Query: 274 NGIFNAILRGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAP 333
G+F IL+ +DF +PWPSIS SAKDLV KML DPK+RITA +VL HPWIK GEAP
Sbjct: 275 EGVFVEILKCKIDFVREPWPSISDSAKDLVEKMLTEDPKRRITAAQVLEHPWIK-GGEAP 333
Query: 334 DTPLDNAVLNRLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSGTITIEE 393
+ P+D+ VL+R+KQFRAMN+ KK+ALKV A LSEEEI GLK +F MDT+ SGTIT E+
Sbjct: 334 EKPIDSTVLSRMKQFRAMNKLKKLALKVSAVSLSEEEIKGLKTLFANMDTNRSGTITYEQ 393
Query: 394 LKQGLAKQGTRLSETEVKQLMEAADADGNGIIDYDEFITATMHMNRLNREEHVYTAFQFF 453
L+ GL++ +RLSETEV+QL+EA+D DGNG IDY EFI+ATMH +L+ +EHV+ AFQ
Sbjct: 394 LQTGLSRLRSRLSETEVQQLVEASDVDGNGTIDYYEFISATMHRYKLHHDEHVHKAFQHL 453
Query: 454 DKDNSGYITIEELEQALHEYNMHDGRDIKEIISEVDADNDGRINYDEFVAMMGKGNPEAN 513
DKD +G+IT +ELE A+ EY M D IKE+ISEVD DNDG+IN++EF AMM G +
Sbjct: 454 DKDKNGHITRDELESAMKEYGMGDEASIKEVISEVDTDNDGKINFEEFRAMMRCGTTQPK 513
Query: 514 TKK 516
K+
Sbjct: 514 GKQ 516
>At2g38910 putative calcium-dependent protein kinase
Length = 583
Score = 600 bits (1548), Expect = e-172
Identities = 297/537 (55%), Positives = 387/537 (71%), Gaps = 38/537 (7%)
Query: 6 SGGTEDPEDKGENNQQNNDANGDSSAT------------TPPPWSK-----PSPQP---- 44
S E K + + +D+NG S+T TPPP K P P+P
Sbjct: 35 SSKDESSRKKNDKSVNGDDSNGHVSSTVDPAPSTLPTPSTPPPPVKMANEEPPPKPITEN 94
Query: 45 -----SKPSKPSA-----------IGPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHK 88
SKP K A I VLGR E++K YS+G++LG+GQFG T LC K
Sbjct: 95 KEDPNSKPQKKEAHMKRMASAGLQIDSVLGRKTENLKDIYSVGRKLGQGQFGTTFLCVDK 154
Query: 89 TTGKQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMEL 148
TGK++ACKTIAKRKL ED+EDVRRE+QIMHHL+G PN+++++GA+ED +VH+VME+
Sbjct: 155 KTGKEFACKTIAKRKLTTPEDVEDVRREIQIMHHLSGHPNVIQIVGAYEDAVAVHVVMEI 214
Query: 149 CAGGELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSP 208
CAGGELFDRII +GHYTE+ AA L R IV ++ CHS+GV+HRDLKPENFL +S DE +
Sbjct: 215 CAGGELFDRIIQRGHYTEKKAAELARIIVGVIEACHSLGVMHRDLKPENFLFVSGDEEAA 274
Query: 209 LKATDFGLSVFYKQGDQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPF 268
LK DFGLSVF+K G+ F D+VGS YY+AP+VL++ Y E D+WS GV++YILL GVPPF
Sbjct: 275 LKTIDFGLSVFFKPGETFTDVVGSPYYVAPEVLRKHYSHECDVWSAGVIIYILLSGVPPF 334
Query: 269 WAESENGIFNAILRGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKE 328
W E+E GIF +L+G +DF S+PWPS+S SAKDLVR+ML DPK+R+T +EVL HPW +
Sbjct: 335 WDETEQGIFEQVLKGDLDFISEPWPSVSESAKDLVRRMLIRDPKKRMTTHEVLCHPWARV 394
Query: 329 DGEAPDTPLDNAVLNRLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSGT 388
DG A D PLD+AVL+RL+QF AMN+ KK+A+KVIA LSEEEI GLK+MFK +DTDNSG
Sbjct: 395 DGVALDKPLDSAVLSRLQQFSAMNKLKKIAIKVIAESLSEEEIAGLKEMFKMIDTDNSGH 454
Query: 389 ITIEELKQGLAKQGTRLSETEVKQLMEAADADGNGIIDYDEFITATMHMNRLNREEHVYT 448
IT+EELK+GL + G L ++E+ LM+AAD D +G IDY EFI A +H+N++ +E+H++T
Sbjct: 455 ITLEELKKGLDRVGADLKDSEILGLMQAADIDNSGTIDYGEFIAAMVHLNKIEKEDHLFT 514
Query: 449 AFQFFDKDNSGYITIEELEQALHEYNMHDGRDIKEIISEVDADNDGRINYDEFVAMM 505
AF +FD+D SGYIT +EL+QA ++ + D + +I+ EVD DNDGRI+Y EFV MM
Sbjct: 515 AFSYFDQDGSGYITRDELQQACKQFGLADVH-LDDILREVDKDNDGRIDYSEFVDMM 570
>At5g23580 calcium-dependent protein kinase (pir||S71196)
Length = 490
Score = 582 bits (1499), Expect = e-166
Identities = 277/454 (61%), Positives = 361/454 (79%), Gaps = 1/454 (0%)
Query: 56 VLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRR 115
VL ++V+ Y +G+ LG+GQFG T LCTHK TG++ ACK+I KRKL +ED +DV R
Sbjct: 10 VLPYKTKNVEDNYFLGQVLGQGQFGTTFLCTHKQTGQKLACKSIPKRKLLCQEDYDDVLR 69
Query: 116 EVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRT 175
E+QIMHHL+ PN+V + A+ED ++VHLVMELC GGELFDRI+ +GHY+ER AA L++T
Sbjct: 70 EIQIMHHLSEYPNVVRIESAYEDTKNVHLVMELCEGGELFDRIVKRGHYSEREAAKLIKT 129
Query: 176 IVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSAYY 235
IV +V CHS+GV+HRDLKPENFL S DE++ LK+TDFGLSVF G+ F ++VGSAYY
Sbjct: 130 IVGVVEACHSLGVVHRDLKPENFLFSSSDEDASLKSTDFGLSVFCTPGEAFSELVGSAYY 189
Query: 236 IAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSI 295
+AP+VL + YGPE D+WS GV+LYILLCG PPFWAESE GIF IL+G ++F +PWPSI
Sbjct: 190 VAPEVLHKHYGPECDVWSAGVILYILLCGFPPFWAESEIGIFRKILQGKLEFEINPWPSI 249
Query: 296 SPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQFK 355
S SAKDL++KML S+PK+R+TA++VL HPWI +D APD PLD AV++RLK+F AMN+ K
Sbjct: 250 SESAKDLIKKMLESNPKKRLTAHQVLCHPWIVDDKVAPDKPLDCAVVSRLKKFSAMNKLK 309
Query: 356 KVALKVIASCLSEEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQGTRLSETEVKQLME 415
K+AL+VIA LSEEEI GLK++FK +DTD SGTIT EELK + + G+ L E+E+++L+
Sbjct: 310 KMALRVIAERLSEEEIGGLKELFKMIDTDKSGTITFEELKDSMRRVGSELMESEIQELLR 369
Query: 416 AADADGNGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYITIEELEQALHEYNM 475
AAD D +G IDY EF+ AT+H+N+L REE++ AF FFDKD SGYITIEEL+QA E+ +
Sbjct: 370 AADVDESGTIDYGEFLAATIHLNKLEREENLVAAFSFFDKDASGYITIEELQQAWKEFGI 429
Query: 476 HDGRDIKEIISEVDADNDGRINYDEFVAMMGKGN 509
+D ++ E+I ++D DNDG+I+Y EFVAMM KGN
Sbjct: 430 NDS-NLDEMIKDIDQDNDGQIDYGEFVAMMRKGN 462
>At1g35670 calcium-dependent protein kinase
Length = 495
Score = 575 bits (1483), Expect = e-164
Identities = 276/468 (58%), Positives = 357/468 (75%), Gaps = 6/468 (1%)
Query: 42 PQPSKPSKPSAIGPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAK 101
P P +PS VL ++ Y +GK+LG+GQFG T+LCT K+T YACK+I K
Sbjct: 5 PNPRRPSNT-----VLPYQTPRLRDHYLLGKKLGQGQFGTTYLCTEKSTSANYACKSIPK 59
Query: 102 RKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAK 161
RKL +ED EDV RE+QIMHHL+ PN+V + G +ED VH+VME+C GGELFDRI++K
Sbjct: 60 RKLVCREDYEDVWREIQIMHHLSEHPNVVRIKGTYEDSVFVHIVMEVCEGGELFDRIVSK 119
Query: 162 GHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYK 221
GH++ER A L++TI+ +V CHS+GV+HRDLKPENFL S +++ LKATDFGLSVFYK
Sbjct: 120 GHFSEREAVKLIKTILGVVEACHSLGVMHRDLKPENFLFDSPKDDAKLKATDFGLSVFYK 179
Query: 222 QGDQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAIL 281
G D+VGS YY+AP+VLK+ YGPE+D+WS GV+LYILL GVPPFWAE+E+GIF IL
Sbjct: 180 PGQYLYDVVGSPYYVAPEVLKKCYGPEIDVWSAGVILYILLSGVPPFWAETESGIFRQIL 239
Query: 282 RGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAV 341
+G +DF SDPWP+IS +AKDL+ KML PK+RI+A+E L HPWI ++ APD PLD AV
Sbjct: 240 QGKLDFKSDPWPTISEAAKDLIYKMLERSPKKRISAHEALCHPWIVDEQAAPDKPLDPAV 299
Query: 342 LNRLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQ 401
L+RLKQF MN+ KK+AL+VIA LSEEEI GLK++FK +DTDNSGTIT EELK GL +
Sbjct: 300 LSRLKQFSQMNKIKKMALRVIAERLSEEEIGGLKELFKMIDTDNSGTITFEELKAGLKRV 359
Query: 402 GTRLSETEVKQLMEAADADGNGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYI 461
G+ L E+E+K LM+AAD D +G IDY EF+ AT+HMN++ REE++ AF +FDKD SGYI
Sbjct: 360 GSELMESEIKSLMDAADIDNSGTIDYGEFLAATLHMNKMEREENLVAAFSYFDKDGSGYI 419
Query: 462 TIEELEQALHEYNMHDGRDIKEIISEVDADNDGRINYDEFVAMMGKGN 509
TI+EL+ A E+ + D + ++I E+D DNDG+I++ EF AMM KG+
Sbjct: 420 TIDELQSACTEFGLCD-TPLDDMIKEIDLDNDGKIDFSEFTAMMRKGD 466
>At4g09570 calmodulin-domain protein kinase CDPK isoform 4 (CPK4)
Length = 501
Score = 574 bits (1480), Expect = e-164
Identities = 276/479 (57%), Positives = 363/479 (75%), Gaps = 6/479 (1%)
Query: 42 PQPSKPSKPSAIGPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAK 101
P P +PS VL ++ Y +GK+LG+GQFG T+LCT K++ YACK+I K
Sbjct: 4 PNPRRPSNS-----VLPYETPRLRDHYLLGKKLGQGQFGTTYLCTEKSSSANYACKSIPK 58
Query: 102 RKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAK 161
RKL +ED EDV RE+QIMHHL+ PN+V + G +ED VH+VME+C GGELFDRI++K
Sbjct: 59 RKLVCREDYEDVWREIQIMHHLSEHPNVVRIKGTYEDSVFVHIVMEVCEGGELFDRIVSK 118
Query: 162 GHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYK 221
G ++ER AA L++TI+ +V CHS+GV+HRDLKPENFL S +++ LKATDFGLSVFYK
Sbjct: 119 GCFSEREAAKLIKTILGVVEACHSLGVMHRDLKPENFLFDSPSDDAKLKATDFGLSVFYK 178
Query: 222 QGDQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAIL 281
G D+VGS YY+AP+VLK+ YGPE+D+WS GV+LYILL GVPPFWAE+E+GIF IL
Sbjct: 179 PGQYLYDVVGSPYYVAPEVLKKCYGPEIDVWSAGVILYILLSGVPPFWAETESGIFRQIL 238
Query: 282 RGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAV 341
+G +DF SDPWP+IS AKDL+ KML+ PK+RI+A+E L HPWI ++ APD PLD AV
Sbjct: 239 QGKIDFKSDPWPTISEGAKDLIYKMLDRSPKKRISAHEALCHPWIVDEHAAPDKPLDPAV 298
Query: 342 LNRLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQ 401
L+RLKQF MN+ KK+AL+VIA LSEEEI GLK++FK +DTDNSGTIT EELK GL +
Sbjct: 299 LSRLKQFSQMNKIKKMALRVIAERLSEEEIGGLKELFKMIDTDNSGTITFEELKAGLKRV 358
Query: 402 GTRLSETEVKQLMEAADADGNGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYI 461
G+ L E+E+K LM+AAD D +G IDY EF+ AT+H+N++ REE++ AF +FDKD SGYI
Sbjct: 359 GSELMESEIKSLMDAADIDNSGTIDYGEFLAATLHINKMEREENLVVAFSYFDKDGSGYI 418
Query: 462 TIEELEQALHEYNMHDGRDIKEIISEVDADNDGRINYDEFVAMMGKGNPEANTKKRRDS 520
TI+EL+QA E+ + D + ++I E+D DNDG+I++ EF AMM KG+ ++ R++
Sbjct: 419 TIDELQQACTEFGLCD-TPLDDMIKEIDLDNDGKIDFSEFTAMMKKGDGVGRSRTMRNN 476
>At5g12480 calcium-dependent protein kinase - like protein
Length = 535
Score = 548 bits (1411), Expect = e-156
Identities = 268/512 (52%), Positives = 359/512 (69%), Gaps = 15/512 (2%)
Query: 1 MGNCCSGGTE--DPEDKGENNQQNNDANGDSSATTPPPWSKPSPQPSKPSKPSAIGPVLG 58
MGNCC + + +G+ +NN + ATT + K S + G
Sbjct: 1 MGNCCGNPSSATNQSKQGKPKNKNNPFYSNEYATTD--------RSGAGFKLSVLKDPTG 52
Query: 59 RPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQ 118
D+ Y +G+E+GRG+FG+T+LCT K TG++YACK+I+K+KL DIEDVRREV+
Sbjct: 53 H---DISLQYDLGREVGRGEFGITYLCTDKETGEKYACKSISKKKLRTAVDIEDVRREVE 109
Query: 119 IMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQ 178
IM H+ PN+V L +FED +VH+VMELC GGELFDRI+A+GHYTERAAA++++TIV+
Sbjct: 110 IMKHMPKHPNVVSLKDSFEDDDAVHIVMELCEGGELFDRIVARGHYTERAAAAVMKTIVE 169
Query: 179 IVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSAYYIAP 238
+V CH GV+HRDLKPENFL +K E S LKA DFGLSVF+K G+QF +IVGS YY+AP
Sbjct: 170 VVQICHKQGVMHRDLKPENFLFANKKETSALKAIDFGLSVFFKPGEQFNEIVGSPYYMAP 229
Query: 239 DVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPS 298
+VL+R YGPE+D+WS GV+LYILLCGVPPFWAE+E G+ AI+R +DF DPWP +S S
Sbjct: 230 EVLRRNYGPEIDVWSAGVILYILLCGVPPFWAETEQGVAQAIIRSVIDFKRDPWPRVSDS 289
Query: 299 AKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQFKKVA 358
AKDLVRKML DPK+R+TA +VL H WI +AP+ L V RLKQF MN+ KK A
Sbjct: 290 AKDLVRKMLEPDPKKRLTAAQVLEHTWILNAKKAPNVSLGETVKARLKQFSVMNKLKKRA 349
Query: 359 LKVIASCLSEEEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQGTRLSETEVKQLMEAAD 418
L+VIA LS EE G+K+ F+ MD + G I +EELK GL K G ++++T+++ LMEA D
Sbjct: 350 LRVIAEHLSVEEAAGIKEAFEMMDVNKRGKINLEELKYGLQKAGQQIADTDLQILMEATD 409
Query: 419 ADGNGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYITIEELEQALHEY--NMH 476
DG+G ++Y EF+ ++H+ ++ +EH++ AF FFD++ SGYI I+EL +AL++ N
Sbjct: 410 VDGDGTLNYSEFVAVSVHLKKMANDEHLHKAFNFFDQNQSGYIEIDELREALNDELDNTS 469
Query: 477 DGRDIKEIISEVDADNDGRINYDEFVAMMGKG 508
I I+ +VD D DGRI+Y+EFVAMM G
Sbjct: 470 SEEVIAAIMQDVDTDKDGRISYEEFVAMMKAG 501
>At1g18890 calcium-dependent protein kinase (ATCDPK1)
Length = 545
Score = 547 bits (1409), Expect = e-156
Identities = 274/490 (55%), Positives = 346/490 (69%), Gaps = 19/490 (3%)
Query: 38 SKPSPQPSKPSKPSAIGPVLG---------RPMEDV---------KATYSMGKELGRGQF 79
SKPS +P KP++ + P G R ++DV Y +G+ELGRG+F
Sbjct: 15 SKPSSKPKKPNRDRKLNPFAGDFTRSPAPIRVLKDVIPMSNQTQISDKYILGRELGRGEF 74
Query: 80 GVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDK 139
G+T+LCT + T + ACK+I+KRKL DIEDVRREV IM L PN+V+L ++ED
Sbjct: 75 GITYLCTDRETHEALACKSISKRKLRTAVDIEDVRREVAIMSTLPEHPNVVKLKASYEDN 134
Query: 140 QSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFL 199
++VHLVMELC GGELFDRI+A+GHYTERAAA++ RTI ++V CHS GV+HRDLKPENFL
Sbjct: 135 ENVHLVMELCEGGELFDRIVARGHYTERAAAAVARTIAEVVMMCHSNGVMHRDLKPENFL 194
Query: 200 LLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLY 259
+K ENSPLKA DFGLSVF+K GD+F +IVGS YY+AP+VLKR YGP VD+WS GV++Y
Sbjct: 195 FANKKENSPLKAIDFGLSVFFKPGDKFTEIVGSPYYMAPEVLKRDYGPGVDVWSAGVIIY 254
Query: 260 ILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYE 319
ILLCGVPPFWAE+E G+ AILRG +DF DPWP IS SAK LV++ML+ DP +R+TA +
Sbjct: 255 ILLCGVPPFWAETEQGVALAILRGVLDFKRDPWPQISESAKSLVKQMLDPDPTKRLTAQQ 314
Query: 320 VLNHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFK 379
VL HPWI+ +AP+ PL + V +RLKQF MN+FKK L+VIA LS +E+ +K MF
Sbjct: 315 VLAHPWIQNAKKAPNVPLGDIVRSRLKQFSMMNRFKKKVLRVIAEHLSIQEVEVIKNMFS 374
Query: 380 GMDTDNSGTITIEELKQGLAKQGTRLSETEVKQLMEAADADGNGIIDYDEFITATMHMNR 439
MD D G IT ELK GL K G++L E E+K LME AD DGNG +DY EF+ +H+ +
Sbjct: 375 LMDDDKDGKITYPELKAGLQKVGSQLGEPEIKMLMEVADVDGNGFLDYGEFVAVIIHLQK 434
Query: 440 LNREEHVYTAFQFFDKDNSGYITIEELEQAL-HEYNMHDGRDIKEIISEVDADNDGRINY 498
+ +E AF FFDKD S YI ++EL +AL E D + +I+ EVD D DGRINY
Sbjct: 435 IENDELFKLAFMFFDKDGSTYIELDELREALADELGEPDASVLSDIMREVDTDKDGRINY 494
Query: 499 DEFVAMMGKG 508
DEFV MM G
Sbjct: 495 DEFVTMMKAG 504
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,810,332
Number of Sequences: 26719
Number of extensions: 603221
Number of successful extensions: 6135
Number of sequences better than 10.0: 1034
Number of HSP's better than 10.0 without gapping: 417
Number of HSP's successfully gapped in prelim test: 620
Number of HSP's that attempted gapping in prelim test: 3596
Number of HSP's gapped (non-prelim): 1576
length of query: 523
length of database: 11,318,596
effective HSP length: 104
effective length of query: 419
effective length of database: 8,539,820
effective search space: 3578184580
effective search space used: 3578184580
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC136507.13


