

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136505.11 + phase: 0
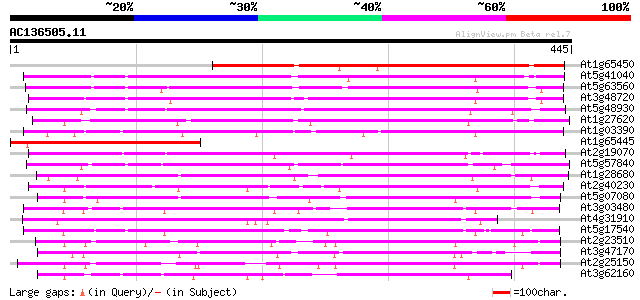
(445 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g65450 unknown protein 330 1e-90
At5g41040 N-hydroxycinnamoyl/benzoyltransferase-like protein 217 1e-56
At5g63560 acyltransferase-like protein 202 3e-52
At3g48720 unknown protein 200 2e-51
At5g48930 anthranilate N-benzoyltransferase 197 1e-50
At1g27620 putative hypersensitivity-related protein 179 4e-45
At1g03390 hypothetical protein 170 2e-42
At1g65445 unknown protein 169 3e-42
At2g19070 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 166 2e-41
At5g57840 N-hydroxycinnamoyl/benzoyltransferase 164 9e-41
At1g28680 anthranilate N-hydroxycinnamoyl/benzoyltransferase lik... 148 5e-36
At2g40230 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 147 1e-35
At5g07080 unknown protein 140 1e-33
At3g03480 putative hypersensitivity-related gene 134 1e-31
At4g31910 unknown protein 133 2e-31
At5g17540 unknown protein 127 1e-29
At2g23510 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 125 6e-29
At3g47170 hypersensitivity-related protein-like protein 122 4e-28
At2g25150 unknown protein 121 7e-28
At3g62160 unknown protein 119 4e-27
>At1g65450 unknown protein
Length = 286
Score = 330 bits (846), Expect = 1e-90
Identities = 169/287 (58%), Positives = 201/287 (69%), Gaps = 14/287 (4%)
Query: 162 IGFTTSHVTFDGLSFKNFLDNLASLATNMKLTATPFHDRQLLAARSPPHVNFPHPEMIKL 221
+G +T+H TFDGLSFK FL+NLASL L+ P +DR LL AR PP V FPH E++K
Sbjct: 1 MGISTNHTTFDGLSFKTFLENLASLLHEKPLSTPPCNDRTLLKARDPPSVAFPHHELVKF 60
Query: 222 DNLPLGIKSGVFEASSEEFDFMVFQLTYEDINNLKEKAK--GNNTSRVISFNVVAAHIWR 279
+ + VFEA+SE DF +F+L+ E I LKE+A N RV FNVV A +WR
Sbjct: 61 QDCET---TTVFEATSEHLDFKIFKLSSEQIKKLKERASETSNGNVRVTGFNVVTALVWR 117
Query: 280 CKALSKSYDPN------RSSIILYAVDIRSRLNPPLPRAYTGNAVLTAYASAKCEELKEG 333
CKALS + + R S ILYAVDIR RLNP LP +YTGNAVLTAYA KC+ L E
Sbjct: 118 CKALSVAAEEGEETNLERESTILYAVDIRGRLNPELPPSYTGNAVLTAYAKEKCKALLEE 177
Query: 334 EFSRLVEMVEEGSRRMSDEYARSIIDWGELYNGFPNGDVLVSSWWRLGFEEVEYPWGKPK 393
F R+VEMV EGS+R++DEYARS IDWGELY GFP+G+VLVSSWW+LGF EVEYPWGKPK
Sbjct: 178 PFGRIVEMVGEGSKRITDEYARSAIDWGELYKGFPHGEVLVSSWWKLGFAEVEYPWGKPK 237
Query: 394 YCCPVVYHRKDIILLFPSFNGGEGGGDGVNIIVALPRDEMEKFKSLF 440
Y CPVVYHRKDI+LLFP +G GV ++ ALP EM KF+ F
Sbjct: 238 YSCPVVYHRKDIVLLFPDI---DGDSKGVYVLAALPSKEMSKFQHWF 281
>At5g41040 N-hydroxycinnamoyl/benzoyltransferase-like protein
Length = 457
Score = 217 bits (552), Expect = 1e-56
Identities = 135/437 (30%), Positives = 233/437 (52%), Gaps = 19/437 (4%)
Query: 12 LDMKVTIHKTSMIFPSKQTENKSMFLSNIDKVLNFYVKTVHFFEANKDFPPQIVTEKLKK 71
+ ++V + +++ P +T FLSN+D+ + V+T++ F++ + + V + +KK
Sbjct: 29 IHLEVHQKEPALVKPESETRKGLYFLSNLDQNIAVIVRTIYCFKSEERGNEEAV-QVIKK 87
Query: 72 ALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLAYPN-QAFA 130
AL LV Y LAGRL ++ E +L +DC EGV FV A + K+++IGD+ P+ +
Sbjct: 88 ALSQVLVHYYPLAGRLTISPE-GKLTVDCTEEGVVFVEAEANCKMDEIGDITKPDPETLG 146
Query: 131 QFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLATNM 190
+ V++ D I ++P Q+T FKCGGF +G +H FDG+ F+++ +A +
Sbjct: 147 KLVYDVVDAKNILEIPPVTAQVTKFKCGGFVLGLCMNHCMFDGIGAMEFVNSWGQVARGL 206
Query: 191 KLTATPFHDRQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASSEEFD-FMVFQLTY 249
LT PF DR +L AR+PP + H E ++++ KS + ++E + F
Sbjct: 207 PLTTPPFSDRTILNARNPPKIENLHQEFEEIED-----KSNINSLYTKEPTLYRSFCFDP 261
Query: 250 EDINNLKEKAKGNNTSRV----ISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSRL 305
E I LK +A N+ S + SF ++A +WR + S ++ + +L+AVD R++
Sbjct: 262 EKIKKLKLQATENSESLLGNSCTSFEALSAFVWRARTKSLKMLSDQKTKLLFAVDGRAKF 321
Query: 306 NPPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWGELYN 365
P LP+ Y GN ++ + + EL E S V +V E + ++D Y RS ID+ E+
Sbjct: 322 EPQLPKGYFGNGIVLTNSICEAGELIEKPLSFAVGLVREAIKMVTDGYMRSAIDYFEVTR 381
Query: 366 GFP--NGDVLVSSWWRLGFEEVEYPWGKPKYCCPVVYHRKDIILLFPSFNGGEGGGDGVN 423
P + +L+++W RLGF ++ WG+P PV K++ L + GE +N
Sbjct: 382 ARPSLSSTLLITTWSRLGFHTTDFGWGEPILSGPVALPEKEVTLF---LSHGE-QRRSIN 437
Query: 424 IIVALPRDEMEKFKSLF 440
+++ LP M+ F+ F
Sbjct: 438 VLLGLPATAMDVFQEQF 454
>At5g63560 acyltransferase-like protein
Length = 426
Score = 202 bits (514), Expect = 3e-52
Identities = 130/436 (29%), Positives = 224/436 (50%), Gaps = 27/436 (6%)
Query: 13 DMKVTIHKTSMIFPSKQTENKSMFLSNIDKVLNFYVKTVHFFEANKDFPPQIVTEKLKKA 72
++ VT + ++ P+ +T +LSN+D+ + VKT ++F++N + E +KK+
Sbjct: 6 ELIVTRKEPVLVSPASETPKGLHYLSNLDQNIAIIVKTFYYFKSNSRSNEESY-EVIKKS 64
Query: 73 LEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQI----GDLAYPNQA 128
L + LV Y AGRL ++ E ++ +DC EGV V A + + +I ++ P +
Sbjct: 65 LSEVLVHYYPAAGRLTISPE-GKIAVDCTGEGVVVVEAEANCGIEKIKKAISEIDQP-ET 122
Query: 129 FAQFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLAT 188
+ V++ I ++P VVQ+T+FKCGGF +G +H FDG++ FL++ A A
Sbjct: 123 LEKLVYDVPGARNILEIPPVVVQVTNFKCGGFVLGLGMNHNMFDGIAAMEFLNSWAETAR 182
Query: 189 NMKLTATPFHDRQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEA-SSEEFDFMVFQL 247
+ L+ PF DR LL R+PP + FPH E L+++ SG + S E+ + F
Sbjct: 183 GLPLSVPPFLDRTLLRPRTPPKIEFPHNEFEDLEDI-----SGTGKLYSDEKLVYKSFLF 237
Query: 248 TYEDINNLKEKAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSRLNP 307
E + LK A+ +T +F + +WR + + P++ +L+A D RSR P
Sbjct: 238 GPEKLERLKIMAETRST----TFQTLTGFLWRARCQALGLKPDQRIKLLFAADGRSRFVP 293
Query: 308 PLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWGELYNGF 367
LP+ Y+GN ++ Y E+ S V +V+ ++D + RS ID+ E+
Sbjct: 294 ELPKGYSGNGIVFTYCVTTAGEVTLNPLSHSVCLVKRAVEMVNDGFMRSAIDYFEVTRAR 353
Query: 368 PN--GDVLVSSWWRLGFEEVEYPWGKPKYCCPVVYHRKDIILLFPSFNGGEGGGD--GVN 423
P+ +L++SW +L F ++ WG+P PV K++IL P G D +N
Sbjct: 354 PSLTATLLITSWAKLSFHTKDFGWGEPVVSGPVGLPEKEVILFLPC------GSDTKSIN 407
Query: 424 IIVALPRDEMEKFKSL 439
+++ LP M+ F+ +
Sbjct: 408 VLLGLPGSAMKVFQGI 423
>At3g48720 unknown protein
Length = 430
Score = 200 bits (508), Expect = 2e-51
Identities = 126/431 (29%), Positives = 225/431 (51%), Gaps = 19/431 (4%)
Query: 16 VTIHKTSMIFPSKQTENKSMFLSNIDKVLNFYVKTVHFFEANKDFPPQIVTEKLKKALED 75
VT +I P +T N +LSN+D+ + VKT++++++ Q +KK+L +
Sbjct: 9 VTRKNPELIPPVSETPNGHYYLSNLDQNIAIIVKTLYYYKSESR-TNQESYNVIKKSLSE 67
Query: 76 ALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLAYPN--QAFAQFV 133
LV Y +AGRL ++ E ++ ++C EGV V A + ++ I + N + + V
Sbjct: 68 VLVHYYPVAGRLTISPE-GKIAVNCTGEGVVVVEAEANCGIDTIKEAISENRMETLEKLV 126
Query: 134 HNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLATNMKLT 193
++ I ++P VVQ+T+FKCGGF +G SH FDG++ FL++ +A + L+
Sbjct: 127 YDVPGARNILEIPPVVVQVTNFKCGGFVLGLGMSHNMFDGVAAAEFLNSWCEMAKGLPLS 186
Query: 194 ATPFHDRQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASSEEFDFMVFQLTYEDIN 253
PF DR +L +R+PP + FPH E ++++ + +++ E+ + F E +
Sbjct: 187 VPPFLDRTILRSRNPPKIEFPHNEFDEIED--ISDTGKIYD--EEKLIYKSFLFEPEKLE 242
Query: 254 NLKEKA-KGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSRLNPPLPRA 312
LK A + NN ++V +F + +W+ + + + P++ +L+A D RSR P LP+
Sbjct: 243 KLKIMAIEENNNNKVSTFQALTGFLWKSRCEALRFKPDQRVKLLFAADGRSRFIPRLPQG 302
Query: 313 YTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWGELYNGFP--NG 370
Y GN ++ EL S V +V+ ++D + RS +D+ E+ P N
Sbjct: 303 YCGNGIVLTGLVTSSGELVGNPLSHSVGLVKRLVELVTDGFMRSAMDYFEVNRTRPSMNA 362
Query: 371 DVLVSSWWRLGFEEVEYPWGKPKYCCPVVYHRKDIILLFPSFNGGEGGGD--GVNIIVAL 428
+L++SW +L ++++ WG+P + PV +++IL PS G D +N+ + L
Sbjct: 363 TLLITSWSKLTLHKLDFGWGEPVFSGPVGLPGREVILFLPS------GDDMKSINVFLGL 416
Query: 429 PRDEMEKFKSL 439
P ME F+ L
Sbjct: 417 PTSAMEVFEEL 427
>At5g48930 anthranilate N-benzoyltransferase
Length = 433
Score = 197 bits (501), Expect = 1e-50
Identities = 133/445 (29%), Positives = 222/445 (49%), Gaps = 31/445 (6%)
Query: 14 MKVTIHKTSMIFPSKQTENKSMFLSNIDKVL-NFYVKTVHFFE---ANKDFPPQIVTEKL 69
MK+ I ++M+ P+ +T +++ SN+D V+ F+ +V+F+ A+ F PQ+ +
Sbjct: 1 MKINIRDSTMVRPATETPITNLWNSNVDLVIPRFHTPSVYFYRPTGASNFFDPQV----M 56
Query: 70 KKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLAYPNQAF 129
K+AL ALV + +AGRLK + + R+EIDCN GV FVVA + ++ GD A P
Sbjct: 57 KEALSKALVPFYPMAGRLKRDDD-GRIEIDCNGAGVLFVVADTPSVIDDFGDFA-PTLNL 114
Query: 130 AQFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLATN 189
Q + I PL V+Q+T FKCGG ++G H DG S +F++ + +A
Sbjct: 115 RQLIPEVDHSAGIHSFPLLVLQVTFFKCGGASLGVGMQHHAADGFSGLHFINTWSDMARG 174
Query: 190 MKLTATPFHDRQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASSEEFDFMVFQLTY 249
+ LT PF DR LL AR PP F H E ++ + + ++ E +F+LT
Sbjct: 175 LDLTIPPFIDRTLLRARDPPQPAFHHVEYQPAPSMKIPLDPS--KSGPENTTVSIFKLTR 232
Query: 250 EDINNLKEKAK-GNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSRLNPP 308
+ + LK K+K NT S+ ++A H+WR ++ ++ + + A D RSRL P
Sbjct: 233 DQLVALKAKSKEDGNTVSYSSYEMLAGHVWRSVGKARGLPNDQETKLYIATDGRSRLRPQ 292
Query: 309 LPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWGELY---- 364
LP Y GN + TA A +L + + RM D Y RS +D+ E+
Sbjct: 293 LPPGYFGNVIFTATPLAVAGDLLSKPTWYAAGQIHDFLVRMDDNYLRSALDYLEMQPDLS 352
Query: 365 ------NGFPNGDVLVSSWWRLGFEEVEYPWGKPKYCCP--VVYHRKDIILLFPSFNGGE 416
+ + ++ ++SW RL + ++ WG+P + P + Y +L P+ +G
Sbjct: 353 ALVRGAHTYKCPNLGITSWVRLPIYDADFGWGRPIFMGPGGIPYEGLSFVLPSPTNDG-- 410
Query: 417 GGGDGVNIIVALPRDEMEKFKSLFY 441
+++ +AL + M+ F+ +
Sbjct: 411 ----SLSVAIALQSEHMKLFEKFLF 431
>At1g27620 putative hypersensitivity-related protein
Length = 442
Score = 179 bits (453), Expect = 4e-45
Identities = 129/436 (29%), Positives = 210/436 (47%), Gaps = 28/436 (6%)
Query: 19 HKTSMIFPSKQTENKSMFLSNIDK--VLNFYVKTVHFFEANKDFPPQIVTEKLKKALEDA 76
++ ++I P T N S++LSN+D L F +K ++ F+ + I LK +L
Sbjct: 12 NQPTLITPLSPTPNHSLYLSNLDDHHFLRFSIKYLYLFQKS------ISPLTLKDSLSRV 65
Query: 77 LVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLA-YPNQAFAQ--FV 133
LV Y AGR++++ E ++LE+DCN EG F A + L+ PN+++ + F
Sbjct: 66 LVDYYPFAGRIRVSDEGSKLEVDCNGEGAVFAEAFMDITCQDFVQLSPKPNKSWRKLLFK 125
Query: 134 HNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLATNMK-L 192
A+ FL D+P V+Q+T +CGG + +H DG+ FL A T+ L
Sbjct: 126 VQAQSFL---DIPPLVIQVTYLRCGGMILCTAINHCLCDGIGTSQFLHAWAHATTSQAHL 182
Query: 193 TATPFHDRQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASS--EEFDFMVFQLTYE 250
PFH R +L R+PP V HP + + KS F+ S + LT+
Sbjct: 183 PTRPFHSRHVLDPRNPPRVTHSHPGFTRTTTVD---KSSTFDISKYLQSQPLAPATLTFN 239
Query: 251 DINNLKEKAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSRLNPPLP 310
+ L+ K + + +F +AA+ WR A S +L++V++R RL P LP
Sbjct: 240 QSHLLRLKKTCAPSLKCTTFEALAANTWRSWAQSLDLPMTMLVKLLFSVNMRKRLTPELP 299
Query: 311 RAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWGE--LYNGFP 368
+ Y GN + A A +K ++L G V+ ++E R++DEY RS ID E
Sbjct: 300 QGYYGNGFVLACAESKVQDLVNGNIYHAVKSIQEAKSRITDEYVRSTIDLLEDKTVKTDV 359
Query: 369 NGDVLVSSWWRLGFEEVEYPWGKPKYCCPVVYHRKDIILLFPSFNGGEGGGDGVNIIVAL 428
+ +++S W +LG EE++ GKP Y P+ DI LF D + + ++L
Sbjct: 360 SCSLVISQWAKLGLEELDLGGGKPMYMGPLT---SDIYCLFLPV---ASDNDAIRVQMSL 413
Query: 429 PRDEMEKFKSLFYTFL 444
P + +++ + FL
Sbjct: 414 PEEVVKRLEYCMVKFL 429
>At1g03390 hypothetical protein
Length = 461
Score = 170 bits (430), Expect = 2e-42
Identities = 131/450 (29%), Positives = 226/450 (50%), Gaps = 30/450 (6%)
Query: 12 LDMKVTIHKTSMIFPSK-----QTENKSMFLSNIDKVLNFYVKT--VHFFEANKDFPPQI 64
L + VTI++ ++ PS Q+ + S++LSN+D ++ V T V+F+ + +
Sbjct: 13 LHIPVTINQQFLVHPSSPTPANQSPHHSLYLSNLDDIIGARVFTPSVYFYPSTNN-RESF 71
Query: 65 VTEKLKKALEDALVAYDFLAGRLKMNTETNRLEIDCNAE-GVGFVVASSEYKLNQIGDLA 123
V ++L+ AL + LV Y L+GRL+ E +LE+ E GV V A+S L +GDL
Sbjct: 72 VLKRLQDALSEVLVPYYPLSGRLR-EVENGKLEVFFGEEQGVLMVSANSSMDLADLGDLT 130
Query: 124 YPNQAFAQFVHN--AKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLD 181
PN A+ + ++ KI ++PL + Q+T F CGGF++G H DG FL
Sbjct: 131 VPNPAWLPLIFRNPGEEAYKILEMPLLIAQVTFFTCGGFSLGIRLCHCICDGFGAMQFLG 190
Query: 182 NLASLATNMKLTA--TPFHDRQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASSEE 239
+ A+ A KL A P DR+ R+PP V +PH E + ++ + + +++ +
Sbjct: 191 SWAATAKTGKLIADPEPVWDRETFKPRNPPMVKYPHHEYLPIEERS-NLTNSLWDTKPLQ 249
Query: 240 FDFMVFQLTYEDINNLKEKAKGNNTSRVIS-FNVVAAHIWR--CKALS-KSYDPNRSSII 295
++++ E +K A+G + + V S F+ +AAHIWR KAL K D N +
Sbjct: 250 ---KCYRISKEFQCRVKSIAQGEDPTLVCSTFDAMAAHIWRSWVKALDVKPLDYNLR--L 304
Query: 296 LYAVDIRSRLNP-PLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYA 354
++V++R+RL L + + GN V A A + E L S+ +V++ R+S++Y
Sbjct: 305 TFSVNVRTRLETLKLRKGFYGNVVCLACAMSSVESLINDSLSKTTRLVQDARLRVSEDYL 364
Query: 355 RSIIDWGELYNGFP---NGDVLVSSWWRLG-FEEVEYPWGKPKYCCPV-VYHRKDIILLF 409
RS++D+ ++ G + ++ W R +E ++ WGKP Y P+ + + +L
Sbjct: 365 RSMVDYVDVKRPKRLEFGGKLTITQWTRFEMYETADFGWGKPVYAGPIDLRPTPQVCVLL 424
Query: 410 PSFNGGEGGGDGVNIIVALPRDEMEKFKSL 439
P G + + + LP + F L
Sbjct: 425 PQGGVESGNDQSMVVCLCLPPTAVHTFTRL 454
>At1g65445 unknown protein
Length = 166
Score = 169 bits (428), Expect = 3e-42
Identities = 84/154 (54%), Positives = 117/154 (75%), Gaps = 3/154 (1%)
Query: 1 MGTTNKETKPLL--DMKVTIHKTSMIFPSKQT-ENKSMFLSNIDKVLNFYVKTVHFFEAN 57
MG++ +E+ PLL D+KVTI ++++IFPS++T E KSMFLSN+D++LNF V+TVHFF N
Sbjct: 1 MGSSYQESPPLLLEDLKVTIKESTLIFPSEETSERKSMFLSNVDQILNFDVQTVHFFRPN 60
Query: 58 KDFPPQIVTEKLKKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLN 117
K+FPP++V+EKL+KAL + AY+FLAGRL+++ + RL++DCN G GFV A+S+Y L
Sbjct: 61 KEFPPEMVSEKLRKALVKLMDAYEFLAGRLRVDPSSGRLDVDCNGAGAGFVTAASDYTLE 120
Query: 118 QIGDLAYPNQAFAQFVHNAKDFLKIGDLPLCVVQ 151
++GDL YPN AFAQ V + L D PL V Q
Sbjct: 121 ELGDLVYPNPAFAQLVTSQLQSLPKDDQPLFVFQ 154
>At2g19070 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 451
Score = 166 bits (421), Expect = 2e-41
Identities = 130/454 (28%), Positives = 204/454 (44%), Gaps = 37/454 (8%)
Query: 16 VTIHKTSMIFPSKQTENKSMFLSNIDKVLNF-YVKTVHFFEANKDFPPQIVTEKLKKALE 74
+T K+ I P++ T + L+ D+V ++ T++F++ + V E LK +L
Sbjct: 4 ITFRKSYTIVPAEPTWSGRFPLAEWDQVGTITHIPTLYFYDKPSESFQGNVVEILKTSLS 63
Query: 75 DALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLAYPNQAFAQFVH 134
LV + +AGRL+ R E++CNAEGV F+ A SE KL+ D + P F +
Sbjct: 64 RVLVHFYPMAGRLRW-LPRGRFELNCNAEGVEFIEAESEGKLSDFKDFS-PTPEFENLMP 121
Query: 135 NAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLATNMKLTA 194
I +PL + Q+T FKCGG ++ SH DG S + + LA L
Sbjct: 122 QVNYKNPIETIPLFLAQVTKFKCGGISLSVNVSHAIVDGQSALHLISEWGRLARGEPLET 181
Query: 195 TPFHDRQLLAARSP--PHVNFPHPEMIKLDNLPLGI-KSGVFEASSEEFDFMVFQLTYED 251
PF DR++L A P P V+ P + + D P I ++ E ++ ++ L+
Sbjct: 182 VPFLDRKILWAGEPLPPFVSPPKFDHKEFDQPPFLIGETDNVEERKKKTIVVMLPLSTSQ 241
Query: 252 INNLKEKAKGNNTSRVIS----FNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSRLNP 307
+ L+ KA G+ S + V H+WRC ++ + P + + + +D RSR+ P
Sbjct: 242 LQKLRSKANGSKHSDPAKGFTRYETVTGHVWRCACKARGHSPEQPTALGICIDTRSRMEP 301
Query: 308 PLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDW------- 360
PLPR Y GNA L A++ EL E ++ + + +++EY I++
Sbjct: 302 PLPRGYFGNATLDVVAASTSGELISNELGFAASLISKAIKNVTNEYVMIGIEYLKNQKDL 361
Query: 361 -------------GELYNGFPNGDVLVSSWWRLGFEEVEYPWGKPKYCCPVVYHRKDIIL 407
G Y G PN V+ SW L +++ WGK Y P + L
Sbjct: 362 KKFQDLHALGSTEGPFY-GNPNLGVV--SWLTLPMYGLDFGWGKEFYTGPGTHDFDGDSL 418
Query: 408 LFPSFNGGEGGGDGVNIIVALPRDEMEKFKSLFY 441
+ P N E G V + L ME FK FY
Sbjct: 419 ILPDQN--EDG--SVILATCLQVAHMEAFKKHFY 448
>At5g57840 N-hydroxycinnamoyl/benzoyltransferase
Length = 443
Score = 164 bits (415), Expect = 9e-41
Identities = 133/455 (29%), Positives = 217/455 (47%), Gaps = 41/455 (9%)
Query: 14 MKVTIHKTSMIFPSKQTENKSMFLSNIDKV-LNFYVKTVHFFE--ANKDFPPQIVTEKLK 70
MK+ + + +++ P+++T +++LSN+D + + ++ T++F++ ++ D P T+ L
Sbjct: 1 MKIRVKQATIVKPAEETPTHNLWLSNLDLIQVRLHMGTLYFYKPCSSSDRPN---TQSLI 57
Query: 71 KALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLAYPNQAFA 130
+AL LV + AGRL+ NT RLE+ CN EGV FV A S+ + IG L + +
Sbjct: 58 EALSKVLVFFYPAAGRLQKNTN-GRLEVQCNGEGVLFVEAESDSTVQDIG-LLTQSLDLS 115
Query: 131 QFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLATNM 190
Q V I PL + Q+T FKCG +G + H + S ++ + A +
Sbjct: 116 QLVPTVDYAGDISSYPLLLFQVTYFKCGTICVGSSIHHTFGEATSLGYIMEAWSLTARGL 175
Query: 191 KLTATPFHDRQLLAARSPPHVNFPHPEMIK--LDNLPLGIKSGVFEASSE-EFDFMVFQL 247
+ TPF DR +L AR+PP FPH E N P+ KS + ++ E + +L
Sbjct: 176 LVKLTPFLDRTVLHARNPPSSVFPHTEYQSPPFHNHPM--KSLAYRSNPESDSAIATLKL 233
Query: 248 TYEDINNLKEKAKGNNTSRVISFNVVAAHIWRCKAL-SKSYDPNRSSIILYAVDIRSRLN 306
T + LK +A+ + S+ ++ V+ AH WRC + ++ S+ + +D R RL
Sbjct: 234 TRLQLKALKARAEIAD-SKFSTYEVLVAHTWRCASFANEDLSEEHSTRLHIIIDGRPRLQ 292
Query: 307 PPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWGELYNG 366
P LP+ Y GN + A + FS VE V R+M +EY RS ID+ E +
Sbjct: 293 PKLPQGYIGNTLFHARPVSPLGAFHRESFSETVERVHREIRKMDNEYLRSAIDYLERH-- 350
Query: 367 FPNGDVLVSSWWRLGFE----------------EVEYPWGKPKYCCPVVYHRKDIILLFP 410
P+ D LV F E+++ WGK Y R +
Sbjct: 351 -PDLDQLVPGEGNTIFSCAANFCIVSLIKQTAYEMDFGWGK------AYYKRASHLNEGK 403
Query: 411 SFNGGEGGGDG-VNIIVALPRDEMEKFKSLFYTFL 444
F G +G + + + L + ++ KF+ LFY FL
Sbjct: 404 GFVTGNPDEEGSMMLTMCLKKTQLSKFRKLFYEFL 438
>At1g28680 anthranilate N-hydroxycinnamoyl/benzoyltransferase like
protein
Length = 451
Score = 148 bits (374), Expect = 5e-36
Identities = 119/444 (26%), Positives = 202/444 (44%), Gaps = 32/444 (7%)
Query: 22 SMIFPSKQ--TENKSMFLSNIDKVLNFYVKTVH---FFEANKDFPPQIVTEKLKKALEDA 76
+++ PS Q + ++++ LS++D N +V + + ++ + + + +L A
Sbjct: 10 ALVQPSHQPLSNDQTLSLSHLDNDNNLHVSFRYLRVYSSSSSTVAGESPSAVVSASLATA 69
Query: 77 LVAYDFLAGRLKMNTETNRLEIDCNA-EGVGFVVASSEYKLNQIGDLAYPNQAFAQFVHN 135
LV Y LAG L+ + NR E+ C+A + V V A+ L +G L P+ F + +
Sbjct: 70 LVHYYPLAGSLRRSASDNRFELLCSAGQSVPLVNATVNCTLESVGYLDGPDPGFVERLV- 128
Query: 136 AKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLATNM-KLTA 194
+ G + C++Q+T F+CGG+ +G + H DGL F + +A LA K++
Sbjct: 129 PDPTREEGMVNPCILQVTMFQCGGWVLGASIHHAICDGLGASLFFNAMAELARGATKISI 188
Query: 195 TPFHDRQ-LLAARSPPHVNFPHPEMIKLDNL--PLGIKSGVFEASSEEFDFMVFQLTYED 251
P DR+ LL R P V P + + LD P G G + F +T +
Sbjct: 189 EPVWDRERLLGPREKPWVGAPVRDFLSLDKDFDPYGQAIGDVKRDC-------FFVTDDS 241
Query: 252 INNLKEKAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSRLNPPLPR 311
++ LK + + +F + A+IWR K + + + +Y+++IR +NPPLP+
Sbjct: 242 LDQLKAQLLEKSGLNFTTFEALGAYIWRAKVRAAKTEEKENVKFVYSINIRRLMNPPLPK 301
Query: 312 AYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWGELYN----GF 367
Y GN + YA K EL E + E++++ SDEY RS ID+ EL++
Sbjct: 302 GYWGNGCVPMYAQIKAGELIEQPIWKTAELIKQSKSNTSDEYVRSFIDFQELHHKDGINA 361
Query: 368 PNGDVLVSSWWRLGFEEVEYPWGKPKYCCPVVYHRKDIILLFPSF--------NGGEGGG 419
G + W LG +++ WG P P+ K + + P F G
Sbjct: 362 GTGVTGFTDWRYLGHSTIDFGWGGPVTVLPL--SNKLLGSMEPCFFLPYSTDAAAGSKKD 419
Query: 420 DGVNIIVALPRDEMEKFKSLFYTF 443
G ++V L M +FK F
Sbjct: 420 SGFKVLVNLRESAMPEFKEAMDKF 443
>At2g40230 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 433
Score = 147 bits (371), Expect = 1e-35
Identities = 124/437 (28%), Positives = 211/437 (47%), Gaps = 27/437 (6%)
Query: 16 VTIHKTSMIFPSKQTENKSMFLSNIDK--VLNFYVKTVHFFEANKDFPPQIVTEKLKKAL 73
V + + ++I PS QT + + LS +D L F ++ + + D P ++ +LK AL
Sbjct: 5 VHVKEATVITPSDQTPSSVLSLSALDSQLFLRFTIEYLLVYPPVSD-PEYSLSGRLKSAL 63
Query: 74 EDALVAYDFLAGRLKMNTETNR-LEIDCNAEGVGFVVASSEYKLNQIGDLAYPNQAFAQF 132
ALV Y +GR++ + LE++C +G F+ A S+ + D P + +
Sbjct: 64 SRALVPYFPFSGRVREKPDGGGGLEVNCRGQGALFLEAVSD--ILTCLDFQKPPRHVTSW 121
Query: 133 --VHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLA--- 187
+ + + P VVQLT + GG A+ +H DG+ FL A L+
Sbjct: 122 RKLLSLHVIDVLAGAPPLVVQLTWLRDGGAALAVGVNHCVSDGIGSAEFLTLFAELSKDS 181
Query: 188 -TNMKLTATPFHDRQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASSEEFDFMVFQ 246
+ +L DRQLL SP + HPE ++ +L + F A +VF+
Sbjct: 182 LSQTELKRKHLWDRQLLMP-SPIRDSLSHPEFNRVPDLCGFVNR--FNAERLVPTSVVFE 238
Query: 247 LTYEDINNLKEKAK--GNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSR 304
+ +N LK+ A G S+ SF V++AH+WR A S + N+ +L++V+IR R
Sbjct: 239 R--QKLNELKKLASRLGEFNSKHTSFEVLSAHVWRSWARSLNLPSNQVLKLLFSVNIRDR 296
Query: 305 LNPPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWGELY 364
+ P LP + GNA + A ++L E S +V++ R+ DEY RS+++
Sbjct: 297 VKPSLPSGFYGNAFVVGCAQTTVKDLTEKGLSYATMLVKQAKERVGDEYVRSVVEAVSKE 356
Query: 365 NGFPN--GDVLVSSWWRLGFEEVEYPWGKPKYCCPVVYHRKDIILLFPSFNGGEGGGDGV 422
P+ G +++S W RLG E++++ GKP + V R ++L P N D V
Sbjct: 357 RASPDSVGVLILSQWSRLGLEKLDFGLGKPVHVGSVCCDRYCLLLPIPEQN------DAV 410
Query: 423 NIIVALPRDEMEKFKSL 439
++VA+P ++ +++L
Sbjct: 411 KVMVAVPSSSVDTYENL 427
>At5g07080 unknown protein
Length = 450
Score = 140 bits (353), Expect = 1e-33
Identities = 122/440 (27%), Positives = 194/440 (43%), Gaps = 43/440 (9%)
Query: 23 MIFPSKQTENKSMFLSNIDK--VLNFYVKTVHFFEANKDFPPQIVTEK---LKKALEDAL 77
++ PSK T + S+ LS +D L KT++ + P V + ++AL DAL
Sbjct: 21 IVKPSKATPDVSLSLSTLDNDPYLETLAKTIYVYAPPS--PNDHVHDPASLFQQALSDAL 78
Query: 78 VAYDFLAGRLKMNTETNRLEIDCN-AEGVGFVVASSEYKLNQIGDLAYPNQAFAQFVHNA 136
V Y LAG+L + +RLE+ C+ AEGV FV A+++ L+ + L + Q V
Sbjct: 79 VYYYPLAGKLHRGSHDHRLELRCSPAEGVPFVRATADCTLSSLNYLKDMDTDLYQLVP-C 137
Query: 137 KDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLATNM-KLTAT 195
GD +Q+T F CGG + SH DG F L LA + +
Sbjct: 138 DVAAPSGDYNPLALQITLFACGGLTLATALSHSLCDGFGASQFFKTLTELAAGKTQPSII 197
Query: 196 PFHDRQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEAS--------SEEFDFMVFQL 247
P DR L + NF + ++ P + G +S S + + +
Sbjct: 198 PVWDRHRLTSN-----NFSLNDQVEEGQAPKLVDFGEACSSAATSPYTPSNDMVCEILNV 252
Query: 248 TYEDINNLKEKAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSRLNP 307
T EDI LKEK G V + ++AAH+WR + + P+ +S+ AV IR + P
Sbjct: 253 TSEDITQLKEKVAGV----VTTLEILAAHVWRARCRALKLSPDGTSLFGMAVGIRRIVEP 308
Query: 308 PLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDE--------YARSIID 359
PLP Y GNA + A + K EL S +V++++E + ++ +
Sbjct: 309 PLPEGYYGNAFVKANVAMKAGELSNSPLSHVVQLIKEAKKAAQEKRYVLEQLRETEKTLK 368
Query: 360 WGELYNGFPNGDVLVSSWWRLG-FEEVEYPWGKPKYCCPVV-YHRKDIILLFPSFNGGEG 417
G +L++ W +LG +E+++ +G P+V + DI + P G
Sbjct: 369 MNVACEGGKGAFMLLTDWRQLGLLDEIDFGYGGSVNIIPLVPKYLPDICIFLPRKQG--- 425
Query: 418 GGDGVNIIVALPRDEMEKFK 437
GV ++V LP+ M FK
Sbjct: 426 ---GVRVLVTLPKSVMVNFK 442
>At3g03480 putative hypersensitivity-related gene
Length = 454
Score = 134 bits (337), Expect = 1e-31
Identities = 119/450 (26%), Positives = 202/450 (44%), Gaps = 48/450 (10%)
Query: 12 LDMKVTIHKTSMIFPSKQTENKSMFLSNID--KVLNFYVKTVHFFEAN--KDFPPQIVTE 67
L KV + ++ P+K T + LS+ID + L F + + F+ N D P +
Sbjct: 15 LSFKVHRQQRELVTPAKPTPRELKPLSDIDDQQGLRFQIPVIFFYRPNLSSDLDP---VQ 71
Query: 68 KLKKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGD---LAY 124
+KKAL DALV Y AGRL+ +L +DC EGV F+ A ++ L ++ + L
Sbjct: 72 VIKKALADALVYYYPFAGRLR-ELSNRKLAVDCTGEGVLFIEAEADVALAELEEADALLP 130
Query: 125 PNQAFAQFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLA 184
P + + + + + + PL +VQ+T KC GF +H DG FL +L
Sbjct: 131 PFPFLEELLFDVEGSSDVLNTPLLLVQVTRLKCCGFIFALRFNHTMTDGAGLSLFLKSLC 190
Query: 185 SLATNMKL-TATPFHDRQLLAARSP----PHVNFPHPEMIKLDNLPLG--IKSGVFEASS 237
LA + + P +R LL + H + + + + +D + G + S F +
Sbjct: 191 ELACGLHAPSVPPVWNRHLLTVSASEARVTHTHREYDDQVGIDVVATGHPLVSRSFFFRA 250
Query: 238 EEFDFMVFQLTYEDINNLKEKAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILY 297
EE + +L D++N SF +++ +WRC+ ++ + DPN +
Sbjct: 251 EEIS-AIRKLLPPDLHN-------------TSFEALSSFLWRCRTIALNPDPNTEMRLTC 296
Query: 298 AVDIRSRL-NPPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARS 356
++ RS+L NPPL Y GN + A A +L E + +++E ++++Y RS
Sbjct: 297 IINSRSKLRNPPLEPGYYGNVFVIPAAIATARDLIEKPLEFALRLIQETKSSVTEDYVRS 356
Query: 357 IIDWGELYNGFP----NGDVLVSSWWRLGFEEVEY-PWGKPKYCCPVVYHRKDIILLFPS 411
+ G P +G+ ++S ++++ PWGKP Y K I LFP
Sbjct: 357 VTAL-MATRGRPMFVASGNYIISDLRHFDLGKIDFGPWGKPVYGGTA----KAGIALFPG 411
Query: 412 FN-----GGEGGGDGVNIIVALPRDEMEKF 436
+ + G G + ++LP ME F
Sbjct: 412 VSFYVPFKNKKGETGTVVAISLPVRAMETF 441
>At4g31910 unknown protein
Length = 458
Score = 133 bits (334), Expect = 2e-31
Identities = 114/407 (28%), Positives = 187/407 (45%), Gaps = 36/407 (8%)
Query: 11 LLDMKVTIHKTSMIFPSKQTENKSMF--LSNIDKVLNFYVKTVHFFEANKDFPPQIVTEK 68
L+ ++ I + ++P Q +K LSN+D+ + +V F++ V
Sbjct: 4 LMATRIDIIQKLNVYPRFQNHDKKKLITLSNLDRQCPLLMYSVFFYKNTTTRDFDSVFSN 63
Query: 69 LKKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVA-SSEYKLNQIGDLAYPNQ 127
LK LE+ + + AGRL ++ +L I CN G V A ++ KL+++GDL N+
Sbjct: 64 LKLGLEETMSVWYPAAGRLGLDGGGCKLNIRCNDGGAVMVEAVATGVKLSELGDLTQYNE 123
Query: 128 AFAQFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLA 187
+ V+ +PL V Q+T F CGG++IG TSH FDG+S F+ AS +
Sbjct: 124 FYENLVYKPSLDGDFSVMPLVVAQVTRFACGGYSIGIGTSHSLFDGISAYEFIHAWASNS 183
Query: 188 -----TNMKLT-------ATPFHDRQLL-----AARSPPHVNFPHP-EMIKLDNLPLGIK 229
+N K+T P HDR+ L A R H ++IK + +
Sbjct: 184 HIHNKSNSKITNKKEDVVIKPVHDRRNLLVNRDAVRETNAAAICHLYQLIKQAMMTYQEQ 243
Query: 230 SGVFEASSEEFDFMVFQLTYEDINNLKEKA-KGNNTSRVISFNVVAAHIWRCKALSKSYD 288
+ E F F+L + I ++K+K+ +G S SF +AAH+W+ + +
Sbjct: 244 NRNLELPDSGFVIKTFELNGDAIESMKKKSLEGFMCS---SFEFLAAHLWKARTRALGLR 300
Query: 289 PNRSSIILYAVDIRSRLNPPLPRAYTGNA-VLTAYASAKCEELKEGEFSRLVEMVEEGSR 347
+ + +AVDIR R PLP ++GNA VL + AS E L+E +V + E +
Sbjct: 301 RDAMVCLQFAVDIRKRTETPLPEGFSGNAYVLASVASTARELLEELTLESIVNKIREAKK 360
Query: 348 RMSDEYARSIIDWGELYNGFPNGDV-------LVSSWWRLGFEEVEY 387
+ Y S + E G +G++ L+S W ++ F V +
Sbjct: 361 SIDQGYINSYM---EALGGSNDGNLPPLKELTLISDWTKMPFHNVGF 404
>At5g17540 unknown protein
Length = 461
Score = 127 bits (319), Expect = 1e-29
Identities = 118/442 (26%), Positives = 203/442 (45%), Gaps = 31/442 (7%)
Query: 12 LDMKVTIHKTSMIFPSKQTENKSMFLSNID--KVLNFYVKTVHFFEANKDFPPQIVTEKL 69
L K+ K ++ P+K T + LS+ID + L F++ T+ F+ N V +
Sbjct: 5 LTFKIYRQKPELVSPAKPTPRELKPLSDIDDQEGLRFHIPTIFFYRHNPTTNSDPVAV-I 63
Query: 70 KKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGD---LAYPN 126
++AL + LV Y AGRL+ +L +DC EGV F+ A ++ L + + L P
Sbjct: 64 RRALAETLVYYYPFAGRLREGPN-RKLAVDCTGEGVLFIEADADVTLVEFEEKDALKPPF 122
Query: 127 QAFAQFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASL 186
F + + N + ++ + PL ++Q+T KCGGF +H D FL +
Sbjct: 123 PCFEELLFNVEGSCEMLNTPLMLMQVTRLKCGGFIFAVRINHAMSDAGGLTLFLKTMCEF 182
Query: 187 ATNMKL-TATPFHDRQLLAARSPPHVNFPHPEMIKLDNLP-LGIKSGVFEASSEEFDFMV 244
T P +R LL+AR V H E D +P +G + G S + +
Sbjct: 183 VRGYHAPTVAPVWERHLLSARVLLRVTHAHREY---DEMPAIGTELG-----SRRDNLVG 234
Query: 245 FQLTYE--DINNLKEKAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIR 302
L + +++ ++ N + + ++ + +WR + ++ D ++ ++ V+ R
Sbjct: 235 RSLFFGPCEMSAIRRLLPPNLVNSSTNMEMLTSFLWRYRTIALRPDQDKEMRLILIVNAR 294
Query: 303 SRL-NPPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWG 361
S+L NPPLPR Y GNA A A EL + + +++E +++EY RS+ D
Sbjct: 295 SKLKNPPLPRGYYGNAFAFPVAIATANELTKKPLEFALRLIKEAKSSVTEEYMRSLADL- 353
Query: 362 ELYNGFP----NGDVLVSSWWRLGFEEVEYP-WGKPKYCCPVVYHRKDI--ILLFPSFNG 414
+ G P +G LVS R+ F ++++ WGKP Y +D+ + SF
Sbjct: 354 MVIKGRPSFSSDGAYLVSD-VRI-FADIDFGIWGKPVYGGIGTAGVEDLPGASFYVSFE- 410
Query: 415 GEGGGDGVNIIVALPRDEMEKF 436
G G+ + V LP M++F
Sbjct: 411 KRNGEIGIVVPVCLPEKAMQRF 432
>At2g23510 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 451
Score = 125 bits (313), Expect = 6e-29
Identities = 119/452 (26%), Positives = 200/452 (43%), Gaps = 60/452 (13%)
Query: 21 TSMIFPSKQTENKSMFLSNIDK--VLNFYVKTVHFFEANK----DFPPQIVTEKLKKALE 74
T M+ PSK +++ LS +D K + F+A D P+ + L++AL
Sbjct: 18 TEMVKPSKHIPQQTLNLSTLDNDPYNEVIYKACYVFKAKNVADDDNRPEAL---LREALS 74
Query: 75 DALVAYDFLAGRLKMNTETNRLEIDCNAEGVG--FVVASSEYKLNQIGDLAYPNQAFAQF 132
D L Y L+G LK +L++ C +G G F VA++ +L+ + +L +
Sbjct: 75 DLLGYYYPLSGSLKRQESDRKLQLSCGGDGGGVPFTVATANVELSSLKNLENIDS----- 129
Query: 133 VHNAKDFLKIGDLPL-----CVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLA 187
A +FL + + + +Q+T F+CGGF +G SH DG + + L LA
Sbjct: 130 -DTALNFLPVLHVDIDGYRPFALQVTKFECGGFILGMAMSHAMCDGYGEGHIMCALTDLA 188
Query: 188 TNMKLT-ATPFHDRQLLAAR----SPPHVNFPHPEMIKLDNLPLGIKSGVFEASSEEFDF 242
K TP +R+ L + PP V P + LP D+
Sbjct: 189 GGKKKPMVTPIWERERLVGKPEDDQPPFV--PGDDTAASPYLPTD-------------DW 233
Query: 243 MVFQLTY--EDINNLKE---KAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILY 297
+ ++T + I LKE K + + +F V+ A++W+ + + + D + +++
Sbjct: 234 VTEKITIRADSIRRLKEATLKEYDFSNETITTFEVIGAYLWKSRVKALNLDRDGVTVLGL 293
Query: 298 AVDIRSRLNPPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDE----- 352
+V IR+ ++PPLP Y GNA + Y E++E S +V++++E R D+
Sbjct: 294 SVGIRNVVDPPLPDGYYGNAYIDMYVPLTAREVEEFTISDIVKLIKEAKRNAHDKDYLQE 353
Query: 353 ---YARSIIDWGELYNGFPNGDVLVSSWWRLG-FEEVEYPWGKPKYCCPVV---YHRKDI 405
II G +G ++ W +G F +++ W +P PVV R
Sbjct: 354 ELANTEKIIKMNLTIKGKKDGLFCLTDWRNIGIFGSMDFGWDEPVNIVPVVPSETARTVN 413
Query: 406 ILLFPSFNGGEGGGDGVNIIVALPRDEMEKFK 437
+ + PS + G GV I+V LPR M KFK
Sbjct: 414 MFMRPSRLESDMVG-GVQIVVTLPRIAMVKFK 444
>At3g47170 hypersensitivity-related protein-like protein
Length = 447
Score = 122 bits (306), Expect = 4e-28
Identities = 116/454 (25%), Positives = 193/454 (41%), Gaps = 73/454 (16%)
Query: 23 MIFPSKQTENKSMFLSNIDKVLNFYV--KTVHFFEAN---KDFPPQIVTEKLKKALEDAL 77
++ PSK T + ++ LS ID N V + F N +D ++ AL +AL
Sbjct: 19 LVKPSKPTPDVTLSLSTIDNDPNNEVILDILCVFSPNPYVRDHTNYHPASFIQLALSNAL 78
Query: 78 VAYDFLAGRLKMNTETNRLEIDCN-AEGVGFVVASSEYKLNQIGDLAYPNQAFAQFV--- 133
V Y LAG+L T L++DC +GV + A++ L+ + L N A +
Sbjct: 79 VYYYPLAGKLHRRTSDQTLQLDCTQGDGVPLIKATASCTLSSLNYLESGNHLEATYQLVP 138
Query: 134 -HNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLATNM-K 191
H+ +G PL + Q+T F CGG IG SH DG+ F + LA +
Sbjct: 139 SHDTLKGCNLGYRPLAL-QITKFTCGGMTIGSVHSHTVCDGIGMAQFSQAILELAAGRAQ 197
Query: 192 LTATPFHDRQLLAARSPPHV-----NFPHPEMIKLDNLPLGIKSGVFEASSEEFDFMVFQ 246
T P DR ++ + + + +P+++ ++ + +E+ +
Sbjct: 198 PTVIPVWDRHMITSNQISTLCKLGNDKNNPKLVDVEK-----DCSSPDTPTEDMVREILN 252
Query: 247 LTYEDINNLK-----------EKAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSII 295
+T EDI LK EK K ++ + V+AA++WR + + +P+ + +
Sbjct: 253 ITSEDITKLKNIIIEDENLTNEKEKN---MKITTVEVLAAYVWRARCRAMKLNPDTITDL 309
Query: 296 LYAVDIRSRLNPPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSD---- 351
+ +V IRS + PPLP Y GNA A + +EL + SRLV ++++ R D
Sbjct: 310 VISVSIRSSIEPPLPEGYYGNAFTHASVALTAKELSKTPISRLVRLIKDAKRAALDNGYV 369
Query: 352 -EYARSI-------IDWGELYNGFPNGDVLVSSWWRLGFEEVEYPWGKPKYCCPVVYHRK 403
E R + + E++ G ++++ W LG + + WG K
Sbjct: 370 CEQLREMENTMKLKLASKEIHGGV---FMMLTDWRHLGLD--QDVWGASK---------- 414
Query: 404 DIILLFPSFNGGEGGGDGVNIIVALPRDEMEKFK 437
G GV ++ LPRD M KFK
Sbjct: 415 ----------AVPGKSGGVRVLTTLPRDAMAKFK 438
>At2g25150 unknown protein
Length = 461
Score = 121 bits (304), Expect = 7e-28
Identities = 111/475 (23%), Positives = 203/475 (42%), Gaps = 76/475 (16%)
Query: 7 ETKPLLDMKVTIHKTSMIFPSKQTENKSMFLSNIDK--VLNFYVKTVHFFEANK---DFP 61
+ KP+L + + ++ PSK T +++ LS +D T++ F+AN D P
Sbjct: 4 QRKPILPLLLEKKPVELVKPSKHTHCETLSLSTLDNDPFNEVMYATIYVFKANGKNLDDP 63
Query: 62 PQIVTEKLKKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGD 121
+ L+KAL + LV Y L+G+L + +L++ EGV F VA+S L+ +
Sbjct: 64 VSL----LRKALSELLVHYYPLSGKLMRSESNGKLQLVYLGEGVPFEVATSTLDLSSLN- 118
Query: 122 LAYPNQAFAQFVHNAKDFLKIGDLP---------LC----VVQLTSFKCGGFAIGFTTSH 168
++ N D + + +P +C +Q+T F CGGF IG +H
Sbjct: 119 ----------YIENLDDQVALRLVPEIEIDYESNVCYHPLALQVTKFACGGFTIGTALTH 168
Query: 169 VTFDGLSFKNFLDNLASLATNMKLTATPFHDRQLLAARSPPHVN--FPHPEMI-KLDNLP 225
DG + L LA A ++ P V + ++ K+DN P
Sbjct: 169 AVCDGYGVAQIIHALTELA----------------AGKTEPSVKSVWQRERLVGKIDNKP 212
Query: 226 LGIKSGVFEASSEEFDFM--------VFQLTYEDINNLKE---KAKGNNTSRVISFNVVA 274
+ + ++ + DI LK+ K ++ V++
Sbjct: 213 GKVPGSHIDGFLATSAYLPTTDVVTETINIRAGDIKRLKDSMMKECEYLKESFTTYEVLS 272
Query: 275 AHIWRCKALSKSYDPNRSSIILYAVDIRSRLNPPLPRAYTGNAVLTAYASAKCEELKEGE 334
++IW+ ++ + +P+ +++ AV IR L+PPLP+ Y GNA + Y EL+E
Sbjct: 273 SYIWKLRSRALKLNPDGITVLGVAVGIRHVLDPPLPKGYYGNAYIDVYVELTVRELEESS 332
Query: 335 FSRLVEMVEEGSRRMSD--------EYARSIIDWGELYNGFPNGDVLVSSWWRLG-FEEV 385
S + V++ + + + ++ ++ G +G ++ W +G F +
Sbjct: 333 ISNIANRVKKAKKTAYEKGYIEEELKNTERLMRDDSMFEGVSDGLFFLTDWRNIGWFGSM 392
Query: 386 EYPWGKPKYCCPVVYHRKDI---ILLFPSFNGGEGGGDGVNIIVALPRDEMEKFK 437
++ W +P P+ + ++L PS + G GV +I+ LPRD M +FK
Sbjct: 393 DFGWNEPVNLRPLTQRESTVHVGMILKPSKSDPSMEG-GVKVIMKLPRDAMVEFK 446
>At3g62160 unknown protein
Length = 428
Score = 119 bits (297), Expect = 4e-27
Identities = 101/395 (25%), Positives = 174/395 (43%), Gaps = 47/395 (11%)
Query: 23 MIFPSKQTENKSMFLSNIDK--VLNFYVKTVHFFEANKDFPPQIVTEKLKKALEDALVAY 80
+I + T + + LS ID VL + +T+H F D +++AL ALV+Y
Sbjct: 12 LIQACETTPSVVLDLSLIDNIPVLRCFARTIHVFTHGPD-----AARAIQEALAKALVSY 66
Query: 81 DFLAGRLKMNTETNRLEIDCNAE-GVGFVVASSEYKLNQIGDLAYPNQAFAQFVHNAKDF 139
L+GRLK +L+IDC + G+ FV A ++ KL +G + N + D
Sbjct: 67 YPLSGRLK-ELNQGKLQIDCTGKTGIWFVDAVTDVKLESVG--YFDNLMEIPYDDLLPDQ 123
Query: 140 LKIGD--LPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLA---TNMKLTA 194
+ D PL +Q+T F CGGF +G H DGL FL + +A TN+ ++
Sbjct: 124 IPKDDDAEPLVQMQVTQFSCGGFVMGLRFCHAICDGLGAAQFLTAVGEIACGQTNLGVSP 183
Query: 195 TPFHD--------RQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASSEEFDFMVFQ 246
D +++ PP PE KL++L I S + E EF
Sbjct: 184 VWHRDFIPQQHRANDVISPAIPPPFPPAFPE-YKLEHLSFDIPSDLIERFKREF------ 236
Query: 247 LTYEDINNLKEKAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSRLN 306
+ + +F V+AA W+ + + + + +++ + R +N
Sbjct: 237 -------------RASTGEICSAFEVIAACFWKLRTEAINLREDTEVKLVFFANCRHLVN 283
Query: 307 PPLPRAYTGNAVLTAYASAKCEEL-KEGEFSRLVEMVEEGSRRMSDEYARSIIDWGE--L 363
PPLP + GN SA+ E+ KE +V+++++ ++ +E+ + + G+
Sbjct: 284 PPLPIGFYGNCFFPVKISAESHEISKEKSVFDVVKLIKQAKTKLPEEFEKFVSGDGDDPF 343
Query: 364 YNGFPNGDVLVSSWWRLGFEEVEYPWGKPKYCCPV 398
+ +S W +LGF +V+Y WG P + P+
Sbjct: 344 APAVGYNTLFLSEWGKLGFNQVDYGWGPPLHVAPI 378
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,289,804
Number of Sequences: 26719
Number of extensions: 460384
Number of successful extensions: 1306
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 64
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1086
Number of HSP's gapped (non-prelim): 81
length of query: 445
length of database: 11,318,596
effective HSP length: 102
effective length of query: 343
effective length of database: 8,593,258
effective search space: 2947487494
effective search space used: 2947487494
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC136505.11


