

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136504.5 + phase: 0
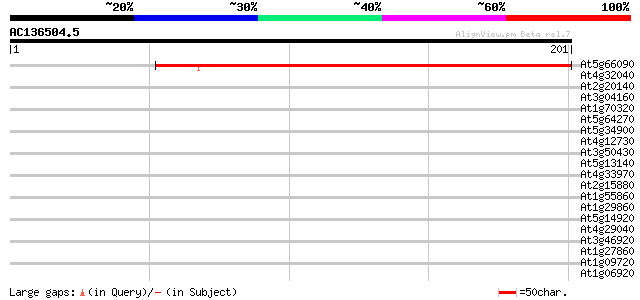
(201 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g66090 unknown protein 214 2e-56
At4g32040 homeodomain containing protein 1 29 1.5
At2g20140 26S proteasome subunit 4 29 1.5
At3g04160 hypothetical protein 28 2.5
At1g70320 hypothetical protein 28 2.5
At5g64270 nuclear protein-like 28 3.3
At5g34900 putative protein 28 3.3
At4g12730 fasciclin-like arabinogalactan protein FLA2 28 3.3
At3g50430 unknown protein 28 3.3
At5g13140 unknown protein 28 4.3
At4g33970 extensin-like protein 28 4.3
At2g15880 unknown protein 28 4.3
At1g55860 ubiquitin-protein ligase 1, putative 28 4.3
At1g29860 putative protein 28 4.3
At5g14920 unknown protein 27 5.6
At4g29040 26S proteasome AAA-ATPase subunit RPT2a 27 5.6
At3g46920 protein kinase - like protein 27 5.6
At1g27860 hypothetical protein 27 5.6
At1g09720 hypothetical protein 27 5.6
At1g06920 hypothetical protein 27 5.6
>At5g66090 unknown protein
Length = 210
Score = 214 bits (545), Expect = 2e-56
Identities = 106/153 (69%), Positives = 131/153 (85%), Gaps = 4/153 (2%)
Query: 53 VKAKPQEPEVAIAT----DSFTQFKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADI 108
VKA+ +E E + + D+F+ KHLLLP+ DR PYLSEGT+QA ATT +LA KYGADI
Sbjct: 58 VKAQAKEAEASPSPVGVGDAFSNVKHLLLPVIDRNPYLSEGTRQAAATTTSLAKKYGADI 117
Query: 109 TVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNL 168
TVVVIDE+++ES EHETQ+S+IRWHLSEGG +++KLLERLG+G K TAIIG+VADEL +
Sbjct: 118 TVVVIDEEKRESSSEHETQVSNIRWHLSEGGFEEFKLLERLGEGKKATAIIGEVADELKM 177
Query: 169 DLVVISMEAIHTKHIDANLLAEFIPCPVMLLPL 201
+LVV+SMEAIH+K+IDANLLAEFIPCPV+LLPL
Sbjct: 178 ELVVMSMEAIHSKYIDANLLAEFIPCPVLLLPL 210
>At4g32040 homeodomain containing protein 1
Length = 383
Score = 29.3 bits (64), Expect = 1.5
Identities = 20/74 (27%), Positives = 30/74 (40%), Gaps = 5/74 (6%)
Query: 4 THYLPP-----LQLVKPSLISPKPSLFFNNPPISITSHFFNPPSTLHTLPSGIIVKAKPQ 58
+H LPP L+ P P F+ P +T+ F N P+TL T S + +
Sbjct: 6 SHLLPPQEDLPLRHFTDQSQQPPPQRHFSETPSLVTASFLNLPTTLTTADSDLAPPHRNG 65
Query: 59 EPEVAIATDSFTQF 72
+ VA + F
Sbjct: 66 DNSVADTNPRWLSF 79
>At2g20140 26S proteasome subunit 4
Length = 443
Score = 29.3 bits (64), Expect = 1.5
Identities = 17/39 (43%), Positives = 24/39 (60%), Gaps = 1/39 (2%)
Query: 149 LGDGSKPTAIIGDVADELNLDLVVI-SMEAIHTKHIDAN 186
LGDG K + VAD+L+ +V I ++A+ TK DAN
Sbjct: 263 LGDGPKLVRELFRVADDLSPSIVFIDEIDAVGTKRYDAN 301
>At3g04160 hypothetical protein
Length = 712
Score = 28.5 bits (62), Expect = 2.5
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 5/64 (7%)
Query: 15 PSLISPKPSLFFNNPPISITSHFF--NPPSTLHTLPSGIIVKAKPQEPEVAIATDSFTQF 72
P +P P+LF++ PP + +FF PP L + IV P P + + + +
Sbjct: 8 PHYQNPNPNLFYHYPPPNSNPNFFFRPPPPPLQNPNNYSIV---PSPPPIRELSGTLSSL 64
Query: 73 KHLL 76
K LL
Sbjct: 65 KSLL 68
>At1g70320 hypothetical protein
Length = 3658
Score = 28.5 bits (62), Expect = 2.5
Identities = 15/54 (27%), Positives = 29/54 (52%), Gaps = 6/54 (11%)
Query: 112 VIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADE 165
++DE + + HET++ +RW + GL ++++ R G G+ I D+ E
Sbjct: 2203 MVDEDEDDF---HETRVIEVRWREALDGLDHFQIVGRSGGGN---GFIDDITAE 2250
>At5g64270 nuclear protein-like
Length = 1269
Score = 28.1 bits (61), Expect = 3.3
Identities = 22/65 (33%), Positives = 34/65 (51%), Gaps = 3/65 (4%)
Query: 91 KQAIATTIALANKYG-ADITVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDY--KLLE 147
KQ + TT+ +ANK G ADI V+++ + ES + +I ++ G D +L E
Sbjct: 795 KQLVETTVEVANKVGVADIVGRVVEDLKDESEQYRRMVMETIDKVVTNLGASDIDARLEE 854
Query: 148 RLGDG 152
L DG
Sbjct: 855 LLIDG 859
>At5g34900 putative protein
Length = 767
Score = 28.1 bits (61), Expect = 3.3
Identities = 15/46 (32%), Positives = 23/46 (49%)
Query: 142 DYKLLERLGDGSKPTAIIGDVADELNLDLVVISMEAIHTKHIDANL 187
D LE+L + KP +I G++ D + +AI H+DA L
Sbjct: 530 DATRLEKLAEWKKPLSIAGNIVDTKWFTTLETPGKAITATHVDAAL 575
>At4g12730 fasciclin-like arabinogalactan protein FLA2
Length = 403
Score = 28.1 bits (61), Expect = 3.3
Identities = 27/87 (31%), Positives = 36/87 (41%), Gaps = 3/87 (3%)
Query: 74 HLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQLSSIRW 133
H + I + P S AT +A ITV+ +D S+ + L IR
Sbjct: 27 HNITRILAKDPDFSTFNHYLSATHLADEINRRQTITVLAVDNSAMSSILSNGYSLYQIRN 86
Query: 134 HLSEGGLKDY---KLLERLGDGSKPTA 157
LS L DY K L ++ DGS TA
Sbjct: 87 ILSLHVLVDYFGTKKLHQITDGSTSTA 113
>At3g50430 unknown protein
Length = 642
Score = 28.1 bits (61), Expect = 3.3
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 1/34 (2%)
Query: 164 DELNLDLV-VISMEAIHTKHIDANLLAEFIPCPV 196
+ L DLV ++ ME +H KH+ N+L E C V
Sbjct: 91 ERLVADLVCLLGMENVHVKHLAGNILVEVSGCLV 124
>At5g13140 unknown protein
Length = 267
Score = 27.7 bits (60), Expect = 4.3
Identities = 14/37 (37%), Positives = 20/37 (53%)
Query: 7 LPPLQLVKPSLISPKPSLFFNNPPISITSHFFNPPST 43
LPPL + P P PSL F NP +++ + + P T
Sbjct: 216 LPPLPTLPPFPSFPFPSLPFGNPNLALPAFDWKNPVT 252
>At4g33970 extensin-like protein
Length = 699
Score = 27.7 bits (60), Expect = 4.3
Identities = 14/50 (28%), Positives = 23/50 (46%), Gaps = 6/50 (12%)
Query: 8 PPLQLVKPSLISPKPSLFFNNPPISITSHFFNPPSTLHTLPSGIIVKAKP 57
PP+ P + SP P ++ PP+ +PP +H+ P V + P
Sbjct: 563 PPVHSPPPPVFSPPPPVYSPPPPV------HSPPPPVHSPPPPAPVHSPP 606
>At2g15880 unknown protein
Length = 727
Score = 27.7 bits (60), Expect = 4.3
Identities = 15/53 (28%), Positives = 24/53 (44%), Gaps = 3/53 (5%)
Query: 8 PPLQLVKPSLISPKPSLFFNNPPISITS---HFFNPPSTLHTLPSGIIVKAKP 57
PP+ P + SP P ++ PP + S F+PP +H+ P + P
Sbjct: 597 PPVHSPPPPVHSPPPPVYSPPPPPPVHSPPPPVFSPPPPVHSPPPPVYSPPPP 649
>At1g55860 ubiquitin-protein ligase 1, putative
Length = 3891
Score = 27.7 bits (60), Expect = 4.3
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 10/66 (15%)
Query: 100 LANKYGADITVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAII 159
L ++Y D+ +DE + HE ++ +RW + GL +++L R G G+ I
Sbjct: 2385 LGDEYNDDM----VDEDDDDF---HENRVIEVRWREALDGLDHFQILGRSGGGN---GFI 2434
Query: 160 GDVADE 165
D+ E
Sbjct: 2435 DDITAE 2440
>At1g29860 putative protein
Length = 282
Score = 27.7 bits (60), Expect = 4.3
Identities = 14/38 (36%), Positives = 22/38 (57%)
Query: 3 FTHYLPPLQLVKPSLISPKPSLFFNNPPISITSHFFNP 40
F+ Y P Q V PS+ +P +L N+P +S +S+ P
Sbjct: 42 FSPYSSPFQSVSPSVNNPYLNLTSNSPVVSSSSNEGEP 79
>At5g14920 unknown protein
Length = 275
Score = 27.3 bits (59), Expect = 5.6
Identities = 20/62 (32%), Positives = 26/62 (41%), Gaps = 3/62 (4%)
Query: 6 YLPPLQLVKPSLISPKPSLFFNNPPISITSHFFNPPSTLHTLPSGIIVKAKPQEPEVAIA 65
Y PP VKP +P PP+ + +NPP+T P+ VK P P V
Sbjct: 157 YKPPTSPVKPPTTTPPVKPPTTTPPVQPPT--YNPPTTPVKPPTAPPVK-PPTPPPVRTR 213
Query: 66 TD 67
D
Sbjct: 214 ID 215
>At4g29040 26S proteasome AAA-ATPase subunit RPT2a
Length = 443
Score = 27.3 bits (59), Expect = 5.6
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query: 149 LGDGSKPTAIIGDVADELNLDLVVI-SMEAIHTKHIDAN 186
LGDG K + VAD+L+ +V I ++A+ TK DA+
Sbjct: 263 LGDGPKLVRELFRVADDLSPSIVFIDEIDAVGTKRYDAH 301
>At3g46920 protein kinase - like protein
Length = 1171
Score = 27.3 bits (59), Expect = 5.6
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 5/51 (9%)
Query: 9 PLQLV--KPSLISPKPSLFFNNPPISITSHFFNPPSTLHTLPSGIIVKAKP 57
P+QL+ SL S +P F + P+S++SH F P + + P + + P
Sbjct: 338 PIQLLPSSTSLFSQQP---FQDSPLSVSSHQFLPAAHMSMAPLNSQISSTP 385
>At1g27860 hypothetical protein
Length = 289
Score = 27.3 bits (59), Expect = 5.6
Identities = 24/100 (24%), Positives = 40/100 (40%), Gaps = 12/100 (12%)
Query: 65 ATDSFTQFKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEH 124
ATD + + LL+ TDR ++ G Q + VV+ E++ E P+
Sbjct: 161 ATDWISLYLKLLILATDRGMFVQTGLPQV------------QILEVVIETEEENEKPPDK 208
Query: 125 ETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVAD 164
+++ GL L +G+ + AII V D
Sbjct: 209 RLNARRAHVYITFTGLPKSPRLVEIGEHVERKAIIRRVID 248
>At1g09720 hypothetical protein
Length = 947
Score = 27.3 bits (59), Expect = 5.6
Identities = 22/63 (34%), Positives = 29/63 (45%), Gaps = 11/63 (17%)
Query: 40 PPSTLHTLPSGIIVKAKPQEPEVAIATDSFTQFKHLLLPITDRKPYLSEGTKQAIATTIA 99
PP LH +PSG + PQ PEV +FK L + RK G Q+ T+ A
Sbjct: 143 PPKHLHLIPSGTNI---PQVPEV-----PKKEFKSQSLMVLSRK---EPGVLQSSETSSA 191
Query: 100 LAN 102
L +
Sbjct: 192 LVS 194
>At1g06920 hypothetical protein
Length = 315
Score = 27.3 bits (59), Expect = 5.6
Identities = 20/70 (28%), Positives = 28/70 (39%), Gaps = 16/70 (22%)
Query: 21 KPSLFFNNPPISITSHFFN----------------PPSTLHTLPSGIIVKAKPQEPEVAI 64
KPS + P S TS+FFN P ++LH + S KP P +I
Sbjct: 44 KPSSESKSLPYSSTSYFFNRSRSRTSFESRILQISPRNSLHNIQSKRKTVYKPSPPSSSI 103
Query: 65 ATDSFTQFKH 74
+ F + H
Sbjct: 104 VSAGFNKTFH 113
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,705,018
Number of Sequences: 26719
Number of extensions: 208147
Number of successful extensions: 758
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 692
Number of HSP's gapped (non-prelim): 91
length of query: 201
length of database: 11,318,596
effective HSP length: 94
effective length of query: 107
effective length of database: 8,807,010
effective search space: 942350070
effective search space used: 942350070
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC136504.5


