

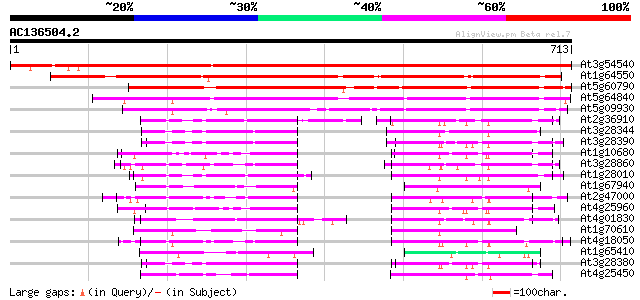
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136504.2 + phase: 0
(713 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g54540 ABC transporter - like protein 1129 0.0
At1g64550 ABC transporter protein, putative 457 e-128
At5g60790 ABC transporter homolog PnATH - like 429 e-120
At5g64840 ABC transporter protein 1-like 249 5e-66
At5g09930 ABC transporter, ATP-binding protein-like 240 2e-63
At2g36910 putative ABC transporter 77 4e-14
At3g28344 P-glycoprotein, 5' partial 76 6e-14
At3g28390 P-glycoprotein, putative 75 2e-13
At1g10680 putative P-glycoprotein-2 emb|CAA71277 73 5e-13
At3g28860 P-glycoprotein, putative 73 6e-13
At1g28010 hypothetical protein 72 8e-13
At1g67940 ABC transporter like protein 72 1e-12
At2g47000 putative ABC transporter 71 2e-12
At4g25960 P-glycoprotein-2 (pgp2) 70 3e-12
At4g01830 putative P-glycoprotein-like protein 70 3e-12
At1g70610 transporter associated with antigen processing-like pr... 70 3e-12
At4g18050 multidrug resistance protein/P-glycoprotein - like 70 4e-12
At1g65410 ABC transporter like protein 69 9e-12
At3g28380 P-glycoprotein, putative 68 2e-11
At4g25450 putative protein 67 3e-11
>At3g54540 ABC transporter - like protein
Length = 723
Score = 1129 bits (2919), Expect = 0.0
Identities = 576/726 (79%), Positives = 644/726 (88%), Gaps = 16/726 (2%)
Query: 1 MGKKKSEDAGPSTKAKPGSKDVSK---KEKFSVSAMLAGMDEKADKPKKASSNKAKPKPA 57
MGKKKS+++ +TK KP KD SK KEK SVSAMLAGMD+K DKPKK SS++ K A
Sbjct: 1 MGKKKSDESAATTKVKPSGKDASKDSKKEKLSVSAMLAGMDQKDDKPKKGSSSRTKA--A 58
Query: 58 PKASAYTDDIDLPPSD---DDESEEEQEEKH-----RPDLKPLEVSIAEKELKKREKKDI 109
PK+++YTD IDLPPSD D ES+EE+ +K + + + LE+S+ +KE KKRE K+
Sbjct: 59 PKSTSYTDGIDLPPSDEEDDGESDEEERQKEARRKLKSEQRHLEISVTDKEQKKREAKER 118
Query: 110 LAAHVAEQAKKEALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELL 169
LA AE AK+EA++DD DAFTVVIGS+ SVL+GDD ADANVKDITIE+FSV+ARGKELL
Sbjct: 119 LALQAAESAKREAMKDDHDAFTVVIGSKTSVLEGDDMADANVKDITIESFSVSARGKELL 178
Query: 170 KNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEA 229
KN SV+ISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGD+K+AL A
Sbjct: 179 KNASVRISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDEKSALNA 238
Query: 230 VVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAES 289
VVSAN EL+K+R++ LQ +SG +G + D +++D GEKLAELY++LQ++GSDAAE+
Sbjct: 239 VVSANEELVKLREEAEALQKSSSGADG-ENVDGEDDDDTGEKLAELYDRLQILGSDAAEA 297
Query: 290 QASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLW 349
QASKILAGLGFTKDMQ R T+SFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLW
Sbjct: 298 QASKILAGLGFTKDMQVRATQSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLW 357
Query: 350 LEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANK 409
LEEYLCRWKKTLVVVSHDRDFLNTVC+EIIHLHD LHFYRGNFD FESGYEQRR+E NK
Sbjct: 358 LEEYLCRWKKTLVVVSHDRDFLNTVCTEIIHLHDQNLHFYRGNFDGFESGYEQRRKEMNK 417
Query: 410 KYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKE-SKSKSKGK-VDEDETQVEVPHKWR 467
K++++DKQ+KAA+RTGN+ QQ+KVKDRAKF AAKE SKSKSKGK VDE+ E P KWR
Sbjct: 418 KFDVYDKQMKAAKRTGNRGQQEKVKDRAKFTAAKEASKSKSKGKTVDEEGPAPEAPRKWR 477
Query: 468 DYSVEFHFPEPTELTPPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKS 527
DYSV FHFPEPTELTPPLLQLIEVSFSYPNR DFRLS+VDVGIDMGTRVAIVGPNGAGKS
Sbjct: 478 DYSVVFHFPEPTELTPPLLQLIEVSFSYPNRPDFRLSNVDVGIDMGTRVAIVGPNGAGKS 537
Query: 528 TLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQE 587
TLLNLLAGDLVP+EGE+RRSQKLRIGRYSQHFVDLLTM ETPVQYLLRLHPDQEG SKQE
Sbjct: 538 TLLNLLAGDLVPTEGEMRRSQKLRIGRYSQHFVDLLTMGETPVQYLLRLHPDQEGFSKQE 597
Query: 588 AVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDA 647
AVRAKLGK+GLPSHNHL+PI KLSGGQKARVVFTSISMS+PHILLLDEPTNHLDMQSIDA
Sbjct: 598 AVRAKLGKFGLPSHNHLSPIAKLSGGQKARVVFTSISMSKPHILLLDEPTNHLDMQSIDA 657
Query: 648 LADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEI 707
LADALDEFTGGVVLVSHDSRLISRVC +EE+SQIWVVEDGTV FPGTFE+YKEDL +EI
Sbjct: 658 LADALDEFTGGVVLVSHDSRLISRVCAEEEKSQIWVVEDGTVNFFPGTFEEYKEDLQREI 717
Query: 708 KAEVDD 713
KAEVD+
Sbjct: 718 KAEVDE 723
>At1g64550 ABC transporter protein, putative
Length = 715
Score = 457 bits (1175), Expect = e-128
Identities = 263/661 (39%), Positives = 402/661 (60%), Gaps = 47/661 (7%)
Query: 53 KPKPAPKASAYTDDIDLPPSDDDESEEEQEEKHRPDLKPLEVSIAEKELKKREKKDILA- 111
KP P ++ A ++ D +++ E P L +++ E+ KK +++ L
Sbjct: 84 KPTPTVRSLAMPVRMNDGMDDGPVKKKKPEPVDGPLLTERDLAKIERRKKKDDRQRELQY 143
Query: 112 -AHVAEQAKKEALRDDRDAFTVVIGSRASVLDGDDGADANVKDITIENFSVAARGKELLK 170
HVAE +A G ++ D G + ++DI ++NF+V+ G++L+
Sbjct: 144 QQHVAEMEAAKA------------GMPTVSVNHDTGGGSAIRDIHMDNFNVSVGGRDLIV 191
Query: 171 NTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIP-VPKNIDVLLVEQEVVGDDKTALEA 229
+ S+ +S G+ YGL+G NG GK+T L+ +A I +P N +L VEQEVVGD TAL+
Sbjct: 192 DGSITLSFGRHYGLVGRNGTGKTTFLRYMAMHAIEGIPTNCQILHVEQEVVGDKTTALQC 251
Query: 230 VVSANVELIKVRQKVADLQNIA--------SGEEGMDKDDTNEEEDAGEKLAELYEQLQL 281
V++ ++E K+ ++ ++Q +A + ++GM DT E + ++L E+Y++L
Sbjct: 252 VLNTDIERTKLLEE--EIQILAKQRETEEPTAKDGMPTKDTVEGDLMSQRLEEIYKRLDA 309
Query: 282 MGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNH 341
+ + AE++A+ ILAGL FT +MQ + T +FSGGWRMRI+LARALF++P LLLLDEPTNH
Sbjct: 310 IDAYTAEARAASILAGLSFTPEMQLKATNTFSGGWRMRIALARALFIEPDLLLLDEPTNH 369
Query: 342 LDLRAVLWLEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYE 401
LDL AVLWLE YL +W KT +VVSH R+FLNTV ++IIHL + KL Y+GN+D FE E
Sbjct: 370 LDLHAVLWLETYLTKWPKTFIVVSHAREFLNTVVTDIIHLQNQKLSTYKGNYDIFERTRE 429
Query: 402 QRRREANKKYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVE 461
++ + K +E ++ R+ +A DK + AK A+ +S+ K+ ++ + +
Sbjct: 430 EQVKNQQKAFESSERS-----RSHMQAFIDKFRYNAKRASLVQSRIKALDRLAHVDQVIN 484
Query: 462 VPHKWRDYSVEFHFPEPTELT-PPLLQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVG 520
P DY +F FP P + PP++ + SF YP +++ GID+ +R+A+VG
Sbjct: 485 DP----DY--KFEFPTPDDKPGPPIISFSDASFGYPG-GPLLFRNLNFGIDLDSRIAMVG 537
Query: 521 PNGAGKSTLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQ 580
PNG GKST+L L++GDL PS G V RS K+R+ +SQH VD L + P+ Y++R +P
Sbjct: 538 PNGIGKSTILKLISGDLQPSSGTVFRSAKVRVAVFSQHHVDGLDLSSNPLLYMMRCYP-- 595
Query: 581 EGLSKQEAVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHL 640
G+ +Q+ +R+ LG G+ + L P+ LSGGQK+RV F I+ +PH+LLLDEP+NHL
Sbjct: 596 -GVPEQK-LRSHLGSLGVTGNLALQPMYTLSGGQKSRVAFAKITFKKPHLLLLDEPSNHL 653
Query: 641 DMQSIDALADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYK 700
D+ +++AL L F GG+ +VSHD LIS D ++WVV DG + F GTF DYK
Sbjct: 654 DLDAVEALIQGLVLFQGGICMVSHDEHLISGSVD-----ELWVVSDGRIAPFHGTFHDYK 708
Query: 701 E 701
+
Sbjct: 709 K 709
>At5g60790 ABC transporter homolog PnATH - like
Length = 595
Score = 429 bits (1104), Expect = e-120
Identities = 236/567 (41%), Positives = 357/567 (62%), Gaps = 43/567 (7%)
Query: 152 KDITIENFSVAARGKELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNID 211
+DI IE+ SV G +L+ ++ +++++G+RYGL+G NG GKSTLL + R+IP+P +D
Sbjct: 67 RDIRIESLSVTFHGYDLIVDSMLELNYGRRYGLLGLNGCGKSTLLTAIGRREIPIPDQMD 126
Query: 212 VLLVEQEVVGDDKTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEK 271
+ + E+ D ++LEAVVS + E +++ ++V L +++ GE+
Sbjct: 127 IYHLSHEIEATDMSSLEAVVSCDEERLRLEKEVEIL--------------VQQDDGGGER 172
Query: 272 LAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPT 331
L +YE+L M ++ AE +A++IL GLGF K+MQ + TK FSGGWRMRI+LARALF+ PT
Sbjct: 173 LQSIYERLDAMDAETAEKRAAEILFGLGFDKEMQAKKTKDFSGGWRMRIALARALFIMPT 232
Query: 332 LLLLDEPTNHLDLRAVLWLEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRG 391
+LLLDEPTNHLDL A +WLEE L + + LVVVSH +DFLN VC+ IIH+ +L +Y G
Sbjct: 233 ILLLDEPTNHLDLEACVWLEESLKNFDRILVVVSHSQDFLNGVCTNIIHMQSKQLKYYTG 292
Query: 392 NFDAFESGYEQRRREANKKYEIFDKQLKAAR----RTGNKAQQDKVKDRAKFAAAKESKS 447
NFD + + K+Y +Q+ + R G+ + AK A +SK
Sbjct: 293 NFDQYCQTRSELEENQMKQYRWEQEQISHMKEYIARFGHGS--------AKLARQAQSKE 344
Query: 448 KSKGKVDEDETQVEVPHKWRDYSVEFHFPEPTELTPPLLQLIEVSFSYPNREDFRL-SDV 506
K+ K++ +V RD + F F + +L PP+LQ +EVSF Y D+ + ++
Sbjct: 345 KTLAKMERGGLTEKVA---RDSVLVFRFADVGKLPPPVLQFVEVSFGY--TPDYLIYKNI 399
Query: 507 DVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMD 566
D G+D+ +RVA+VGPNGAGKSTLL L+ G+L P+EG VRR L+I +Y QH + L ++
Sbjct: 400 DFGVDLDSRVALVGPNGAGKSTLLKLMTGELHPTEGMVRRHNHLKIAQYHQHLAEKLDLE 459
Query: 567 ETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMS 626
+ Y++R P E +E +RA +G++GL + P+ LS GQ++RV+F ++
Sbjct: 460 LPALLYMMREFPGTE----EEKMRAAIGRFGLTGKAQVMPMKNLSDGQRSRVIFAWLAYK 515
Query: 627 RPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLVSHDSRLISRVCDDEERSQIWVVED 686
+P++LLLDEPTNHLD+++ID+LA+AL+E+ GG+VLVSHD RLI++V +IWV E
Sbjct: 516 QPNMLLLDEPTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVA-----HEIWVCEK 570
Query: 687 GTVRNFPGTFEDYKEDLLKEIKAEVDD 713
+ + G D+K L + KA ++D
Sbjct: 571 QCITKWNGDIMDFKRHL--KAKAGLED 595
>At5g64840 ABC transporter protein 1-like
Length = 692
Score = 249 bits (635), Expect = 5e-66
Identities = 176/634 (27%), Positives = 318/634 (49%), Gaps = 54/634 (8%)
Query: 106 KKDI--LAAHVAEQAKKEALRDDRDAFTVVIGSRASVLDGD---------DGADANVKDI 154
++DI + A V+ + + +++ +D + + S D D +GA +
Sbjct: 39 RRDISTIRAQVSTISLETSVKQRQDEIESLFSKQPSQQDSDRKRNGKSSKNGASGISSGV 98
Query: 155 TIENFSVAARGKELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIP-------VP 207
+EN + +G +LK+ + ++ G++ GL+G NG GK+T L+++ ++ P
Sbjct: 99 KLENIRKSYKGVTVLKDVTWEVKRGEKVGLVGVNGAGKTTQLRIITGQEEPDSGNVIKAK 158
Query: 208 KNIDVLLVEQEV-VGDDKTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEE 266
N+ V + QE V KT E ++A E +++ +K+ +Q G DD +
Sbjct: 159 PNMKVAFLSQEFEVSMSKTVREEFMTAFKEEMEITEKLEKVQKAIEGSV----DDLDLMG 214
Query: 267 DAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARAL 326
++ L + Q + D+ +++ SK++ LGF + R SFSGGW+MR+SL + L
Sbjct: 215 RLLDEFDLLQRRAQAVNLDSVDAKISKLMPELGFAPEDADRLVASFSGGWQMRMSLGKIL 274
Query: 327 FVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKL 386
P LLLLDEPTNHLDL + WLE YL + +V++SHDR FL+ +C++I+
Sbjct: 275 LQDPDLLLLDEPTNHLDLDTIEWLEGYLQKQDVPMVIISHDRAFLDQLCTKIVETEMGVS 334
Query: 387 HFYRGNFDAFESGYEQRRREANKKYEIFDKQLKAARRTGNKAQQDKVKDR-AKFAAAKES 445
+ GN+ + + N +E K + D KD A+ A S
Sbjct: 335 RTFEGNYSQYVISKAEWIETQNAAWEKQQKDI------------DSTKDLIARLGAGANS 382
Query: 446 --KSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEPTELTPPLLQLIEVSFSYPNREDFRL 503
S ++ K+++ + Q + ++ ++ FPE ++ + + F + ++ F+
Sbjct: 383 GRASTAEKKLEKLQEQELIEKPFQRKQMKIRFPERGTSGRSVVNVKNIDFGFEDKMLFK- 441
Query: 504 SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVRRSQKLRIGRY-SQHFVDL 562
++ I+ G ++AI+GPNG GKSTLL L+ G P +GEV + + Y Q+ ++
Sbjct: 442 -KANLSIERGEKIAILGPNGCGKSTLLKLIMGLEKPVKGEVILGEHNVLPNYFEQNQAEV 500
Query: 563 LTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTS 622
L +D+T ++ + D + + ++ LG+ + + LSGG+KAR+ F
Sbjct: 501 LDLDKTVLETVCEAAEDW----RSDDIKGLLGRCNFKADMLDRKVSLLSGGEKARLAFCK 556
Query: 623 ISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLVSHDSRLISRVCDDEERSQIW 682
++ +L+LDEPTNHLD+ S + L +A++E+ G V+ VSHD I ++ +++
Sbjct: 557 FMVTPSTLLVLDEPTNHLDIPSKEMLEEAINEYQGTVIAVSHDRYFIKQIV-----NRVI 611
Query: 683 VVEDGTVRNFPGTFEDYKEDLL----KEIKAEVD 712
VEDG + ++ G + Y E L KE++ E +
Sbjct: 612 EVEDGCLEDYAGDYNYYLEKNLDARTKELEREAE 645
>At5g09930 ABC transporter, ATP-binding protein-like
Length = 674
Score = 240 bits (612), Expect = 2e-63
Identities = 175/578 (30%), Positives = 296/578 (50%), Gaps = 46/578 (7%)
Query: 144 DDGADANVKDITIENFSVAARGKELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRK 203
++GA + + +EN S + G +LK+ + ++ G++ GLIG NG GK+T L+++ ++
Sbjct: 74 NNGASSISSGVRLENISKSYEGITVLKDVTWEVKKGEKVGLIGVNGAGKTTQLRIITGQE 133
Query: 204 IP-------VPKNIDVLLVEQEV-VGDDKTALEAVVSANVELIKVRQKVADLQNIASGEE 255
P N+ V + QE V KT E + E +++ +K+ +LQ EE
Sbjct: 134 EPDSGNVIWAKPNLKVAFLSQEFEVSMGKTVKEEFMCTFKEEMEIARKLENLQKAI--EE 191
Query: 256 GMDKDDTNEEEDAGEKLAE---LYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSF 312
+D + E G+ L E L + Q + D+ ++ SK+++ LGF + R SF
Sbjct: 192 AVD-----DLELMGKLLDEFDLLQRRAQEVDLDSIHAKISKLMSELGFVSEDADRLVASF 246
Query: 313 SGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWKKTLVVVSHDRDFLN 372
S GW+MR+SL + L P LLLLDEPTNHLDL + WLE YL + +V++SHDR FL+
Sbjct: 247 SSGWQMRMSLGKILLQNPDLLLLDEPTNHLDLDTIEWLEGYLIKQDVPMVIISHDRAFLD 306
Query: 373 TVCSEIIHLHDLKLHFYRGNFDAFESGYEQRRREANKKYEIFDKQLKAARRTGNKAQQDK 432
+C++I+ G F+ Y Q K E+ + Q A + + + K
Sbjct: 307 QLCTKIVETE-------MGVSRTFDGNYSQ---YVISKAELVEAQYAAWEKQQKEIEATK 356
Query: 433 VKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEPTELTPPLLQLIEVS 492
A A ++ S K+ E+E +E P + + ++ FPE ++ + +
Sbjct: 357 DLISRLSAGANSGRASSAEKLQEEEL-IEKPFQRK--QMKIRFPECGLSGRSVVTVKNLV 413
Query: 493 FSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVRRSQKLRI 552
F + ++ F + ++ I+ G +VAI+GPNG GKSTLL L+ G P GEV + +
Sbjct: 414 FGFDDKMLF--NKANLAIERGEKVAIIGPNGCGKSTLLKLIMGLEKPMRGEVILGEHNVL 471
Query: 553 GRY-SQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLPSHNHLTPIVKLS 611
Y Q+ + +D+T ++ ++ D + + ++A LG+ + + LS
Sbjct: 472 PNYFEQNQAEAQDLDKTVIETVVEAAVDW----RIDDIKALLGRCNFKADMLDRKVSLLS 527
Query: 612 GGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLVSHDSRLISR 671
GG+KAR+ F + +L+LDEPTNHLD+ S + L +A++E+ G V+ VSHD I +
Sbjct: 528 GGEKARLAFCKFMVKPSTLLVLDEPTNHLDIPSKEMLEEAINEYKGTVITVSHDRYFIKQ 587
Query: 672 VCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEIKA 709
+ +++ V DG + ++ G DY L K ++A
Sbjct: 588 IV-----NRVIEVRDGGLMDYAG---DYNYFLEKNVEA 617
>At2g36910 putative ABC transporter
Length = 1286
Score = 76.6 bits (187), Expect = 4e-14
Identities = 68/240 (28%), Positives = 118/240 (48%), Gaps = 42/240 (17%)
Query: 485 LLQLIEVSFSYPNREDFR-LSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGE 543
L++L V FSYP+R D + L++ + + G +A+VG +G+GKST+++L+ P+ G+
Sbjct: 367 LVELKNVDFSYPSRPDVKILNNFCLSVPAGKTIALVGSSGSGKSTVVSLIERFYDPNSGQ 426
Query: 544 VR------RSQKLR-----IGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAK 592
V ++ KLR IG SQ T + + +L PD + + +EA R
Sbjct: 427 VLLDGQDLKTLKLRWLRQQIGLVSQEPALFATSIK---ENILLGRPDADQVEIEEAARVA 483
Query: 593 LGKYGLPSHNHLTPI------------VKLSGGQKARVVFTSISMSRPHILLLDEPTNHL 640
+H+ + + ++LSGGQK R+ + P ILLLDE T+ L
Sbjct: 484 ------NAHSFIIKLPDGFDTQVGERGLQLSGGQKQRIAIARAMLKNPAILLLDEATSAL 537
Query: 641 DMQSIDALADALDEFTGG--VVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFED 698
D +S + +ALD F G ++++H R+ + + V++ G+V GT ++
Sbjct: 538 DSESEKLVQEALDRFMIGRTTLIIAH------RLSTIRKADLVAVLQQGSVSEI-GTHDE 590
Score = 69.7 bits (169), Expect = 5e-12
Identities = 68/245 (27%), Positives = 117/245 (47%), Gaps = 30/245 (12%)
Query: 467 RDYSVEFHFPEPTELTPPL---LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPN 522
R +E P+ T + L ++L + FSYP+R D ++ D+ + G +A+VGP+
Sbjct: 1002 RKTEIEPDDPDTTPVPDRLRGEVELKHIDFSYPSRPDIQIFRDLSLRARAGKTLALVGPS 1061
Query: 523 GAGKSTLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFV--DLLTMDETPVQYLLRLHPD- 579
G GKS++++L+ PS G V K I +Y+ + + + + P + ++ +
Sbjct: 1062 GCGKSSVISLIQRFYEPSSGRVMIDGK-DIRKYNLKAIRKHIAIVPQEPCLFGTTIYENI 1120
Query: 580 -------QEGLSKQEAVRAKLGKY--GLPSHNHLTPI----VKLSGGQKARVVFTSISMS 626
E Q A A K+ LP + T + V+LSGGQK R+ +
Sbjct: 1121 AYGHECATEAEIIQAATLASAHKFISALP-EGYKTYVGERGVQLSGGQKQRIAIARALVR 1179
Query: 627 RPHILLLDEPTNHLDMQSIDALADALDEFTGG--VVLVSHDSRLISRVCDDEERSQIWVV 684
+ I+LLDE T+ LD +S ++ +ALD+ G ++V+H R+ I V+
Sbjct: 1180 KAEIMLLDEATSALDAESERSVQEALDQACSGRTSIVVAH------RLSTIRNAHVIAVI 1233
Query: 685 EDGTV 689
+DG V
Sbjct: 1234 DDGKV 1238
Score = 57.4 bits (137), Expect = 3e-08
Identities = 74/284 (26%), Positives = 123/284 (43%), Gaps = 37/284 (13%)
Query: 167 ELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTA 226
++L N + + GK L+G +G GKST++ L+ + P + VLL Q++
Sbjct: 384 KILNNFCLSVPAGKTIALVGSSGSGKSTVVSLI--ERFYDPNSGQVLLDGQDL------- 434
Query: 227 LEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDA 286
++L +RQ++ + S E + E G A+ E +
Sbjct: 435 ------KTLKLRWLRQQIG----LVSQEPALFATSIKENILLGRPDADQVEIEEAARVAN 484
Query: 287 AESQASKILAGLGFTKDMQ-GRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLR 345
A S K+ G D Q G SGG + RI++ARA+ P +LLLDE T+ LD
Sbjct: 485 AHSFIIKLPDGF----DTQVGERGLQLSGGQKQRIAIARAMLKNPAILLLDEATSALDSE 540
Query: 346 AVLWLEEYLCRWK--KTLVVVSHDRDFLNTVCSEIIHLHDLKLHFYRGNFDAFESGYEQR 403
+ ++E L R+ +T ++++H L+T I DL +G+ + E
Sbjct: 541 SEKLVQEALDRFMIGRTTLIIAHR---LST-----IRKADLVAVLQQGSVSEIGTHDELF 592
Query: 404 RREANKKYEIFDKQLKAARRTGNKAQQDKVKDRAKFAAAKESKS 447
+ N Y K +AA T A + K A+ ++A+ S S
Sbjct: 593 SKGENGVYAKLIKMQEAAHET---AMSNARKSSARPSSARNSVS 633
Score = 48.5 bits (114), Expect = 1e-05
Identities = 50/202 (24%), Positives = 92/202 (44%), Gaps = 24/202 (11%)
Query: 167 ELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTA 226
++ ++ S++ GK L+GP+G GKS+++ L+ ++ P + V++ +++
Sbjct: 1040 QIFRDLSLRARAGKTLALVGPSGCGKSSVISLI--QRFYEPSSGRVMIDGKDI------- 1090
Query: 227 LEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDA 286
L +R+ +A I E + E G + A E +Q +
Sbjct: 1091 ------RKYNLKAIRKHIA----IVPQEPCLFGTTIYENIAYGHECATEAEIIQAATLAS 1140
Query: 287 AESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRA 346
A S + G K G SGG + RI++ARAL + ++LLDE T+ LD +
Sbjct: 1141 AHKFISALPEGY---KTYVGERGVQLSGGQKQRIAIARALVRKAEIMLLDEATSALDAES 1197
Query: 347 VLWLEEYLCR--WKKTLVVVSH 366
++E L + +T +VV+H
Sbjct: 1198 ERSVQEALDQACSGRTSIVVAH 1219
>At3g28344 P-glycoprotein, 5' partial
Length = 626
Score = 76.3 bits (186), Expect = 6e-14
Identities = 68/219 (31%), Positives = 106/219 (48%), Gaps = 28/219 (12%)
Query: 479 TELTPPLLQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDL 537
TE ++ ++V FSYP R D + + + I+ G AIVGP+G+GKST++ L+
Sbjct: 381 TERITGQVEFLDVDFSYPTRPDVIIFKNFSIKIEEGKSTAIVGPSGSGKSTIIGLIERFY 440
Query: 538 VPSEGEVR------RSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQ--EAV 589
P +G V+ RS LR R + L++ + T +R + G+S + EA
Sbjct: 441 DPLKGIVKIDGRDIRSYHLRSLR---RHIALVSQEPTLFAGTIRENIIYGGVSDKIDEAE 497
Query: 590 RAKLGKYGLPSHNHLTPI------------VKLSGGQKARVVFTSISMSRPHILLLDEPT 637
+ K +H+ +T + V+LSGGQK R+ + P +LLLDE T
Sbjct: 498 IIEAAK-AANAHDFITSLTEGYDTYCGDRGVQLSGGQKQRIAIARAVLKNPSVLLLDEAT 556
Query: 638 NHLDMQSIDALADALDEFTGG--VVLVSHDSRLISRVCD 674
+ LD QS + DAL+ G V+++H I CD
Sbjct: 557 SALDSQSERVVQDALERVMVGRTSVVIAHRLSTIQN-CD 594
Score = 53.1 bits (126), Expect = 5e-07
Identities = 54/201 (26%), Positives = 93/201 (45%), Gaps = 22/201 (10%)
Query: 168 LLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTAL 227
+ KN S+KI GK ++GP+G GKST++ L + + D L ++ G D
Sbjct: 405 IFKNFSIKIEEGKSTAIVGPSGSGKSTIIGL-------IERFYDPLKGIVKIDGRD---- 453
Query: 228 EAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAA 287
+ L +R+ +A + S E + E G ++ E + + AA
Sbjct: 454 ----IRSYHLRSLRRHIA----LVSQEPTLFAGTIRENIIYGGVSDKIDEAEIIEAAKAA 505
Query: 288 ESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAV 347
+ G+ R + SGG + RI++ARA+ P++LLLDE T+ LD ++
Sbjct: 506 NAHDFITSLTEGYDTYCGDRGVQ-LSGGQKQRIAIARAVLKNPSVLLLDEATSALDSQSE 564
Query: 348 LWLEEYLCRWK--KTLVVVSH 366
+++ L R +T VV++H
Sbjct: 565 RVVQDALERVMVGRTSVVIAH 585
>At3g28390 P-glycoprotein, putative
Length = 1225
Score = 74.7 bits (182), Expect = 2e-13
Identities = 72/227 (31%), Positives = 110/227 (47%), Gaps = 43/227 (18%)
Query: 491 VSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVR---- 545
V F+YP R D + + + I+ G AIVGP+G+GKST+++L+ P +G V+
Sbjct: 985 VDFAYPTRPDVIIFQNFSIDIEDGKSTAIVGPSGSGKSTIISLIERFYDPLKGIVKIDGR 1044
Query: 546 --RSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQ-------EAVRAKLGKY 596
RS LR R QH + L++ + T +R + G S + EA +A
Sbjct: 1045 DIRSCHLRSLR--QH-IALVSQEPTLFAGTIRENIMYGGASNKIDESEIIEAAKAA---- 1097
Query: 597 GLPSHNHLTPI------------VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQS 644
+H+ +T + V+LSGGQK R+ + P +LLLDE T+ LD QS
Sbjct: 1098 --NAHDFITSLSNGYDTCCGDRGVQLSGGQKQRIAIARAVLKNPSVLLLDEATSALDSQS 1155
Query: 645 IDALADALDEFTGG--VVLVSHDSRLISRVCDDEERSQIWVVEDGTV 689
+ DAL+ G V+++H I + CD I V+E+G V
Sbjct: 1156 ESVVQDALERLMVGRTSVVIAHRLSTIQK-CD-----TIAVLENGAV 1196
Score = 64.7 bits (156), Expect = 2e-10
Identities = 67/247 (27%), Positives = 117/247 (47%), Gaps = 36/247 (14%)
Query: 480 ELTPPLLQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLV 538
E T ++ V F+YP+R + + D+ + + G VA+VG +G+GKST+++LL
Sbjct: 341 EKTRGEVEFNHVKFTYPSRPETPIFDDLCLRVPSGKTVALVGGSGSGKSTVISLLQRFYD 400
Query: 539 PSEGEVRRS----QKLRIGRYSQHFVDLLTMDETPVQYLLRLHPD----QEGLSKQEAVR 590
P GE+ KL++ ++ + + L++ + PV + + + +E S E V
Sbjct: 401 PIAGEILIDGLPINKLQV-KWLRSQMGLVSQE--PVLFATSIKENILFGKEDASMDEVVE 457
Query: 591 AKLGKYGLPSHNHLTPI------------VKLSGGQKARVVFTSISMSRPHILLLDEPTN 638
A +H+ ++ V+LSGGQK R+ + P ILLLDE T+
Sbjct: 458 AAKASN---AHSFISQFPNSYQTQVGERGVQLSGGQKQRIAIARAIIKSPIILLLDEATS 514
Query: 639 HLDMQSIDALADALDEFTGG--VVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTF 696
LD +S + +ALD + G ++++H R+ I VV +G + G+
Sbjct: 515 ALDSESERVVQEALDNASIGRTTIVIAH------RLSTIRNADVICVVHNGRIIE-TGSH 567
Query: 697 EDYKEDL 703
E+ E L
Sbjct: 568 EELLEKL 574
Score = 53.1 bits (126), Expect = 5e-07
Identities = 53/201 (26%), Positives = 92/201 (45%), Gaps = 22/201 (10%)
Query: 168 LLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTAL 227
+ +N S+ I GK ++GP+G GKST++ L + + D L ++ G D
Sbjct: 997 IFQNFSIDIEDGKSTAIVGPSGSGKSTIISL-------IERFYDPLKGIVKIDGRD---- 1045
Query: 228 EAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAA 287
+ L +RQ +A + S E + E G ++ E + + AA
Sbjct: 1046 ----IRSCHLRSLRQHIA----LVSQEPTLFAGTIRENIMYGGASNKIDESEIIEAAKAA 1097
Query: 288 ESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAV 347
+ G+ R + SGG + RI++ARA+ P++LLLDE T+ LD ++
Sbjct: 1098 NAHDFITSLSNGYDTCCGDRGVQ-LSGGQKQRIAIARAVLKNPSVLLLDEATSALDSQSE 1156
Query: 348 LWLEEYLCRWK--KTLVVVSH 366
+++ L R +T VV++H
Sbjct: 1157 SVVQDALERLMVGRTSVVIAH 1177
Score = 48.5 bits (114), Expect = 1e-05
Identities = 52/197 (26%), Positives = 87/197 (43%), Gaps = 28/197 (14%)
Query: 174 VKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNI--DVLLVEQEVVGDDKTALEAVV 231
+++ GK L+G +G GKST++ LL P+ I D L + + V ++ + +V
Sbjct: 370 LRVPSGKTVALVGGSGSGKSTVISLLQRFYDPIAGEILIDGLPINKLQVKWLRSQM-GLV 428
Query: 232 SANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQA 291
S L K +NI G+E D+ E A + + S S
Sbjct: 429 SQEPVLFATSIK----ENILFGKEDASMDEVVEAAKASNAHSFI--------SQFPNSYQ 476
Query: 292 SKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLE 351
+++ G SGG + RI++ARA+ P +LLLDE T+ LD + ++
Sbjct: 477 TQV-----------GERGVQLSGGQKQRIAIARAIIKSPIILLLDEATSALDSESERVVQ 525
Query: 352 EYL--CRWKKTLVVVSH 366
E L +T +V++H
Sbjct: 526 EALDNASIGRTTIVIAH 542
>At1g10680 putative P-glycoprotein-2 emb|CAA71277
Length = 1227
Score = 73.2 bits (178), Expect = 5e-13
Identities = 61/228 (26%), Positives = 115/228 (49%), Gaps = 37/228 (16%)
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
++L V FSYP+R D + SD ++ + G +A+VG +G+GKS++L+L+ P+ G +
Sbjct: 982 IELKGVHFSYPSRPDVTIFSDFNLLVPSGKSMALVGQSGSGKSSVLSLVLRFYDPTAGII 1041
Query: 545 ----RRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPD----QEGLSKQEAVRA-KLGK 595
+ +KL++ +H + + + P + ++ + +EG S+ E + A KL
Sbjct: 1042 MIDGQDIKKLKLKSLRRH---IGLVQQEPALFATTIYENILYGKEGASESEVMEAAKLAN 1098
Query: 596 YGLPSHNHLTPI------------VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQ 643
+H+ ++ + +++SGGQ+ R+ + P ILLLDE T+ LD++
Sbjct: 1099 ----AHSFISSLPEGYSTKVGERGIQMSGGQRQRIAIARAVLKNPEILLLDEATSALDVE 1154
Query: 644 SIDALADALDEFTGG--VVLVSHDSRLISRVCDDEERSQIWVVEDGTV 689
S + ALD V+V+H R+ + I V++DG +
Sbjct: 1155 SERVVQQALDRLMRDRTTVVVAH------RLSTIKNSDMISVIQDGKI 1196
Score = 65.9 bits (159), Expect = 8e-11
Identities = 58/198 (29%), Positives = 96/198 (48%), Gaps = 29/198 (14%)
Query: 490 EVSFSYPNREDFRLSD-VDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVR--- 545
+V+F+YP+R D + D ++ I G VA+VG +G+GKST+++L+ P++G V
Sbjct: 365 DVTFTYPSRPDVVIFDKLNFVIPAGKVVALVGGSGSGKSTMISLIERFYEPTDGAVMLDG 424
Query: 546 --------RSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYG 597
+ + IG +Q V T + Y ++ + +E A
Sbjct: 425 NDIRYLDLKWLRGHIGLVNQEPVLFATTIRENIMY------GKDDATSEEITNAAKLSEA 478
Query: 598 LPSHNHL-----TPI----VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDAL 648
+ N+L T + ++LSGGQK R+ + + P ILLLDE T+ LD +S +
Sbjct: 479 ISFINNLPEGFETQVGERGIQLSGGQKQRISISRAIVKNPSILLLDEATSALDAESEKIV 538
Query: 649 ADALDEFTGG--VVLVSH 664
+ALD G V+V+H
Sbjct: 539 QEALDRVMVGRTTVVVAH 556
Score = 57.8 bits (138), Expect = 2e-08
Identities = 66/242 (27%), Positives = 114/242 (46%), Gaps = 39/242 (16%)
Query: 137 RASVLDGDDGADANVKDITIE----NFSVAARGK-ELLKNTSVKISHGKRYGLIGPNGMG 191
R + + GD G + + + TIE +FS +R + + ++ + GK L+G +G G
Sbjct: 963 RRTQVVGDTGEELSNVEGTIELKGVHFSYPSRPDVTIFSDFNLLVPSGKSMALVGQSGSG 1022
Query: 192 KSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELIKVRQKVADL---- 247
KS++L L+ P +++++ G D L+ + S + V+Q+ A
Sbjct: 1023 KSSVLSLVLRFYDPTA---GIIMID----GQDIKKLK-LKSLRRHIGLVQQEPALFATTI 1074
Query: 248 -QNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTKDMQG 306
+NI G+EG + + E KLA + + S E ++K+ G
Sbjct: 1075 YENILYGKEGASESEVME----AAKLANAHSFI----SSLPEGYSTKV-----------G 1115
Query: 307 RPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRW--KKTLVVV 364
SGG R RI++ARA+ P +LLLDE T+ LD+ + +++ L R +T VVV
Sbjct: 1116 ERGIQMSGGQRQRIAIARAVLKNPEILLLDEATSALDVESERVVQQALDRLMRDRTTVVV 1175
Query: 365 SH 366
+H
Sbjct: 1176 AH 1177
Score = 53.5 bits (127), Expect = 4e-07
Identities = 63/228 (27%), Positives = 104/228 (44%), Gaps = 30/228 (13%)
Query: 143 GDDGADANVKDITIENFSVAARGKELL-KNTSVKISHGKRYGLIGPNGMGKSTLLKLLAW 201
G+ D KD+T F+ +R ++ + I GK L+G +G GKST++ L+
Sbjct: 355 GNVNGDILFKDVT---FTYPSRPDVVIFDKLNFVIPAGKVVALVGGSGSGKSTMISLI-- 409
Query: 202 RKIPVPKNIDVLLVEQEVVGDDKTALEAVVS-ANVELIKVRQKVADLQNIASGEEGMDKD 260
+ P + V+L ++ D L + N E + + + NI G++ D
Sbjct: 410 ERFYEPTDGAVMLDGNDIRYLDLKWLRGHIGLVNQEPVLFATTIRE--NIMYGKD----D 463
Query: 261 DTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRI 320
T+EE KL+E + + + E+Q G SGG + RI
Sbjct: 464 ATSEEITNAAKLSEAISFINNL-PEGFETQV--------------GERGIQLSGGQKQRI 508
Query: 321 SLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWK--KTLVVVSH 366
S++RA+ P++LLLDE T+ LD + ++E L R +T VVV+H
Sbjct: 509 SISRAIVKNPSILLLDEATSALDAESEKIVQEALDRVMVGRTTVVVAH 556
>At3g28860 P-glycoprotein, putative
Length = 1252
Score = 72.8 bits (177), Expect = 6e-13
Identities = 68/231 (29%), Positives = 107/231 (45%), Gaps = 25/231 (10%)
Query: 477 EPTELTPPLLQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNL--- 532
+P E ++ V F+YP+R D + D ++ I G A+VG +G+GKS+++ +
Sbjct: 1001 DPVETIRGDIEFRHVDFAYPSRPDVMVFRDFNLRIRAGHSQALVGASGSGKSSVIAMIER 1060
Query: 533 ----LAGDLVPSEGEVRR----SQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLS 584
LAG ++ ++RR S +L+IG Q + Y + E +
Sbjct: 1061 FYDPLAGKVMIDGKDIRRLNLKSLRLKIGLVQQEPALFAATIFDNIAYGKDGATESEVID 1120
Query: 585 KQEAVRAKLGKYGLPSHNHLTPI----VKLSGGQKARVVFTSISMSRPHILLLDEPTNHL 640
A A GLP + TP+ V+LSGGQK R+ + P +LLLDE T+ L
Sbjct: 1121 AARAANAHGFISGLPE-GYKTPVGERGVQLSGGQKQRIAIARAVLKNPTVLLLDEATSAL 1179
Query: 641 DMQSIDALADALDEFTGG--VVLVSHDSRLISRVCDDEERSQIWVVEDGTV 689
D +S L +AL+ G V+V+H I V I V++DG +
Sbjct: 1180 DAESECVLQEALERLMRGRTTVVVAHRLSTIRGV------DCIGVIQDGRI 1224
Score = 71.2 bits (173), Expect = 2e-12
Identities = 65/237 (27%), Positives = 114/237 (47%), Gaps = 38/237 (16%)
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
++ +V+FSYP+R D + + ++ G VA+VG +G+GKST+++L+ P+ G++
Sbjct: 365 IEFKDVTFSYPSRPDVMIFRNFNIFFPSGKTVAVVGGSGSGKSTVVSLIERFYDPNSGQI 424
Query: 545 RRS----QKLRIGRYSQHFVDLLTMDE-----TPVQYLLRLHPDQEGLSKQEAVRAKLGK 595
+ L++ ++ + + L+ + T ++ +L PD + + A A
Sbjct: 425 LLDGVEIKTLQL-KFLREQIGLVNQEPALFATTILENILYGKPDATMVEVEAAASAA--- 480
Query: 596 YGLPSHNHLTPI------------VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQ 643
+H+ +T + V+LSGGQK R+ + P ILLLDE T+ LD
Sbjct: 481 ---NAHSFITLLPKGYDTQVGERGVQLSGGQKQRIAIARAMLKDPKILLLDEATSALDAS 537
Query: 644 SIDALADALDEFTGG--VVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFED 698
S + +ALD G V+V+H R+C I V++ G V GT E+
Sbjct: 538 SESIVQEALDRVMVGRTTVVVAH------RLCTIRNVDSIAVIQQGQVVE-TGTHEE 587
Score = 54.7 bits (130), Expect = 2e-07
Identities = 69/255 (27%), Positives = 114/255 (44%), Gaps = 51/255 (20%)
Query: 134 IGSRASVLDGD-----DGADANVK-----DITIENFSVAARGKE---LLKNTSVKISHGK 180
+GS SVLD D ADA+ DI + A + + ++ +++I G
Sbjct: 980 VGSVFSVLDRQTRIDPDDADADPVETIRGDIEFRHVDFAYPSRPDVMVFRDFNLRIRAGH 1039
Query: 181 RYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELIKV 240
L+G +G GKS+++ + + + D L + + G D L + S +++ V
Sbjct: 1040 SQALVGASGSGKSSVIAM-------IERFYDPLAGKVMIDGKDIRRLN-LKSLRLKIGLV 1091
Query: 241 RQKVA-----DLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKIL 295
+Q+ A NIA G++G + + + A + A +
Sbjct: 1092 QQEPALFAATIFDNIAYGKDGATESEVIDA--------------------ARAANAHGFI 1131
Query: 296 AGL--GFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEY 353
+GL G+ K G SGG + RI++ARA+ PT+LLLDE T+ LD + L+E
Sbjct: 1132 SGLPEGY-KTPVGERGVQLSGGQKQRIAIARAVLKNPTVLLLDEATSALDAESECVLQEA 1190
Query: 354 LCRWK--KTLVVVSH 366
L R +T VVV+H
Sbjct: 1191 LERLMRGRTTVVVAH 1205
Score = 54.3 bits (129), Expect = 2e-07
Identities = 65/236 (27%), Positives = 107/236 (44%), Gaps = 38/236 (16%)
Query: 141 LDGD--DGADANV--KDITIENFSVAARGKELL-KNTSVKISHGKRYGLIGPNGMGKSTL 195
LDG D N+ KD+T FS +R ++ +N ++ GK ++G +G GKST+
Sbjct: 353 LDGKCLDQVHGNIEFKDVT---FSYPSRPDVMIFRNFNIFFPSGKTVAVVGGSGSGKSTV 409
Query: 196 LKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELIKVRQKV---ADLQNIAS 252
+ L+ + P + +LL E+ KT + + L+ + L+NI
Sbjct: 410 VSLI--ERFYDPNSGQILLDGVEI----KTLQLKFLREQIGLVNQEPALFATTILENILY 463
Query: 253 GEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSF 312
G+ D T E +A A + + L+ G+ + R +
Sbjct: 464 GKP----DATMVEVEAAASAANAHSFITLLPK--------------GYDTQVGERGVQ-L 504
Query: 313 SGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWK--KTLVVVSH 366
SGG + RI++ARA+ P +LLLDE T+ LD + ++E L R +T VVV+H
Sbjct: 505 SGGQKQRIAIARAMLKDPKILLLDEATSALDASSESIVQEALDRVMVGRTTVVVAH 560
>At1g28010 hypothetical protein
Length = 1247
Score = 72.4 bits (176), Expect = 8e-13
Identities = 64/240 (26%), Positives = 109/240 (44%), Gaps = 27/240 (11%)
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
++ VSF+YP R + + ++++ + G +A+VGP+G+GKST++ L+ PS G +
Sbjct: 1006 IEFRNVSFAYPTRPEIAIFKNLNLRVSAGKSLAVVGPSGSGKSTVIGLIMRFYDPSNGNL 1065
Query: 545 R------RSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPD----QEGLSKQEAVRAKLG 594
+S LR R L + + P + +H + E S+ E + A
Sbjct: 1066 CIDGHDIKSVNLRSLRKK-----LALVQQEPALFSTSIHENIKYGNENASEAEIIEAAKA 1120
Query: 595 K-----YGLPSHNHLTPI----VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSI 645
++T + V+LSGGQK RV + P +LLLDE T+ LD +
Sbjct: 1121 ANAHEFISRMEEGYMTHVGDKGVQLSGGQKQRVAIARAVLKDPSVLLLDEATSALDTSAE 1180
Query: 646 DALADALDEFTGG--VVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDL 703
+ +ALD+ G +LV+H I + + VVE G+ R + + + L
Sbjct: 1181 KQVQEALDKLMKGRTTILVAHRLSTIRKADTIVVLHKGKVVEKGSHRELVSKSDGFYKKL 1240
Score = 66.6 bits (161), Expect = 5e-11
Identities = 61/221 (27%), Positives = 103/221 (46%), Gaps = 22/221 (9%)
Query: 486 LQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVR 545
++ VSF+YP+R + ++ I G A VGP+G+GKST+++++ P GE+
Sbjct: 373 IEFCGVSFAYPSRPNMVFENLSFTIHSGKTFAFVGPSGSGKSTIISMVQRFYEPRSGEIL 432
Query: 546 ------RSQKLRIGRYSQHFVDL-LTMDETPVQYLLRLHPDQEGLSK-QEAVRAKLGKYG 597
++ KL+ R V + T + + L ++ + + EA +A
Sbjct: 433 LDGNDIKNLKLKWLREQMGLVSQEPALFATTIASNILLGKEKANMDQIIEAAKAANADSF 492
Query: 598 LPS--HNHLTPI----VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADA 651
+ S + + T + +LSGGQK R+ + P ILLLDE T+ LD +S + A
Sbjct: 493 IKSLPNGYNTQVGEGGTQLSGGQKQRIAIARAVLRNPKILLLDEATSALDAESEKIVQQA 552
Query: 652 LDEF--TGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVR 690
LD ++++H I V +I V+ DG VR
Sbjct: 553 LDNVMEKRTTIVIAHRLSTIRNV------DKIVVLRDGQVR 587
Score = 62.4 bits (150), Expect = 9e-10
Identities = 58/219 (26%), Positives = 103/219 (46%), Gaps = 27/219 (12%)
Query: 153 DITIENFSVAARGKE---LLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKN 209
DI N S A + + KN ++++S GK ++GP+G GKST++ L+ + P N
Sbjct: 1005 DIEFRNVSFAYPTRPEIAIFKNLNLRVSAGKSLAVVGPSGSGKSTVIGLIM--RFYDPSN 1062
Query: 210 IDVLLVEQEVVGDDKTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAG 269
++ + ++ +V L +R+K+A +Q E + +E G
Sbjct: 1063 GNLCIDGHDI-------------KSVNLRSLRKKLALVQQ----EPALFSTSIHENIKYG 1105
Query: 270 EKLAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQ 329
+ A E ++ + A S++ G G SGG + R+++ARA+
Sbjct: 1106 NENASEAEIIEAAKAANAHEFISRMEEGY---MTHVGDKGVQLSGGQKQRVAIARAVLKD 1162
Query: 330 PTLLLLDEPTNHLDLRAVLWLEEYLCRWKK--TLVVVSH 366
P++LLLDE T+ LD A ++E L + K T ++V+H
Sbjct: 1163 PSVLLLDEATSALDTSAEKQVQEALDKLMKGRTTILVAH 1201
Score = 62.0 bits (149), Expect = 1e-09
Identities = 54/228 (23%), Positives = 108/228 (46%), Gaps = 25/228 (10%)
Query: 158 NFSVAARGKELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQ 217
+F+ +R + +N S I GK + +GP+G GKST++ ++ ++ P++ ++LL
Sbjct: 379 SFAYPSRPNMVFENLSFTIHSGKTFAFVGPSGSGKSTIISMV--QRFYEPRSGEILLDGN 436
Query: 218 EVVGDDKTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYE 277
++ N++L +R+++ + S E + G++ A + +
Sbjct: 437 DI-------------KNLKLKWLREQMG----LVSQEPALFATTIASNILLGKEKANMDQ 479
Query: 278 QLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDE 337
++ + A+S + G G SGG + RI++ARA+ P +LLLDE
Sbjct: 480 IIEAAKAANADSFIKSLPNGYN---TQVGEGGTQLSGGQKQRIAIARAVLRNPKILLLDE 536
Query: 338 PTNHLDLRAVLWLEEYL--CRWKKTLVVVSHDRDFLNTVCSEIIHLHD 383
T+ LD + +++ L K+T +V++H + V +I+ L D
Sbjct: 537 ATSALDAESEKIVQQALDNVMEKRTTIVIAHRLSTIRNV-DKIVVLRD 583
>At1g67940 ABC transporter like protein
Length = 263
Score = 72.0 bits (175), Expect = 1e-12
Identities = 69/213 (32%), Positives = 103/213 (47%), Gaps = 38/213 (17%)
Query: 161 VAARGKELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVV 220
VA G +LK ++ I G G+IGP+G GKST L+ L ++ P V L +++
Sbjct: 37 VADDGSRILKGVTIDIPKGMIVGVIGPSGSGKSTFLRSL--NRLWEPPESTVFLDGEDIT 94
Query: 221 GDDKTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDA-GEKLA--ELYE 277
NV++I +R++V L + +G D+ + GEKL+ E+Y+
Sbjct: 95 -------------NVDVIALRRRVGMLFQLPVLFQGTVADNVRYGPNLRGEKLSDEEVYK 141
Query: 278 QLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDE 337
L L DA S A K A L S G R++LAR L +P +LLLDE
Sbjct: 142 LLSLADLDA--SFAKKTGAEL--------------SVGQAQRVALARTLANEPEVLLLDE 185
Query: 338 PTNHLDLRAVLWLEEYLCRWKK----TLVVVSH 366
PT+ LD + +E+ + + KK T V+VSH
Sbjct: 186 PTSALDPISTENIEDVIVKLKKQRGITTVIVSH 218
Score = 55.1 bits (131), Expect = 1e-07
Identities = 52/185 (28%), Positives = 79/185 (42%), Gaps = 14/185 (7%)
Query: 503 LSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEG-------EVRRSQKLRIGRY 555
L V + I G V ++GP+G+GKST L L P E ++ + + R
Sbjct: 45 LKGVTIDIPKGMIVGVIGPSGSGKSTFLRSLNRLWEPPESTVFLDGEDITNVDVIALRRR 104
Query: 556 SQHFVDLLTMDETPVQYLLRLHPDQEG--LSKQEAVRAKLGKYGLPSHNHLTPIVKLSGG 613
L + + V +R P+ G LS +E + L L + +LS G
Sbjct: 105 VGMLFQLPVLFQGTVADNVRYGPNLRGEKLSDEEVYKL-LSLADLDASFAKKTGAELSVG 163
Query: 614 QKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTG----GVVLVSHDSRLI 669
Q RV + P +LLLDEPT+ LD S + + D + + V+VSH + I
Sbjct: 164 QAQRVALARTLANEPEVLLLDEPTSALDPISTENIEDVIVKLKKQRGITTVIVSHSIKQI 223
Query: 670 SRVCD 674
+V D
Sbjct: 224 QKVAD 228
>At2g47000 putative ABC transporter
Length = 1286
Score = 71.2 bits (173), Expect = 2e-12
Identities = 75/259 (28%), Positives = 113/259 (42%), Gaps = 66/259 (25%)
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
++L VSF YP R D ++ D+ + I G VA+VG +G+GKST++ LL P GE+
Sbjct: 1042 IELRHVSFKYPARPDVQIFQDLCLSIRAGKTVALVGESGSGKSTVIALLQRFYDPDSGEI 1101
Query: 545 RRSQKLRIGRYSQHFVDLLTMDETPVQYL-LRLHPDQEGLSKQ------EAVRAKL--GK 595
T+D ++ L L+ Q GL Q E +RA + GK
Sbjct: 1102 -------------------TLDGVEIKSLRLKWLRQQTGLVSQEPILFNETIRANIAYGK 1142
Query: 596 YGLPSHNHLTPI-------------------------VKLSGGQKARVVFTSISMSRPHI 630
G S + + ++LSGGQK RV + P +
Sbjct: 1143 GGDASESEIVSSAELSNAHGFISGLQQGYDTMVGERGIQLSGGQKQRVAIARAIVKDPKV 1202
Query: 631 LLLDEPTNHLDMQSIDALADALDEFTGGVVLVSHDSRLIS-RVCDDEERSQIWVVEDGTV 689
LLLDE T+ LD +S + DALD V+V+ + +++ R+ + I VV++G +
Sbjct: 1203 LLLDEATSALDAESERVVQDALDR-----VMVNRTTIVVAHRLSTIKNADVIAVVKNGVI 1257
Query: 690 RNFPGTFEDYKEDLLKEIK 708
E K D L IK
Sbjct: 1258 ------VEKGKHDTLINIK 1270
Score = 63.9 bits (154), Expect = 3e-10
Identities = 72/235 (30%), Positives = 108/235 (45%), Gaps = 31/235 (13%)
Query: 136 SRASVLDGDDGADANVKDITIENFSVAARGKE-LLKNTSVKISHGKRYGLIGPNGMGKST 194
+ VLD G D +KD+ F+ AR E + + S+ IS G L+G +G GKST
Sbjct: 372 TNGKVLDDIKG-DIELKDVY---FTYPARPDEQIFRGFSLFISSGTTVALVGQSGSGKST 427
Query: 195 LLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVS-ANVELIKVRQKVADLQNIASG 253
++ L+ + P+ DVL+ + + + + + E + + D NIA G
Sbjct: 428 VVSLI--ERFYDPQAGDVLIDGINLKEFQLKWIRSKIGLVSQEPVLFTASIKD--NIAYG 483
Query: 254 EEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFS 313
+E D T EE A +LA A K+ GL M G S
Sbjct: 484 KE----DATTEEIKAAAELAN------------ASKFVDKLPQGLD---TMVGEHGTQLS 524
Query: 314 GGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCR--WKKTLVVVSH 366
GG + RI++ARA+ P +LLLDE T+ LD + ++E L R +T VVV+H
Sbjct: 525 GGQKQRIAVARAILKDPRILLLDEATSALDAESERVVQEALDRIMVNRTTVVVAH 579
Score = 63.9 bits (154), Expect = 3e-10
Identities = 61/198 (30%), Positives = 95/198 (47%), Gaps = 21/198 (10%)
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLL-------AGDL 537
++L +V F+YP R D ++ + I GT VA+VG +G+GKST+++L+ AGD+
Sbjct: 384 IELKDVYFTYPARPDEQIFRGFSLFISSGTTVALVGQSGSGKSTVVSLIERFYDPQAGDV 443
Query: 538 VPSEGEVRRSQ----KLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKL 593
+ ++ Q + +IG SQ V + + Y +E + E A
Sbjct: 444 LIDGINLKEFQLKWIRSKIGLVSQEPVLFTASIKDNIAYGKEDATTEEIKAAAELANASK 503
Query: 594 GKYGLPSHNHLTPIV-----KLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDAL 648
LP L +V +LSGGQK R+ + P ILLLDE T+ LD +S +
Sbjct: 504 FVDKLPQG--LDTMVGEHGTQLSGGQKQRIAVARAILKDPRILLLDEATSALDAESERVV 561
Query: 649 ADALDEF--TGGVVLVSH 664
+ALD V+V+H
Sbjct: 562 QEALDRIMVNRTTVVVAH 579
Score = 55.1 bits (131), Expect = 1e-07
Identities = 68/266 (25%), Positives = 117/266 (43%), Gaps = 40/266 (15%)
Query: 118 AKKEALRDDRDAFTVVIGSRASVLDGDDGADANVK----------DITIENFSV---AAR 164
++ +L D V S +++D + D +V+ DI + + S A
Sbjct: 996 SQSSSLSPDSSKADVAAASIFAIMDRESKIDPSVESGRVLDNVKGDIELRHVSFKYPARP 1055
Query: 165 GKELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDD- 223
++ ++ + I GK L+G +G GKST++ LL ++ P + ++ L E+
Sbjct: 1056 DVQIFQDLCLSIRAGKTVALVGESGSGKSTVIALL--QRFYDPDSGEITLDGVEIKSLRL 1113
Query: 224 KTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMG 283
K + + E I + + NIA G+ G D +E E +
Sbjct: 1114 KWLRQQTGLVSQEPILFNETIR--ANIAYGKGG----DASESE---------------IV 1152
Query: 284 SDAAESQASKILAGLGFTKD-MQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHL 342
S A S A ++GL D M G SGG + R+++ARA+ P +LLLDE T+ L
Sbjct: 1153 SSAELSNAHGFISGLQQGYDTMVGERGIQLSGGQKQRVAIARAIVKDPKVLLLDEATSAL 1212
Query: 343 DLRAVLWLEEYLCR--WKKTLVVVSH 366
D + +++ L R +T +VV+H
Sbjct: 1213 DAESERVVQDALDRVMVNRTTIVVAH 1238
>At4g25960 P-glycoprotein-2 (pgp2)
Length = 1233
Score = 70.5 bits (171), Expect = 3e-12
Identities = 56/202 (27%), Positives = 102/202 (49%), Gaps = 29/202 (14%)
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
++L V FSYP+R D + D D+ + G +A+VG +G+GKS++++L+ P+ G+V
Sbjct: 990 IELKGVHFSYPSRPDVVIFRDFDLIVRAGKSMALVGQSGSGKSSVISLILRFYDPTAGKV 1049
Query: 545 ----RRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPD----QEGLSKQEAVRAKLGKY 596
+ +KL + +H + + + P + ++ + EG S+ E V + +
Sbjct: 1050 MIEGKDIKKLDLKALRKH---IGLVQQEPALFATTIYENILYGNEGASQSEVVESAMLAN 1106
Query: 597 GLPSHNHLTPI------------VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQS 644
+H+ +T + V++SGGQ+ R+ + P ILLLDE T+ LD++S
Sbjct: 1107 ---AHSFITSLPEGYSTKVGERGVQMSGGQRQRIAIARAILKNPAILLLDEATSALDVES 1163
Query: 645 IDALADALDEFTGG--VVLVSH 664
+ ALD V+V+H
Sbjct: 1164 ERVVQQALDRLMANRTTVVVAH 1185
Score = 68.6 bits (166), Expect = 1e-11
Identities = 65/226 (28%), Positives = 112/226 (48%), Gaps = 27/226 (11%)
Query: 486 LQLIEVSFSYPNREDFRLSD-VDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
+Q + +FSYP+R D + D +++ I G VA+VG +G+GKST+++L+ P G V
Sbjct: 361 IQFKDATFSYPSRPDVVIFDRLNLAIPAGKIVALVGGSGSGKSTVISLIERFYEPISGAV 420
Query: 545 ----RRSQKLRIGRYSQHFVDLLTMDETPVQYLLR---LHPDQEGLSKQEAVRAKLGKYG 597
+L I ++ + + L+ + +R L+ + +++ AKL +
Sbjct: 421 LLDGNNISELDI-KWLRGQIGLVNQEPALFATTIRENILYGKDDATAEEITRAAKLSE-A 478
Query: 598 LPSHNHL-----TPI----VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDAL 648
+ N+L T + ++LSGGQK R+ + + P ILLLDE T+ LD +S ++
Sbjct: 479 ISFINNLPEGFETQVGERGIQLSGGQKQRIAISRAIVKNPSILLLDEATSALDAESEKSV 538
Query: 649 ADALDEFTGG--VVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNF 692
+ALD G V+V+H R+ I VV +G + F
Sbjct: 539 QEALDRVMVGRTTVVVAH------RLSTVRNADIIAVVHEGKIVEF 578
Score = 55.1 bits (131), Expect = 1e-07
Identities = 60/237 (25%), Positives = 108/237 (45%), Gaps = 29/237 (12%)
Query: 137 RASVLDGDDGADANVKDITIE----NFSVAARGKELL-KNTSVKISHGKRYGLIGPNGMG 191
R + + G+ + N + TIE +FS +R ++ ++ + + GK L+G +G G
Sbjct: 971 RKTQIVGETSEELNNVEGTIELKGVHFSYPSRPDVVIFRDFDLIVRAGKSMALVGQSGSG 1030
Query: 192 KSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSANVELIKVRQKVADLQNIA 251
KS+++ L+ + P V++ +++ D AL + V+ +NI
Sbjct: 1031 KSSVISLIL--RFYDPTAGKVMIEGKDIKKLDLKALRKHIGL-VQQEPALFATTIYENIL 1087
Query: 252 SGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKS 311
G EG + + E LA + + + E ++K+ G
Sbjct: 1088 YGNEGASQSEVVESA----MLANAHSFI----TSLPEGYSTKV-----------GERGVQ 1128
Query: 312 FSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRW--KKTLVVVSH 366
SGG R RI++ARA+ P +LLLDE T+ LD+ + +++ L R +T VVV+H
Sbjct: 1129 MSGGQRQRIAIARAILKNPAILLLDEATSALDVESERVVQQALDRLMANRTTVVVAH 1185
Score = 48.9 bits (115), Expect = 1e-05
Identities = 56/197 (28%), Positives = 89/197 (44%), Gaps = 26/197 (13%)
Query: 173 SVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVS 232
++ I GK L+G +G GKST++ L+ + P + VLL + D L +
Sbjct: 383 NLAIPAGKIVALVGGSGSGKSTVISLI--ERFYEPISGAVLLDGNNISELDIKWLRGQIG 440
Query: 233 -ANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQA 291
N E + +NI G++ D T EE KL+E + + + E+Q
Sbjct: 441 LVNQEPALFATTIR--ENILYGKD----DATAEEITRAAKLSEAISFINNL-PEGFETQV 493
Query: 292 SKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLE 351
G SGG + RI+++RA+ P++LLLDE T+ LD + ++
Sbjct: 494 --------------GERGIQLSGGQKQRIAISRAIVKNPSILLLDEATSALDAESEKSVQ 539
Query: 352 EYLCRWK--KTLVVVSH 366
E L R +T VVV+H
Sbjct: 540 EALDRVMVGRTTVVVAH 556
>At4g01830 putative P-glycoprotein-like protein
Length = 1230
Score = 70.5 bits (171), Expect = 3e-12
Identities = 66/232 (28%), Positives = 114/232 (48%), Gaps = 28/232 (12%)
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
++L +SF+Y R D ++ D+ + I G VA+VG +G+GKST+++LL P G +
Sbjct: 985 IELCHISFTYQTRPDVQVFRDLCLSIRAGQTVALVGESGSGKSTVISLLQRFYDPDSGHI 1044
Query: 545 R----RSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKL--GKYGL 598
+KLR+ ++ + + L+ + +R + G +EA A++
Sbjct: 1045 TLDGVELKKLRL-KWLRQQMGLVGQEPVLFNDTIRAN-IAYGKGGEEATEAEIIAASELA 1102
Query: 599 PSHNHLTPI------------VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSID 646
+H ++ I ++LSGGQK RV + P ILLLDE T+ LD +S
Sbjct: 1103 NAHRFISSIQKGYDTVVGERGIQLSGGQKQRVAIARAIVKEPKILLLDEATSALDAESER 1162
Query: 647 ALADALDEFTGGVVLVSHDSRLIS-RVCDDEERSQIWVVEDGTVRNFPGTFE 697
+ DALD V+V+ + +++ R+ + I VV++G + GT E
Sbjct: 1163 VVQDALDR-----VMVNRTTIVVAHRLSTIKNADVIAVVKNGVIAE-KGTHE 1208
Score = 61.2 bits (147), Expect = 2e-09
Identities = 81/289 (28%), Positives = 122/289 (42%), Gaps = 50/289 (17%)
Query: 159 FSVAARGKE-LLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQ 217
FS AR KE + S+ I G L+G +G GKST++ L+ + P + VL+
Sbjct: 360 FSYPARPKEEVFGGFSLLIPSGTTTALVGESGSGKSTVISLI--ERFYDPNSGQVLIDGV 417
Query: 218 EVVGDDKTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYE 277
++ + + V V + ++NI G+EG T EE A KLA
Sbjct: 418 DLKEFQLKWIRGKIGL-VSQEPVLFSSSIMENIGYGKEGA----TVEEIQAASKLAN--- 469
Query: 278 QLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDE 337
A K+ GL + + G SGG + RI++ARA+ P +LLLDE
Sbjct: 470 ---------AAKFIDKLPLGL---ETLVGEHGTQLSGGQKQRIAIARAILKDPRILLLDE 517
Query: 338 PTNHLDLRAVLWLEEYLCR--WKKTLVVVSH------DRDFL-----NTVCSEIIHLHDL 384
T+ LD + ++E L R +T V+V+H + D + + E H L
Sbjct: 518 ATSALDAESERVVQEALDRIMVNRTTVIVAHRLSTVRNADIIAVIHRGKIVEEGSHSELL 577
Query: 385 KLHFYRGNFDAFESGYEQ--RRREAN---KKYEIFDKQLKAARRTGNKA 428
K H E Y Q R +E N K+ EI D + + GN +
Sbjct: 578 KDH---------EGAYSQLLRLQEINKESKRLEISDGSISSGSSRGNNS 617
Score = 58.2 bits (139), Expect = 2e-08
Identities = 59/202 (29%), Positives = 94/202 (46%), Gaps = 29/202 (14%)
Query: 486 LQLIEVSFSYPNR-EDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
++L +V FSYP R ++ + I GT A+VG +G+GKST+++L+ P+ G+V
Sbjct: 353 IELRDVCFSYPARPKEEVFGGFSLLIPSGTTTALVGESGSGKSTVISLIERFYDPNSGQV 412
Query: 545 R-----------RSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQE----AV 589
+ + +IG SQ V + + Y +EG + +E +
Sbjct: 413 LIDGVDLKEFQLKWIRGKIGLVSQEPVLFSSSIMENIGY------GKEGATVEEIQAASK 466
Query: 590 RAKLGKYGLPSHNHLTPIV-----KLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQS 644
A K+ L +V +LSGGQK R+ + P ILLLDE T+ LD +S
Sbjct: 467 LANAAKFIDKLPLGLETLVGEHGTQLSGGQKQRIAIARAILKDPRILLLDEATSALDAES 526
Query: 645 IDALADALDEF--TGGVVLVSH 664
+ +ALD V+V+H
Sbjct: 527 ERVVQEALDRIMVNRTTVIVAH 548
Score = 53.1 bits (126), Expect = 5e-07
Identities = 52/203 (25%), Positives = 94/203 (45%), Gaps = 24/203 (11%)
Query: 167 ELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTA 226
++ ++ + I G+ L+G +G GKST++ LL P +I +
Sbjct: 1001 QVFRDLCLSIRAGQTVALVGESGSGKSTVISLLQRFYDPDSGHITL-------------- 1046
Query: 227 LEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDA 286
VEL K+R K Q G+E + +DT A K E + +++ +
Sbjct: 1047 ------DGVELKKLRLKWLRQQMGLVGQEPVLFNDTIRANIAYGKGGEEATEAEIIAASE 1100
Query: 287 AESQASKILAGLGFTKD-MQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLR 345
+ A + ++ + D + G SGG + R+++ARA+ +P +LLLDE T+ LD
Sbjct: 1101 L-ANAHRFISSIQKGYDTVVGERGIQLSGGQKQRVAIARAIVKEPKILLLDEATSALDAE 1159
Query: 346 AVLWLEEYLCR--WKKTLVVVSH 366
+ +++ L R +T +VV+H
Sbjct: 1160 SERVVQDALDRVMVNRTTIVVAH 1182
>At1g70610 transporter associated with antigen processing-like
protein (TAP1)
Length = 700
Score = 70.5 bits (171), Expect = 3e-12
Identities = 55/173 (31%), Positives = 89/173 (50%), Gaps = 14/173 (8%)
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
++ ++VSFSYP+R++ + +V++ + G VAIVG +G+GKSTL+NLL P+ G++
Sbjct: 455 IEFVDVSFSYPSRDEVAVVQNVNISVHPGEVVAIVGLSGSGKSTLVNLLLQLYEPTSGQI 514
Query: 545 R-----------RSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEG-LSKQEAVRAK 592
+ + RIG Q T + ++Y + QE +S + A
Sbjct: 515 LLDGVPLKELDVKWLRQRIGYVGQEPKLFRTDISSNIKYGCDRNISQEDIISAAKQAYAH 574
Query: 593 LGKYGLPS-HNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQS 644
LP+ +N + LSGGQK R+ + P IL+LDE T+ LD +S
Sbjct: 575 DFITALPNGYNTIVDDDLLSGGQKQRIAIARAILRDPRILILDEATSALDAES 627
Score = 47.8 bits (112), Expect = 2e-05
Identities = 52/228 (22%), Positives = 100/228 (43%), Gaps = 54/228 (23%)
Query: 158 NFSVAARGK-ELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIP---------VP 207
+FS +R + +++N ++ + G+ ++G +G GKSTL+ LL P VP
Sbjct: 461 SFSYPSRDEVAVVQNVNISVHPGEVVAIVGLSGSGKSTLVNLLLQLYEPTSGQILLDGVP 520
Query: 208 -KNIDVLLVEQEV--VGDDKTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNE 264
K +DV + Q + VG + +S+N++ G D++ + E
Sbjct: 521 LKELDVKWLRQRIGYVGQEPKLFRTDISSNIKY------------------GCDRNISQE 562
Query: 265 EEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLAR 324
+ + K A ++ + + + +L SGG + RI++AR
Sbjct: 563 DIISAAKQAYAHDFITALPNGYNTIVDDDLL-----------------SGGQKQRIAIAR 605
Query: 325 ALFVQPTLLLLDEPTNHLD------LRAVLWLEEYLCRWKKTLVVVSH 366
A+ P +L+LDE T+ LD ++ VL K++++V++H
Sbjct: 606 AILRDPRILILDEATSALDAESEHNVKGVLRSIGNDSATKRSVIVIAH 653
>At4g18050 multidrug resistance protein/P-glycoprotein - like
Length = 1323
Score = 70.1 bits (170), Expect = 4e-12
Identities = 63/237 (26%), Positives = 115/237 (47%), Gaps = 29/237 (12%)
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
++ VSF YP R D ++ D+ + I G VA+VG +G+GKST+++++ P G++
Sbjct: 1035 IEFRHVSFRYPMRPDVQIFRDLCLTIPSGKTVALVGESGSGKSTVISMIERFYNPDSGKI 1094
Query: 545 RRSQ-KLRIGRYSQHFVDLLTMDETPVQYLLRLHPD-----QEGLSKQEAVRAKLGKYGL 598
Q +++ + S + + + P+ + + + G +++E + A
Sbjct: 1095 LIDQVEIQTFKLSWLRQQMGLVSQEPILFNETIRSNIAYGKTGGATEEEIIAAAKAAN-- 1152
Query: 599 PSHNHLTPI------------VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSID 646
+HN ++ + V+LSGGQK R+ + P ILLLDE T+ LD +S
Sbjct: 1153 -AHNFISSLPQGYDTSVGERGVQLSGGQKQRIAIARAILKDPKILLLDEATSALDAESER 1211
Query: 647 ALADALDEFTGGVVLVSHDSRLIS-RVCDDEERSQIWVVEDGTVRNFPGTFEDYKED 702
+ DALD V+V+ + +++ R+ + I VV++G + G E ED
Sbjct: 1212 VVQDALDR-----VMVNRTTVVVAHRLTTIKNADVIAVVKNGVIAE-KGRHETLDED 1262
Score = 65.9 bits (159), Expect = 8e-11
Identities = 71/261 (27%), Positives = 111/261 (42%), Gaps = 36/261 (13%)
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
++L +V F YP R D ++ + + + G VA+VG +G+GKST+++L+ P G+V
Sbjct: 355 IELKDVYFRYPARPDVQIFAGFSLFVPNGKTVALVGQSGSGKSTVISLIERFYDPESGQV 414
Query: 545 -------RRSQ----KLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKL 593
++ Q + +IG SQ V T + + Y DQE + E A
Sbjct: 415 LIDNIDLKKLQLKWIRSKIGLVSQEPVLFATTIKENIAYGKEDATDQEIRTAIELANAAK 474
Query: 594 GKYGLPSHNHLTPIV-----KLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDAL 648
LP L +V ++SGGQK R+ + P ILLLDE T+ LD +S +
Sbjct: 475 FIDKLPQG--LDTMVGEHGTQMSGGQKQRLAIARAILKNPKILLLDEATSALDAESERIV 532
Query: 649 ADALDEF-----------------TGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRN 691
DAL T V+ V H +++ + DE VR
Sbjct: 533 QDALVNLMSNRTTVVVAHRLTTIRTADVIAVVHQGKIVEKGTHDEMIQDPEGAYSQLVRL 592
Query: 692 FPGTFEDYKEDLLKEIKAEVD 712
G+ E+ E E +V+
Sbjct: 593 QEGSKEEATESERPETSLDVE 613
Score = 54.7 bits (130), Expect = 2e-07
Identities = 67/236 (28%), Positives = 107/236 (44%), Gaps = 39/236 (16%)
Query: 139 SVLDGDDGADANVKDITIENFSVAARGK-ELLKNTSVKISHGKRYGLIGPNGMGKSTLLK 197
SVL+ D D +KD+ F AR ++ S+ + +GK L+G +G GKST++
Sbjct: 346 SVLE-DIRGDIELKDVY---FRYPARPDVQIFAGFSLFVPNGKTVALVGQSGSGKSTVIS 401
Query: 198 LLAWRKIP-----VPKNIDVLLVEQEVVGDDKTALEAVVSANVELIKVRQKVADLQNIAS 252
L+ P + NID+ ++ + + + +VS L K +NIA
Sbjct: 402 LIERFYDPESGQVLIDNIDLKKLQLKWI----RSKIGLVSQEPVLFATTIK----ENIAY 453
Query: 253 GEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSF 312
G+E D T++E +LA A K+ GL M G
Sbjct: 454 GKE----DATDQEIRTAIELAN------------AAKFIDKLPQGLD---TMVGEHGTQM 494
Query: 313 SGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRW--KKTLVVVSH 366
SGG + R+++ARA+ P +LLLDE T+ LD + +++ L +T VVV+H
Sbjct: 495 SGGQKQRLAIARAILKNPKILLLDEATSALDAESERIVQDALVNLMSNRTTVVVAH 550
Score = 52.4 bits (124), Expect = 9e-07
Identities = 53/203 (26%), Positives = 94/203 (46%), Gaps = 25/203 (12%)
Query: 167 ELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTA 226
++ ++ + I GK L+G +G GKST++ ++ + P + +L+ + E+ +
Sbjct: 1051 QIFRDLCLTIPSGKTVALVGESGSGKSTVISMI--ERFYNPDSGKILIDQVEIQTFKLSW 1108
Query: 227 LEAVVS-ANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSD 285
L + + E I + + NIA G+ G T EE A K A + + +
Sbjct: 1109 LRQQMGLVSQEPILFNETIRS--NIAYGKTG---GATEEEIIAAAKAANAHNFISSLPQ- 1162
Query: 286 AAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLR 345
G+ + R + SGG + RI++ARA+ P +LLLDE T+ LD
Sbjct: 1163 -------------GYDTSVGERGVQ-LSGGQKQRIAIARAILKDPKILLLDEATSALDAE 1208
Query: 346 AVLWLEEYLCR--WKKTLVVVSH 366
+ +++ L R +T VVV+H
Sbjct: 1209 SERVVQDALDRVMVNRTTVVVAH 1231
>At1g65410 ABC transporter like protein
Length = 345
Score = 68.9 bits (167), Expect = 9e-12
Identities = 63/247 (25%), Positives = 107/247 (42%), Gaps = 55/247 (22%)
Query: 166 KELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVL------LVEQEV 219
K +LK S KI HG+ G+IGP+G GKST+LK++A P + + L+ E
Sbjct: 97 KHILKGVSFKIRHGEAVGVIGPSGTGKSTILKIMAGLLAPDKGEVYIRGKKRAGLISDEE 156
Query: 220 VGDDKTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQL 279
+ + L +A + + VR+ V L LYE+
Sbjct: 157 ISGLRIGLVFQSAALFDSLSVRENVGFL---------------------------LYERS 189
Query: 280 QLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALF-------VQPTL 332
++ + +E ++ LA +G K ++ R SGG + R++LAR+L ++P +
Sbjct: 190 KMSENQISE-LVTQTLAAVGL-KGVENRLPSELSGGMKKRVALARSLIFDTTKEVIEPEV 247
Query: 333 LLLDEPTNHLDLRAVLWLEEYLCRWKKT-------------LVVVSHDRDFLNTVCSEII 379
LL DEPT LD A +E+ + T +VV+H + ++
Sbjct: 248 LLYDEPTAGLDPIASTVVEDLIRSVHMTDEDAVGKPGKIASYLVVTHQHSTIQRAVDRLL 307
Query: 380 HLHDLKL 386
L++ K+
Sbjct: 308 FLYEGKI 314
Score = 62.0 bits (149), Expect = 1e-09
Identities = 62/207 (29%), Positives = 84/207 (39%), Gaps = 37/207 (17%)
Query: 503 LSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFV-- 560
L V I G V ++GP+G GKST+L ++AG L P +GEV K R G S +
Sbjct: 100 LKGVSFKIRHGEAVGVIGPSGTGKSTILKIMAGLLAPDKGEVYIRGKKRAGLISDEEISG 159
Query: 561 -------------DLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLPSHNHLTPI 607
D L++ E V +LL E V L GL + P
Sbjct: 160 LRIGLVFQSAALFDSLSVREN-VGFLLYERSKMSENQISELVTQTLAAVGLKGVENRLP- 217
Query: 608 VKLSGGQKARVVF-------TSISMSRPHILLLDEPTNHLDMQSIDALADAL------DE 654
+LSGG K RV T+ + P +LL DEPT LD + + D + DE
Sbjct: 218 SELSGGMKKRVALARSLIFDTTKEVIEPEVLLYDEPTAGLDPIASTVVEDLIRSVHMTDE 277
Query: 655 FTGG-------VVLVSHDSRLISRVCD 674
G ++V+H I R D
Sbjct: 278 DAVGKPGKIASYLVVTHQHSTIQRAVD 304
>At3g28380 P-glycoprotein, putative
Length = 1240
Score = 68.2 bits (165), Expect = 2e-11
Identities = 62/217 (28%), Positives = 100/217 (45%), Gaps = 38/217 (17%)
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
+ + V F+YP R D + + + ID G AIVG +G+GKST++ L+ P +G V
Sbjct: 995 ITFLNVDFAYPTRPDVVIFENFSIEIDEGKSTAIVGTSGSGKSTIIGLIERFYDPLKGTV 1054
Query: 545 R------RSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQ-------EAVRA 591
+ RS LR R ++ L++ + +R + G S + EA +A
Sbjct: 1055 KIDGRDIRSYHLRSLR---KYISLVSQEPMLFAGTIRENIMYGGTSDKIDESEIIEAAKA 1111
Query: 592 KLGKYGLPSHNHLTPI------------VKLSGGQKARVVFTSISMSRPHILLLDEPTNH 639
+H+ +T + V+LSGGQK R+ + P +LLLDE T+
Sbjct: 1112 A------NAHDFITSLSNGYDTNCGDKGVQLSGGQKQRIAIARAVLKNPSVLLLDEATSA 1165
Query: 640 LDMQSIDALADALDEFTGG--VVLVSHDSRLISRVCD 674
LD +S + DAL+ G ++++H I CD
Sbjct: 1166 LDSKSERVVQDALERVMVGRTSIMIAHRLSTIQN-CD 1201
Score = 57.4 bits (137), Expect = 3e-08
Identities = 53/197 (26%), Positives = 92/197 (45%), Gaps = 29/197 (14%)
Query: 491 VSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVRRS-- 547
V F+Y +R + + D+ + I G VA+VG +G+GKST+++LL P GE+
Sbjct: 364 VKFTYLSRPETTIFDDLCLKIPAGKTVALVGGSGSGKSTVISLLQRFYDPIAGEILIDGV 423
Query: 548 --QKLRIGRYSQHFVDLLTMDETPVQYLLRLHPD----QEGLSKQEAVRAKLGKYGLPSH 601
KL++ + + + PV + + + +E S E V A +H
Sbjct: 424 SIDKLQVNWLRSQ---MGLVSQEPVLFATSITENILFGKEDASLDEVVEAAKASN---AH 477
Query: 602 NHLTPI------------VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALA 649
++ V++SGGQK R+ + P ILLLDE T+ LD +S +
Sbjct: 478 TFISQFPLGYKTQVGERGVQMSGGQKQRIAIARAIIKSPKILLLDEATSALDSESERVVQ 537
Query: 650 DALDEFTGG--VVLVSH 664
++LD + G ++++H
Sbjct: 538 ESLDNASIGRTTIVIAH 554
Score = 52.4 bits (124), Expect = 9e-07
Identities = 53/195 (27%), Positives = 85/195 (43%), Gaps = 24/195 (12%)
Query: 174 VKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTALEAVVSA 233
+KI GK L+G +G GKST++ LL P+ I + + V DK + + S
Sbjct: 382 LKIPAGKTVALVGGSGSGKSTVISLLQRFYDPIAGEILI-----DGVSIDKLQVNWLRSQ 436
Query: 234 NVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAAESQASK 293
+ S E + E G++ A L E ++ + A + S+
Sbjct: 437 --------------MGLVSQEPVLFATSITENILFGKEDASLDEVVEAAKASNAHTFISQ 482
Query: 294 ILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEY 353
G K G SGG + RI++ARA+ P +LLLDE T+ LD + ++E
Sbjct: 483 FPLGY---KTQVGERGVQMSGGQKQRIAIARAIIKSPKILLLDEATSALDSESERVVQES 539
Query: 354 L--CRWKKTLVVVSH 366
L +T +V++H
Sbjct: 540 LDNASIGRTTIVIAH 554
Score = 47.0 bits (110), Expect = 4e-05
Identities = 48/201 (23%), Positives = 92/201 (44%), Gaps = 22/201 (10%)
Query: 168 LLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTAL 227
+ +N S++I GK ++G +G GKST++ L + + D L ++ G D
Sbjct: 1012 IFENFSIEIDEGKSTAIVGTSGSGKSTIIGL-------IERFYDPLKGTVKIDGRD---- 1060
Query: 228 EAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEKLAELYEQLQLMGSDAA 287
+ L +R+ ++ + S E + E G ++ E + + AA
Sbjct: 1061 ----IRSYHLRSLRKYIS----LVSQEPMLFAGTIRENIMYGGTSDKIDESEIIEAAKAA 1112
Query: 288 ESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAV 347
+ G+ + G SGG + RI++ARA+ P++LLLDE T+ LD ++
Sbjct: 1113 NAHDFITSLSNGYDTNC-GDKGVQLSGGQKQRIAIARAVLKNPSVLLLDEATSALDSKSE 1171
Query: 348 LWLEEYLCRWK--KTLVVVSH 366
+++ L R +T ++++H
Sbjct: 1172 RVVQDALERVMVGRTSIMIAH 1192
>At4g25450 putative protein
Length = 280
Score = 67.0 bits (162), Expect = 3e-11
Identities = 59/227 (25%), Positives = 110/227 (47%), Gaps = 31/227 (13%)
Query: 485 LLQLIEVSFSYPNREDFRLSD-VDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGE 543
+L ++V F+YP R D ++ D + + ++ GT A+VG +GAGKST++ LLA P++G
Sbjct: 35 ILSSVDVHFAYPLRPDVKVLDGLSLTLNSGTVTALVGSSGAGKSTIVQLLARFYEPTQGR 94
Query: 544 VR-RSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPD------QEGLSKQEAVRAKLGKY 596
+ + +R+ S+ + +++ PV + L + + E +SK + ++A
Sbjct: 95 ITVGGEDVRMFDKSEWAKVVSIVNQEPVLFSLSVAENIAYGLPNEHVSKDDIIKA---AK 151
Query: 597 GLPSHNHLTPIVK------------LSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQS 644
+H+ + + + LSGGQ+ RV + IL+LDE T+ LD S
Sbjct: 152 AANAHDFIISLPQGYDTLVGERGGLLSGGQRQRVAIARSLLKNAPILILDEATSALDAVS 211
Query: 645 IDALADALDEFTGG--VVLVSHDSRLISRVCDDEERSQIWVVEDGTV 689
+ AL+ ++++H R+ + +QI V DG +
Sbjct: 212 ERLVQSALNRLMKDRTTLVIAH------RLSTVQSANQIAVCSDGKI 252
Score = 52.8 bits (125), Expect = 7e-07
Identities = 60/205 (29%), Positives = 94/205 (45%), Gaps = 28/205 (13%)
Query: 167 ELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTA 226
++L S+ ++ G L+G +G GKST+++LLA P I V ++V DK+
Sbjct: 52 KVLDGLSLTLNSGTVTALVGSSGAGKSTIVQLLARFYEPTQGRITV--GGEDVRMFDKSE 109
Query: 227 LEAVVS-ANVELIKVRQKVADLQNIASG--EEGMDKDDTNEEEDAGEKLAELYEQLQLMG 283
VVS N E + VA+ NIA G E + KDD +
Sbjct: 110 WAKVVSIVNQEPVLFSLSVAE--NIAYGLPNEHVSKDDI------------------IKA 149
Query: 284 SDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLD 343
+ AA + I G+ + G SGG R R+++AR+L +L+LDE T+ LD
Sbjct: 150 AKAANAHDFIISLPQGYDT-LVGERGGLLSGGQRQRVAIARSLLKNAPILILDEATSALD 208
Query: 344 LRAVLWLEEYLCRWKK--TLVVVSH 366
+ ++ L R K T +V++H
Sbjct: 209 AVSERLVQSALNRLMKDRTTLVIAH 233
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,627,758
Number of Sequences: 26719
Number of extensions: 771396
Number of successful extensions: 5796
Number of sequences better than 10.0: 304
Number of HSP's better than 10.0 without gapping: 158
Number of HSP's successfully gapped in prelim test: 151
Number of HSP's that attempted gapping in prelim test: 4402
Number of HSP's gapped (non-prelim): 1074
length of query: 713
length of database: 11,318,596
effective HSP length: 106
effective length of query: 607
effective length of database: 8,486,382
effective search space: 5151233874
effective search space used: 5151233874
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC136504.2


