

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136502.1 + phase: 0
(64 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
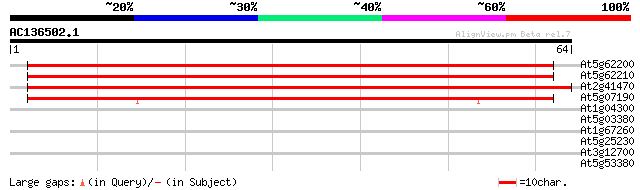
At5g62200 unknown protein 78 9e-16
At5g62210 embryo-specific protein - like predicted GPI-anchored ... 75 8e-15
At2g41470 unknown protein 68 1e-12
At5g07190 embryo-specific protein 3 (ATS3) 63 2e-11
At1g04300 unknown protein 30 0.17
At5g03380 farnesylated protein - like 28 0.65
At1g67260 hypothetical protein 26 4.2
At5g25230 translation Elongation Factor 2 - like protein 25 7.2
At3g12700 unknown protein 25 7.2
At5g53380 putative protein 25 9.4
>At5g62200 unknown protein
Length = 190
Score = 77.8 bits (190), Expect = 9e-16
Identities = 34/60 (56%), Positives = 42/60 (69%)
Query: 3 SFDIVGQCLNDICKLYLHRVGSNGWIPTTVTAYNYGYPPVKFYYNTYVPENVDYGFNHCN 62
+F I G C IC +YL+R G +GWIP TV Y++G V F YNTYVPE+V YGFN+CN
Sbjct: 100 TFQINGPCTYQICYVYLYRSGPDGWIPNTVKIYSHGSKAVTFPYNTYVPESVWYGFNYCN 159
>At5g62210 embryo-specific protein - like predicted GPI-anchored
protein
Length = 223
Score = 74.7 bits (182), Expect = 8e-15
Identities = 31/60 (51%), Positives = 42/60 (69%)
Query: 3 SFDIVGQCLNDICKLYLHRVGSNGWIPTTVTAYNYGYPPVKFYYNTYVPENVDYGFNHCN 62
+F + G+CLN IC +Y++R G++GWIP TV Y G VKF +N VPEN+ YG N+CN
Sbjct: 94 TFQVKGKCLNTICSVYIYRSGTDGWIPETVEIYKEGSKSVKFDFNKNVPENIWYGNNYCN 153
>At2g41470 unknown protein
Length = 365
Score = 67.8 bits (164), Expect = 1e-12
Identities = 29/62 (46%), Positives = 38/62 (60%)
Query: 3 SFDIVGQCLNDICKLYLHRVGSNGWIPTTVTAYNYGYPPVKFYYNTYVPENVDYGFNHCN 62
++ I G C+ D+C LYL R GS+GW P V Y V FYYN ++P +V YGFN CN
Sbjct: 100 TYKISGPCMRDVCYLYLLRQGSDGWKPENVKIYGSSIRSVTFYYNLFLPNSVWYGFNVCN 159
Query: 63 EV 64
+
Sbjct: 160 GI 161
>At5g07190 embryo-specific protein 3 (ATS3)
Length = 213
Score = 63.2 bits (152), Expect = 2e-11
Identities = 29/62 (46%), Positives = 42/62 (66%), Gaps = 2/62 (3%)
Query: 3 SFDIVGQCLND-ICKLYLHRVGSNGWIPTTVTAYNYGYPPVKFYYNTYVPE-NVDYGFNH 60
+F + GQCLND IC LY++R G +GW+P ++ Y+ G VKF ++ VP+ N YG N+
Sbjct: 88 TFQVRGQCLNDPICSLYINRNGPDGWVPESIEIYSEGSKSVKFDFSKSVPQLNTWYGHNN 147
Query: 61 CN 62
CN
Sbjct: 148 CN 149
>At1g04300 unknown protein
Length = 1082
Score = 30.4 bits (67), Expect = 0.17
Identities = 13/44 (29%), Positives = 22/44 (49%)
Query: 6 IVGQCLNDICKLYLHRVGSNGWIPTTVTAYNYGYPPVKFYYNTY 49
IVG L + H S+G +PTT+ + +Y P Y +++
Sbjct: 851 IVGNSLGSSSSSFNHHPSSHGVVPTTLPSSSYSQAPTSSYQSSF 894
>At5g03380 farnesylated protein - like
Length = 392
Score = 28.5 bits (62), Expect = 0.65
Identities = 16/39 (41%), Positives = 18/39 (46%), Gaps = 4/39 (10%)
Query: 23 GSNGWIPTTVTAYN-YGYPPVKFYYNTYVPENVDYGFNH 60
G G + + YN YGYPP Y Y PE YG H
Sbjct: 283 GDGGAMDVKKSEYNGYGYPPQPMY---YYPEGQVYGQQH 318
>At1g67260 hypothetical protein
Length = 359
Score = 25.8 bits (55), Expect = 4.2
Identities = 7/14 (50%), Positives = 13/14 (92%)
Query: 46 YNTYVPENVDYGFN 59
YNT++P+N+DY ++
Sbjct: 316 YNTFLPQNLDYSYD 329
>At5g25230 translation Elongation Factor 2 - like protein
Length = 973
Score = 25.0 bits (53), Expect = 7.2
Identities = 12/24 (50%), Positives = 12/24 (50%)
Query: 23 GSNGWIPTTVTAYNYGYPPVKFYY 46
GSNGWI T N P K YY
Sbjct: 37 GSNGWITTINENQNIVLPEDKKYY 60
>At3g12700 unknown protein
Length = 439
Score = 25.0 bits (53), Expect = 7.2
Identities = 18/60 (30%), Positives = 25/60 (41%), Gaps = 3/60 (5%)
Query: 3 SFDIVGQCLNDICKLYLHRVGSNGWIPT--TVTAYNYGYPPVKFYYNTYVPENVDYGFNH 60
SF VG CL CK+ L + S PT T +Y+Y Y + E + G +
Sbjct: 132 SFKTVG-CLTQTCKVDLMNLFSLTTCPTPSTPCSYDYRYADGSAAQGVFAKETITVGLTN 190
>At5g53380 putative protein
Length = 483
Score = 24.6 bits (52), Expect = 9.4
Identities = 11/31 (35%), Positives = 16/31 (51%), Gaps = 3/31 (9%)
Query: 20 HRVGSNGWIPTTVTAYNYGYPPVKFYYNTYV 50
HR G+ GW ++ A Y V+ +NT V
Sbjct: 183 HRFGNKGWYSRSINAVYYA---VRLIWNTIV 210
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.144 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,811,542
Number of Sequences: 26719
Number of extensions: 66785
Number of successful extensions: 151
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 143
Number of HSP's gapped (non-prelim): 10
length of query: 64
length of database: 11,318,596
effective HSP length: 40
effective length of query: 24
effective length of database: 10,249,836
effective search space: 245996064
effective search space used: 245996064
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC136502.1


