

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136472.17 - phase: 0
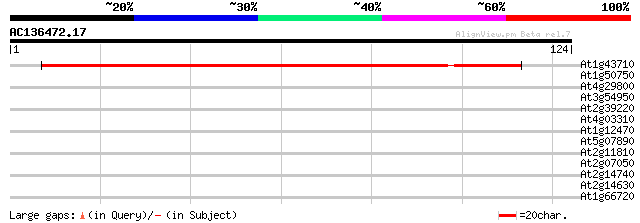
(124 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g43710 histidine decarboxylase like protein 166 2e-42
At1g50750 hypothetical protein 29 0.52
At4g29800 unknown protein 28 0.88
At3g54950 putative protein 28 1.2
At2g39220 similar to latex allergen from Hevea brasiliensis 27 2.0
At4g03310 hypothetical protein 27 3.4
At1g12470 hypothetical protein 26 4.4
At5g07890 putative protein 26 5.7
At2g11810 MGDG synthase type C (mgdC) 26 5.7
At2g07050 cycloartenol synthase 25 7.5
At2g14740 putative vacuolar sorting receptor 25 9.8
At2g14630 En/Spm-like transposon protein 25 9.8
At1g66720 hypothetical protein 25 9.8
>At1g43710 histidine decarboxylase like protein
Length = 482
Score = 166 bits (421), Expect = 2e-42
Identities = 79/106 (74%), Positives = 94/106 (88%), Gaps = 1/106 (0%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
F+KEVQKCLRNAHY KDRL EAGI AMLNELSSTVVFERP DEEF+R+WQLAC+G+IAHV
Sbjct: 373 FQKEVQKCLRNAHYLKDRLREAGISAMLNELSSTVVFERPKDEEFVRRWQLACQGDIAHV 432
Query: 68 VVMPNVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCL 113
VVMP+VTIEKLD+FL +LV+ R W+E G+ QP C+AS+VG N+C+
Sbjct: 433 VVMPSVTIEKLDNFLKDLVKHRLIWYEDGS-QPPCLASEVGTNNCI 477
>At1g50750 hypothetical protein
Length = 816
Score = 29.3 bits (64), Expect = 0.52
Identities = 19/70 (27%), Positives = 32/70 (45%), Gaps = 1/70 (1%)
Query: 39 SSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEYGTF 98
+ T + E P DE + + +VV P T + DD L ++ + A F+ G+
Sbjct: 583 AGTNMDESPFDENNSSEPPFCADRGVVDIVVSPPETRQACDDEL-DVNESNADMFDDGSK 641
Query: 99 QPYCIASDVG 108
QP C+ + G
Sbjct: 642 QPKCLLHEDG 651
>At4g29800 unknown protein
Length = 525
Score = 28.5 bits (62), Expect = 0.88
Identities = 13/26 (50%), Positives = 15/26 (57%)
Query: 89 RATWFEYGTFQPYCIASDVGENSCLA 114
RATW E GTF P S G+ C+A
Sbjct: 298 RATWAEPGTFDPVRTCSVDGKTRCVA 323
>At3g54950 putative protein
Length = 488
Score = 28.1 bits (61), Expect = 1.2
Identities = 12/26 (46%), Positives = 16/26 (61%)
Query: 89 RATWFEYGTFQPYCIASDVGENSCLA 114
RATW E G F+P + S G+ C+A
Sbjct: 261 RATWAEPGVFEPVEMKSVDGQTKCVA 286
>At2g39220 similar to latex allergen from Hevea brasiliensis
Length = 499
Score = 27.3 bits (59), Expect = 2.0
Identities = 12/26 (46%), Positives = 16/26 (61%)
Query: 89 RATWFEYGTFQPYCIASDVGENSCLA 114
RATW E G F+P + S G+ C+A
Sbjct: 274 RATWAEPGVFEPVEMRSVDGKTRCVA 299
>At4g03310 hypothetical protein
Length = 562
Score = 26.6 bits (57), Expect = 3.4
Identities = 11/30 (36%), Positives = 19/30 (62%)
Query: 30 GIGAMLNELSSTVVFERPHDEEFIRKWQLA 59
G+G++ NE SS V +PH++ + +LA
Sbjct: 331 GVGSLHNEASSATVGPQPHNDPVVLSEKLA 360
>At1g12470 hypothetical protein
Length = 950
Score = 26.2 bits (56), Expect = 4.4
Identities = 11/33 (33%), Positives = 20/33 (60%), Gaps = 2/33 (6%)
Query: 34 MLNELSSTVVFERPHDEEFIRKWQLACKGNIAH 66
M+NE+ T+ F +P D ++ W L + N+A+
Sbjct: 912 MINEI--TLPFIKPEDSQYSTSWDLRSETNLAN 942
>At5g07890 putative protein
Length = 389
Score = 25.8 bits (55), Expect = 5.7
Identities = 20/94 (21%), Positives = 40/94 (42%), Gaps = 17/94 (18%)
Query: 9 EKEVQKCLRNAHYFKDRLIEAG-----IGAMLNELSSTVVFERPHDEEFIRKWQLACKGN 63
EKE+ C + Y +D+LI + L++L + R +EE C
Sbjct: 64 EKELLNCYKEIDYLRDQLIFRSKEVNYLNEHLHDLEFKLAESRNLEEEVNSLRDELCMSK 123
Query: 64 IAHVVVM------------PNVTIEKLDDFLNEL 85
H++++ ++T+EKL++ ++ L
Sbjct: 124 SEHLLLLQELESKEIELQCSSLTLEKLEETISSL 157
>At2g11810 MGDG synthase type C (mgdC)
Length = 465
Score = 25.8 bits (55), Expect = 5.7
Identities = 23/93 (24%), Positives = 39/93 (41%), Gaps = 15/93 (16%)
Query: 19 AHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKL 78
A+Y K+ IEAG+ ++ +V H ++ KWQ K I V+ T +
Sbjct: 160 AYYAKE--IEAGLMEYKPDIIISVHPLMQHIPLWVMKWQGLHKKVIFVTVITDLNTCHR- 216
Query: 79 DDFLNELVQKRATWFEYGTFQPYCIASDVGENS 111
TWF +G + YC + +V + +
Sbjct: 217 ------------TWFHHGVSRCYCPSKEVAKRA 237
>At2g07050 cycloartenol synthase
Length = 759
Score = 25.4 bits (54), Expect = 7.5
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 6/61 (9%)
Query: 44 FERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQ---KRATWFEYGTFQP 100
F + H + + + Q + + I+ V +P V IE DD E+V+ KR F Y T Q
Sbjct: 53 FVQKHSADLLMRLQFSRENLISPV--LPQVKIEDTDDVTEEMVETTLKRGLDF-YSTIQA 109
Query: 101 Y 101
+
Sbjct: 110 H 110
>At2g14740 putative vacuolar sorting receptor
Length = 628
Score = 25.0 bits (53), Expect = 9.8
Identities = 15/59 (25%), Positives = 29/59 (48%), Gaps = 8/59 (13%)
Query: 61 KGNIAHVVVMPNVTIE------KLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCL 113
KG+ V ++P + + KL+ + +++ + FE T C+++DV N CL
Sbjct: 365 KGSRGDVTILPTLVVNNRQYRGKLEK--SAVLKALCSGFEETTEPAICLSTDVESNECL 421
>At2g14630 En/Spm-like transposon protein
Length = 133
Score = 25.0 bits (53), Expect = 9.8
Identities = 11/30 (36%), Positives = 19/30 (62%)
Query: 30 GIGAMLNELSSTVVFERPHDEEFIRKWQLA 59
G+G++ NE SS V +PH++ + +LA
Sbjct: 20 GVGSLHNEASSATVGPQPHNDLVVLLEKLA 49
>At1g66720 hypothetical protein
Length = 352
Score = 25.0 bits (53), Expect = 9.8
Identities = 16/53 (30%), Positives = 26/53 (48%), Gaps = 1/53 (1%)
Query: 31 IGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVV-VMPNVTIEKLDDFL 82
IGA LN+L+ V E+ E F +A +G + ++ TIE +D +
Sbjct: 233 IGASLNDLAQQGVIEKDKVESFNITLYIAEEGELRQIIEENGKFTIEAFEDII 285
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.138 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,807,356
Number of Sequences: 26719
Number of extensions: 99983
Number of successful extensions: 301
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 294
Number of HSP's gapped (non-prelim): 13
length of query: 124
length of database: 11,318,596
effective HSP length: 87
effective length of query: 37
effective length of database: 8,994,043
effective search space: 332779591
effective search space used: 332779591
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 53 (25.0 bits)
Medicago: description of AC136472.17


