

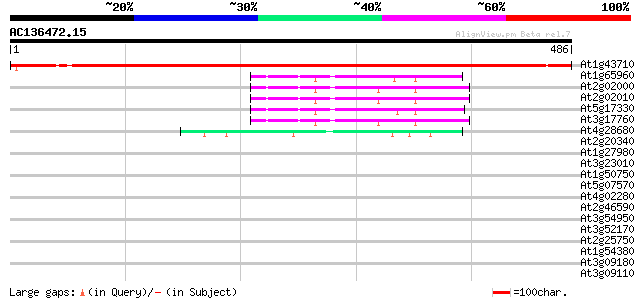
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136472.15 + phase: 0
(486 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g43710 histidine decarboxylase like protein 785 0.0
At1g65960 unknown protein 59 5e-09
At2g02000 glutamate decarboxylase like protein 58 1e-08
At2g02010 putative glutamate decarboxylase 55 1e-07
At5g17330 glutamate decarboxylase 1 (GAD 1) (sp|Q42521) 54 3e-07
At3g17760 glutamate decarboxylase, putative 52 6e-07
At4g28680 aromatic amino-acid decarboxylase - like protein 47 2e-05
At2g20340 tyrosine decarboxylase like protein 40 0.003
At1g27980 unknown protein 37 0.019
At3g23010 disease resistance protein, putative 33 0.46
At1g50750 hypothetical protein 33 0.46
At5g07570 glycine/proline-rich protein 31 1.8
At4g02280 putative sucrose synthetase 30 2.3
At2g46590 DOF zinc finger like protein 30 2.3
At3g54950 putative protein 30 3.0
At3g52170 putative protein 30 3.9
At2g25750 unknown protein 29 6.7
At1g54380 unknown protein 29 6.7
At3g09180 unknown protein 28 8.7
At3g09110 hypothetical protein 28 8.7
>At1g43710 histidine decarboxylase like protein
Length = 482
Score = 785 bits (2028), Expect = 0.0
Identities = 380/488 (77%), Positives = 430/488 (87%), Gaps = 8/488 (1%)
Query: 1 MVGS--ADVLVNGLSTNGAVELLPDDFDVSAIIKDPVPPVVAADNGIGKEEAKINGGKEK 58
MVGS +D ++ + +++L DDFD +A++ +P+PP V NGIG ++ GG +
Sbjct: 1 MVGSLESDQTLSMATLIEKLDILSDDFDPTAVVTEPLPPPVT--NGIGADKG---GGGGE 55
Query: 59 REIVLGRNIHTTCLEVTEPEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDF 118
RE+VLGRNIHTT L VTEPE +DE TGD++A+MASVLARYRK+L ERTK HLGYPYNLDF
Sbjct: 56 REMVLGRNIHTTSLAVTEPEVNDEFTGDKEAYMASVLARYRKTLVERTKNHLGYPYNLDF 115
Query: 119 DYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGT 178
DYGAL QLQHFSINNLGDPFIESNYGVHSR FEVGVLDWFARLWE+E+++YWGYITNCGT
Sbjct: 116 DYGALGQLQHFSINNLGDPFIESNYGVHSRPFEVGVLDWFARLWEIERDDYWGYITNCGT 175
Query: 179 EGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKL 238
EGNLHGILVGRE+ PDGILYASRESHYS+FKAARMYRMECEKV+TL SGEIDCDD + KL
Sbjct: 176 EGNLHGILVGREMFPDGILYASRESHYSVFKAARMYRMECEKVDTLMSGEIDCDDLRKKL 235
Query: 239 LRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKR 298
L ++DKPAI+NVNIGTTVKGAVDDLDLVI+ LEE GFS DRFYIH DGALFGLMMPFVKR
Sbjct: 236 LANKDKPAILNVNIGTTVKGAVDDLDLVIKTLEECGFSHDRFYIHCDGALFGLMMPFVKR 295
Query: 299 APKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNG 358
APKVTF KPIGSVSVSGHKFVGCPMPCGVQITR+EHI LS NVEYLASRDATIMGSRNG
Sbjct: 296 APKVTFNKPIGSVSVSGHKFVGCPMPCGVQITRMEHIKVLSSNVEYLASRDATIMGSRNG 355
Query: 359 HAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDE 418
HAP+FLWYTLNRKGY+GFQKEVQKCLRNAHY KDRL EAGI AMLNELSSTVVFERP DE
Sbjct: 356 HAPLFLWYTLNRKGYKGFQKEVQKCLRNAHYLKDRLREAGISAMLNELSSTVVFERPKDE 415
Query: 419 EFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGEN 478
EF+R+WQLAC+G+IAHVVVMP+VTIEKLD+FL +LV+ R W+EDG+ QP C+AS+VG N
Sbjct: 416 EFVRRWQLACQGDIAHVVVMPSVTIEKLDNFLKDLVKHRLIWYEDGS-QPPCLASEVGTN 474
Query: 479 SCLCAQHK 486
+C+C HK
Sbjct: 475 NCICPAHK 482
>At1g65960 unknown protein
Length = 494
Score = 59.3 bits (142), Expect = 5e-09
Identities = 52/193 (26%), Positives = 95/193 (48%), Gaps = 14/193 (7%)
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V L+ G D KA + ++ + + +G+T+ G +D+ D
Sbjct: 168 KFARYFEVELKEVN-LSEGYYVMDPDKAAEMVDENTICVAAI-LGSTLNGEFEDVKRLND 225
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKP-IGSVSVSGHKFVGCPM 323
L+++K EE G++ IHVD A G + PF+ + F+ P + S++VSGHK+
Sbjct: 226 LLVKKNEETGWNTP---IHVDAASGGFIAPFIYPELEWDFRLPLVKSINVSGHKYGLVYA 282
Query: 324 PCGVQITRL--EHINALSRNVEYLASRDA--TIMGSRNGHAPIFLWYTLNRKGYRGFQKE 379
G + R + L ++ YL + T+ S+ I +Y L R G+ G++
Sbjct: 283 GIGWVVWRAAEDLPEELIFHINYLGADQPTFTLNFSKGSSQIIAQYYQLIRLGFEGYKNV 342
Query: 380 VQKCLRNAHYFKD 392
++ C+ N K+
Sbjct: 343 MENCIENMVVLKE 355
>At2g02000 glutamate decarboxylase like protein
Length = 500
Score = 57.8 bits (138), Expect = 1e-08
Identities = 52/199 (26%), Positives = 98/199 (49%), Gaps = 14/199 (7%)
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V+ L G D KA + ++ ++ + +G+T+ G +D+ D
Sbjct: 169 KFARYFEVELKEVK-LREGYYVMDPDKAVEMVDENTICVVAI-LGSTLTGEFEDVKLLND 226
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKP-IGSVSVSGHKF--VGC 321
L+++K ++ G+ IHVD A G + PF+ + F+ P + S++VSGHK+ V
Sbjct: 227 LLVEKNKKTGWDTP---IHVDAASGGFIAPFLYPDLEWDFRLPLVKSINVSGHKYGLVYA 283
Query: 322 PMPCGVQITRLEHINALSRNVEYLASRDA--TIMGSRNGHAPIFLWYTLNRKGYRGFQKE 379
+ V T+ + + L ++ YL + T+ S+ I +Y L R G+ G++
Sbjct: 284 GIGWVVWRTKTDLPDELIFHINYLGADQPTFTLNFSKGSSQVIAQYYQLIRLGFEGYRNV 343
Query: 380 VQKCLRNAHYFKDRLIEAG 398
+ C N + L + G
Sbjct: 344 MDNCRENMMVLRQGLEKTG 362
>At2g02010 putative glutamate decarboxylase
Length = 493
Score = 54.7 bits (130), Expect = 1e-07
Identities = 51/199 (25%), Positives = 95/199 (47%), Gaps = 14/199 (7%)
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V L D KA + ++ + + +G+T+ G +D+ D
Sbjct: 169 KFARYFEVELKEVN-LREDYYVMDPVKAVEMVDENTICVAAI-LGSTLTGEFEDVKLLND 226
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKP-IGSVSVSGHKF--VGC 321
L+++K ++ G+ IHVD A G + PF+ + F+ P + S++VSGHK+ V
Sbjct: 227 LLVEKNKQTGWDTP---IHVDAASGGFIAPFLYPELEWDFRLPLVKSINVSGHKYGLVYA 283
Query: 322 PMPCGVQITRLEHINALSRNVEYLASRDA--TIMGSRNGHAPIFLWYTLNRKGYRGFQKE 379
+ V T+ + + L ++ YL + T+ S+ I +Y L R G+ G++
Sbjct: 284 GIGWVVWRTKTDLPDELIFHINYLGADQPTFTLNFSKGSSQVIAQYYQLIRLGFEGYRNV 343
Query: 380 VQKCLRNAHYFKDRLIEAG 398
+ C N + L + G
Sbjct: 344 MDNCRENMMVLRQGLEKTG 362
>At5g17330 glutamate decarboxylase 1 (GAD 1) (sp|Q42521)
Length = 502
Score = 53.5 bits (127), Expect = 3e-07
Identities = 51/195 (26%), Positives = 95/195 (48%), Gaps = 14/195 (7%)
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V+ L+ G D +A + ++ + + +G+T+ G +D+ D
Sbjct: 169 KFARYFEVELKEVK-LSEGYYVMDPQQAVDMVDENTICVAAI-LGSTLNGEFEDVKLLND 226
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKP-IGSVSVSGHKFVGCPM 323
L+++K +E G+ IHVD A G + PF+ + F+ P + S++VSGHK+
Sbjct: 227 LLVEKNKETGWDTP---IHVDAASGGFIAPFLYPELEWDFRLPLVKSINVSGHKYGLVYA 283
Query: 324 PCGVQITRLEH--INALSRNVEYLASRDA--TIMGSRNGHAPIFLWYTLNRKGYRGFQKE 379
G I R + L ++ YL + T+ S+ I +Y L R G+ G++
Sbjct: 284 GIGWVIWRNKEDLPEELIFHINYLGADQPTFTLNFSKGSSQVIAQYYQLIRLGHEGYRNV 343
Query: 380 VQKCLRNAHYFKDRL 394
++ C N ++ L
Sbjct: 344 MENCRENMIVLREGL 358
>At3g17760 glutamate decarboxylase, putative
Length = 494
Score = 52.4 bits (124), Expect = 6e-07
Identities = 51/199 (25%), Positives = 96/199 (47%), Gaps = 14/199 (7%)
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V+ L+ D KA + ++ + + +G+T+ G +D+ D
Sbjct: 168 KFARYFEVELKEVK-LSEDYYVMDPAKAVEMVDENTICVAAI-LGSTLTGEFEDVKQLND 225
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKP-IGSVSVSGHKF--VGC 321
L+ +K E G+ IHVD A G + PF+ + F+ P + S++VSGHK+ V
Sbjct: 226 LLAEKNAETGWETP---IHVDAASGGFIAPFLYPDLEWDFRLPWVKSINVSGHKYGLVYA 282
Query: 322 PMPCGVQITRLEHINALSRNVEYLASRDA--TIMGSRNGHAPIFLWYTLNRKGYRGFQKE 379
+ V T+ + L ++ YL + T+ S+ I +Y R G+ G++
Sbjct: 283 GVGWVVWRTKDDLPEELVFHINYLGADQPTFTLNFSKGSSQIIAQYYQFIRLGFEGYKNI 342
Query: 380 VQKCLRNAHYFKDRLIEAG 398
++ C+ NA ++ + G
Sbjct: 343 MENCMDNARRLREGIEMTG 361
>At4g28680 aromatic amino-acid decarboxylase - like protein
Length = 545
Score = 47.0 bits (110), Expect = 2e-05
Identities = 63/278 (22%), Positives = 107/278 (37%), Gaps = 39/278 (14%)
Query: 149 QFEVGVLDWFARLWELEKN-----EYWGYITNCGTEGNLHGIL---------VGREVLPD 194
+ E+ VLDW A+L +L + G I G E L +L VG+ +LP
Sbjct: 176 ELEIIVLDWLAKLLQLPDHFLSTGNGGGVIQGTGCEAVLVVVLAARDRILKKVGKTLLPQ 235
Query: 195 GILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDK--------PA 246
++Y S ++H S KA + + E + L + + L P
Sbjct: 236 LVVYGSDQTHSSFRKACLIGGIHEENIRLLKTDSSTNYGMPPESLEEAISHDLAKGFIPF 295
Query: 247 IINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKK 306
I +GTT AVD L + ++ G ++HVD A G + + +
Sbjct: 296 FICATVGTTSSAAVDPLVPLGNIAKKYG-----IWLHVDAAYAGNACICPEYRKFIDGIE 350
Query: 307 PIGSVSVSGHKFVGCPMPCGVQIT--RLEHINALSRNVEYL---ASRDATIMGSRNGHAP 361
S +++ HK++ C R I+AL N EYL S+ T++ ++
Sbjct: 351 NADSFNMNAHKWLFANQTCSPLWVKDRYSLIDALKTNPEYLEFKVSKKDTVVNYKDWQIS 410
Query: 362 IF-------LWYTLNRKGYRGFQKEVQKCLRNAHYFKD 392
+ LW L G + ++ + A +F+D
Sbjct: 411 LSRRFRSLKLWMVLRLYGSENLRNFIRDHVNLAKHFED 448
>At2g20340 tyrosine decarboxylase like protein
Length = 490
Score = 40.0 bits (92), Expect = 0.003
Identities = 73/354 (20%), Positives = 134/354 (37%), Gaps = 58/354 (16%)
Query: 77 PEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDYGALSQLQHFSINNLGD 136
PE D++ D A + + ++ + YP N G L ++ + +G
Sbjct: 61 PETLDQVLDDVRAKILPGVTHWQSP-----SFFAYYPSNSSVA-GFLGEMLSAGLGIVGF 114
Query: 137 PFIESNYGVHSRQFEVGVLDWFARLWEL-----EKNEYWGYITNCGTEGNLHGIL----- 186
++ S + E+ VLDW A+L L K G I +E L ++
Sbjct: 115 SWVTSPAAT---ELEMIVLDWVAKLLNLPEQFMSKGNGGGVIQGSASEAVLVVLIAARDK 171
Query: 187 ----VGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSG-------------EI 229
VG+ L ++Y+S ++H ++ KA ++ + E L + E
Sbjct: 172 VLRSVGKNALEKLVVYSSDQTHSALQKACQIAGIHPENCRVLTTDSSTNYALRPESLQEA 231
Query: 230 DCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALF 289
D +A L+ P + N+GTT AVD L + + G + HVD A
Sbjct: 232 VSRDLEAGLI-----PFFLCANVGTTSSTAVDPLAALGKIANSNG-----IWFHVDAAYA 281
Query: 290 GLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEH--INALSRNVEYL-- 345
G + + + S +++ HK+ C + + + ALS N E+L
Sbjct: 282 GSACICPEYRQYIDGVETADSFNMNAHKWFLTNFDCSLLWVKDQDSLTLALSTNPEFLKN 341
Query: 346 -ASRDATIMGSRNGHAP-------IFLWYTLNRKGYRGFQKEVQKCLRNAHYFK 391
AS+ ++ ++ P + LW L G + ++ ++ A F+
Sbjct: 342 KASQANLVVDYKDWQIPLGRRFRSLKLWMVLRLYGSETLKSYIRNHIKLAKEFE 395
>At1g27980 unknown protein
Length = 544
Score = 37.4 bits (85), Expect = 0.019
Identities = 76/321 (23%), Positives = 131/321 (40%), Gaps = 60/321 (18%)
Query: 171 GYITNCGTEGNLHGILVGREVL--------PDGILYASRESHYSIFKAARMYRMECEKVE 222
G +T+ GTE + + R+ + P+ I+ S S Y KAA+ ++++ +V
Sbjct: 200 GNMTSGGTESIVLAVKSSRDYMKYKKGITRPEMIIPESGHSAYD--KAAQYFKIKLWRVP 257
Query: 223 TLNSGEIDCDDFKAKLL---RHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDR 279
+D DF+A + RH IN N V A +I +EE G
Sbjct: 258 ------VD-KDFRADVKATRRH------INRNTIMIVGSAPGFPHGIIDPIEELGQLALS 304
Query: 280 FYI--HVDGALFGLMMPFVKR----APKVTFK-KPIGSVSVSGHKFVGCPMPCGVQITRL 332
+ I HVD L G ++PF ++ P F + + S+SV HK+ P + R
Sbjct: 305 YGICFHVDLCLGGFVLPFARKLGYQIPPFDFSVQGVTSISVDVHKYGLAPKGTSTVLYRN 364
Query: 333 EHINALSRNVEYLASRD--------ATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCL 384
I R +++A + TI GSR G W + G G+ + K +
Sbjct: 365 HEI----RKHQFVAVTEWSGGLYVSPTIAGSRPGSLVAGAWAAMMSLGEEGYLQNTSKIM 420
Query: 385 RNAHYFKDRLIEAGIGAMLNELSSTVV---------FERPHDEEFIRKWQLAC--KGNIA 433
+ ++ + E ++ + T+V FE +D + W L + N
Sbjct: 421 EASKRLEEGVREIHELFVIGKPDMTIVAFGSKALDIFE-VNDIMSSKGWHLNALQRPNSI 479
Query: 434 HV-VVMPNVTIEKLDDFLNEL 453
H+ + + +V + +DDFL +L
Sbjct: 480 HICITLQHVPV--VDDFLRDL 498
>At3g23010 disease resistance protein, putative
Length = 595
Score = 32.7 bits (73), Expect = 0.46
Identities = 29/99 (29%), Positives = 43/99 (43%), Gaps = 21/99 (21%)
Query: 249 NVNIGT------TVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKV 302
NVN T KG D D + + FS +RF H+ G++ GL+
Sbjct: 403 NVNFSTYDSIDLVYKGVETDFDRIFEGFNAIDFSGNRFSGHIPGSI-GLL---------- 451
Query: 303 TFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRN 341
+ +++SG+ F G P IT LE ++ LSRN
Sbjct: 452 ---SELRLLNLSGNAFTGNIPPSLANITNLESLD-LSRN 486
>At1g50750 hypothetical protein
Length = 816
Score = 32.7 bits (73), Expect = 0.46
Identities = 20/70 (28%), Positives = 33/70 (46%), Gaps = 1/70 (1%)
Query: 407 SSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTF 466
+ T + E P DE + + +VV P T + DD L ++ + A F+DG+
Sbjct: 583 AGTNMDESPFDENNSSEPPFCADRGVVDIVVSPPETRQACDDEL-DVNESNADMFDDGSK 641
Query: 467 QPYCIASDVG 476
QP C+ + G
Sbjct: 642 QPKCLLHEDG 651
>At5g07570 glycine/proline-rich protein
Length = 1504
Score = 30.8 bits (68), Expect = 1.8
Identities = 26/82 (31%), Positives = 37/82 (44%), Gaps = 8/82 (9%)
Query: 2 VGSADVLVNGLSTNGAVELLPDD---FDVSAIIKDPVPPVVAADNGIGKEEAKINGGKEK 58
VG++D G S GA + P D FD+ + DP PVV A +G +E NGG
Sbjct: 1284 VGASD---RGASGEGASDGCPPDVGPFDICLLSIDP--PVVGASDGGPPDEGASNGGPPD 1338
Query: 59 REIVLGRNIHTTCLEVTEPEAD 80
+ G + + L + P D
Sbjct: 1339 TDSFGGDPLCASPLGIDPPIVD 1360
>At4g02280 putative sucrose synthetase
Length = 809
Score = 30.4 bits (67), Expect = 2.3
Identities = 35/132 (26%), Positives = 51/132 (38%), Gaps = 25/132 (18%)
Query: 58 KREIVLGRNIHTTCLE---------VTEPEADDEITGD---RDAHMASVLARYRKSLTE- 104
K E+V G N CLE V P I + H++SV+ R + L
Sbjct: 121 KEELVDGPNSDPFCLELDFEPFNANVPRPSRSSSIGNGVQFLNRHLSSVMFRNKDCLEPL 180
Query: 105 ----RTKYHLGYPYNLDFDYGALSQLQ--------HFSINNLGDPFIESNYGVHSRQFEV 152
R + G+P L+ ++S+LQ H S + PF E Y + FE
Sbjct: 181 LDFLRVHKYKGHPLMLNDRIQSISRLQIQLSKAEDHISKLSQETPFSEFEYALQGMGFEK 240
Query: 153 GVLDWFARLWEL 164
G D R+ E+
Sbjct: 241 GWGDTAGRVLEM 252
>At2g46590 DOF zinc finger like protein
Length = 357
Score = 30.4 bits (67), Expect = 2.3
Identities = 44/188 (23%), Positives = 80/188 (42%), Gaps = 25/188 (13%)
Query: 245 PAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTF 304
P + + I KG+ DL+L+ + QD+ + HV + F L MP ++ +T
Sbjct: 145 PILFSNQIHNKSKGSSQDLNLLSFPV-----MQDQHHHHVHMSQF-LQMPKMEGNGNITH 198
Query: 305 KK-PIGSVSVSGHKFVGCP----MPCGVQITRLEHINAL---------SRNVEYLASRDA 350
++ P S SV G + GV ++ IN+ S V Y +S
Sbjct: 199 QQQPSSSSSVYGSSSSPVSALELLRTGVNVSSRSGINSSFMPSGSMMDSNTVLYTSSGFP 258
Query: 351 TIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTV 410
T++ + P L ++ + +G + L + H+ + R++ G + ELSS++
Sbjct: 259 TMVD----YKPSNLSFSTDHQGLGHNSNNRSEALHSDHHQQGRVLFP-FGDQMKELSSSI 313
Query: 411 VFERPHDE 418
E HD+
Sbjct: 314 TQEVDHDD 321
>At3g54950 putative protein
Length = 488
Score = 30.0 bits (66), Expect = 3.0
Identities = 37/150 (24%), Positives = 54/150 (35%), Gaps = 35/150 (23%)
Query: 350 ATIMGSRNGHAPIF----LWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNE 405
A + GSR+G+ PIF W L R K L R++ G G
Sbjct: 153 AMLFGSRDGNRPIFKADDTWQFLTRNA---------KGLYGGAGILKRVLRTGSGCC--- 200
Query: 406 LSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQK--------- 456
S T ++ E F +L K + V++ P ++ FL
Sbjct: 201 -SGTAKLKKVMKESFS---ELTLKDTLKPVLI-PCYDLKSSGPFLFSRADALETDGYDFR 255
Query: 457 -----RATWFEDGTFQPYCIASDVGENSCL 481
RATW E G F+P + S G+ C+
Sbjct: 256 LSEVCRATWAEPGVFEPVEMKSVDGQTKCV 285
>At3g52170 putative protein
Length = 490
Score = 29.6 bits (65), Expect = 3.9
Identities = 17/44 (38%), Positives = 27/44 (60%), Gaps = 6/44 (13%)
Query: 1 MVGSADVLVNGLSTNGAVELLPDDFDVSAIIKDPVPPVVAADNG 44
++G D+L+ G NG+V+ D S+I+ DPVPP+ + NG
Sbjct: 109 VLGPGDLLLEG---NGSVQ---DQSLSSSILMDPVPPLSLSPNG 146
>At2g25750 unknown protein
Length = 282
Score = 28.9 bits (63), Expect = 6.7
Identities = 22/78 (28%), Positives = 33/78 (42%), Gaps = 6/78 (7%)
Query: 133 NLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGR--- 189
N G+ F+ + G Q V D F W +K Y+T+ T G+ GI++ R
Sbjct: 137 NSGNSFVVMSKGTQYLQQSYKVSDSFPFKWINKKWREGFYVTSMATAGSKWGIVMSRGAS 196
Query: 190 ---EVLPDGILYASRESH 204
+V+ LY S H
Sbjct: 197 FSDQVIELDFLYPSEGIH 214
>At1g54380 unknown protein
Length = 515
Score = 28.9 bits (63), Expect = 6.7
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 1/57 (1%)
Query: 233 DFKAKLLRHQDKPAIINVNIGTTVK-GAVDDLDLVIQKLEEAGFSQDRFYIHVDGAL 288
DFK +L D A + N T++ GAV + +Q+++E+ FS+ VDGAL
Sbjct: 27 DFKKQLNSEIDPQASSSQNDAITMEDGAVSVNNRDLQEIKESSFSKGSEQYRVDGAL 83
>At3g09180 unknown protein
Length = 402
Score = 28.5 bits (62), Expect = 8.7
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 2/63 (3%)
Query: 234 FKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMM 293
F A RH D P + + +++ + DL L+ +KLEE+G + + +GL M
Sbjct: 59 FVASQPRHTDMPLQLFLREDASMRQHLQDLRLIGKKLEESGVLTESLRSRSNS--WGLHM 116
Query: 294 PFV 296
P V
Sbjct: 117 PLV 119
>At3g09110 hypothetical protein
Length = 452
Score = 28.5 bits (62), Expect = 8.7
Identities = 24/92 (26%), Positives = 36/92 (39%), Gaps = 11/92 (11%)
Query: 368 LNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLA 427
LN GY GF K ++ LI+ G +L L E P + F+RK ++
Sbjct: 187 LNDFGYSGFDK-----------LQEMLIDVGFEEILTLLGCLFTSEAPLTDTFLRKHCMS 235
Query: 428 CKGNIAHVVVMPNVTIEKLDDFLNELVQKRAT 459
K + +V + D+ L V R T
Sbjct: 236 RKRKMLTPLVQESSVAGAADNLLTLKVYVRKT 267
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.139 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,263,747
Number of Sequences: 26719
Number of extensions: 492264
Number of successful extensions: 1163
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 1154
Number of HSP's gapped (non-prelim): 24
length of query: 486
length of database: 11,318,596
effective HSP length: 103
effective length of query: 383
effective length of database: 8,566,539
effective search space: 3280984437
effective search space used: 3280984437
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC136472.15


