

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136472.10 + phase: 0
(427 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
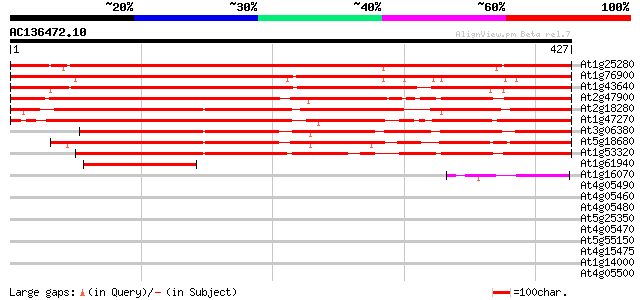
Score E
Sequences producing significant alignments: (bits) Value
At1g25280 unknown protein 629 0.0
At1g76900 unknown protein 618 e-177
At1g43640 unknown protein (At1g43640) 588 e-168
At2g47900 Tub family like protein 477 e-135
At2g18280 tubby-like protein 2 415 e-116
At1g47270 unknown protein 400 e-112
At3g06380 unknown protein 372 e-103
At5g18680 tub family-like protein 360 e-100
At1g53320 tubby like protein (Tub family protein) 338 3e-93
At1g61940 hypothetical protein, 5' partial 118 5e-27
At1g16070 unknown protein 59 5e-09
At4g05490 N7 -like protein 37 0.016
At4g05460 N7 like-protein 37 0.027
At4g05480 N7 -like protein 36 0.046
At5g25350 leucine-rich repeats containing protein 35 0.10
At4g05470 N7 -like protein 35 0.10
At5g55150 putative protein 34 0.18
At4g15475 unknown protein 30 1.9
At1g14000 putative protein kinase 30 2.5
At4g05500 30 3.3
>At1g25280 unknown protein
Length = 445
Score = 629 bits (1622), Expect = 0.0
Identities = 328/448 (73%), Positives = 374/448 (83%), Gaps = 24/448 (5%)
Query: 1 MSFRSIVRDVRDSFGSLSRRSFDVKLTGHHHRGKSHGSV-------QDLHDQPLVIQNSC 53
MSFR IV+D+RD FGSLSRRSFD +L+ H +GK+ GS +DL P+++Q S
Sbjct: 1 MSFRGIVQDLRDGFGSLSRRSFDFRLSSLH-KGKAQGSSFREYSSSRDLLS-PVIVQTSR 58
Query: 54 WANLPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKDIVRSPEFCGKLTFPV 113
WANLPPELLFDVI+RLEESE+ WP+RKHVVACASVC+SWR MC++IV PE CGKLTFPV
Sbjct: 59 WANLPPELLFDVIKRLEESESNWPARKHVVACASVCRSWRAMCQEIVLGPEICGKLTFPV 118
Query: 114 SLKQPGPRDGIIQCFIKRDKSNLTYHLFLCLSPALLVENGKFLLSAKRTRRTTYTEYVIS 173
SLKQPGPRD +IQCFIKRDKS LT+HLFLCLSPALLVENGKFLLSAKRTRRTT TEY+IS
Sbjct: 119 SLKQPGPRDAMIQCFIKRDKSKLTFHLFLCLSPALLVENGKFLLSAKRTRRTTRTEYIIS 178
Query: 174 MDADNISRSSNTYIGKLRSNFLGTKFIIYDTQ-PPCTSAHICPPGTGKTSRRFSKKVSPK 232
MDADNISRSSN+Y+GKLRSNFLGTKF++YDTQ PP TS+ SR S++VSPK
Sbjct: 179 MDADNISRSSNSYLGKLRSNFLGTKFLVYDTQPPPNTSSSALITDRTSRSRFHSRRVSPK 238
Query: 233 VPSGSYNIAQVTYELNVLGTRGPRKMHCIMHSIPNSALDAGGSVPGQPELI---HRSLED 289
VPSGSYNIAQ+TYELNVLGTRGPR+MHCIM+SIP S+L+ GGSVP QPE + SL+D
Sbjct: 239 VPSGSYNIAQITYELNVLGTRGPRRMHCIMNSIPISSLEPGGSVPNQPEKLVPAPYSLDD 298
Query: 290 SFRS-ISFSK-SLDH-SIEFSSARFSEIGGSCNE-DDDGKMRPLVLKNKPPRWHEQLQCW 345
SFRS ISFSK S DH S++FSS+RFSE+G SC++ +++ RPL+LKNK PRWHEQLQCW
Sbjct: 299 SFRSNISFSKSSFDHRSLDFSSSRFSEMGISCDDNEEEASFRPLILKNKQPRWHEQLQCW 358
Query: 346 CLNFRGRVTVASVKNFQLIASTQP------PAGAPTPSQPAPPEHDKIILQFGKVGKDMF 399
CLNFRGRVTVASVKNFQL+A+ QP A APT S PA PE DK+ILQFGKVGKDMF
Sbjct: 359 CLNFRGRVTVASVKNFQLVAARQPQPQGTGAAAAPT-SAPAHPEQDKVILQFGKVGKDMF 417
Query: 400 TMDYRYPLSAFQAFAICLSSFDTKLACE 427
TMDYRYPLSAFQAFAICLSSFDTKLACE
Sbjct: 418 TMDYRYPLSAFQAFAICLSSFDTKLACE 445
>At1g76900 unknown protein
Length = 455
Score = 618 bits (1594), Expect = e-177
Identities = 331/456 (72%), Positives = 370/456 (80%), Gaps = 30/456 (6%)
Query: 1 MSFRSIVRDVRDSFGSLSRRSFDVKLTG-HHHRGKSHGSVQDLHDQPLV--IQNSCWANL 57
MSFRSIVRDVRDS GSLSRRSFD KL+ + GKS GSVQD H++ LV IQ + WANL
Sbjct: 1 MSFRSIVRDVRDSIGSLSRRSFDFKLSSLNKEGGKSRGSVQDSHEEQLVVTIQETPWANL 60
Query: 58 PPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKDIVRSPEFCGKLTFPVSLKQ 117
PPELL DVI+RLEESE+ WP+R+HVVACASVC+SWR+MCK+IV+SPE GK+TFPVSLKQ
Sbjct: 61 PPELLRDVIKRLEESESVWPARRHVVACASVCRSWRDMCKEIVQSPELSGKITFPVSLKQ 120
Query: 118 PGPRDGIIQCFIKRDKSNLTYHLFLCLSPALLVENGKFLLSAKRTRRTTYTEYVISMDAD 177
PGPRD +QCFIKRDKSNLTYHL+LCLSPALLVENGKFLLSAKR RRTTYTEYVISM AD
Sbjct: 121 PGPRDATMQCFIKRDKSNLTYHLYLCLSPALLVENGKFLLSAKRIRRTTYTEYVISMHAD 180
Query: 178 NISRSSNTYIGKLRSNFLGTKFIIYDTQPPCTS--AHICPPGTGKTSRRFSKKVSPKVPS 235
ISRSSNTYIGK+RSNFLGTKFIIYDTQP S A P G + R +SK+VSPKVPS
Sbjct: 181 TISRSSNTYIGKIRSNFLGTKFIIYDTQPAYNSNIARAVQP-VGLSRRFYSKRVSPKVPS 239
Query: 236 GSYNIAQVTYELNVLGTRGPRKMHCIMHSIPNSALDAGGSVPGQPELI--HRSLEDSFRS 293
GSY IAQV+YELNVLGTRGPR+MHC M+SIP S+L GG+VPGQP++I L++SFRS
Sbjct: 240 GSYKIAQVSYELNVLGTRGPRRMHCAMNSIPASSLAEGGTVPGQPDIIVPRSILDESFRS 299
Query: 294 ISFSKS----LDHSIEFSSARFSEIGGSCNED-----DDGKMR---PLVLKNKPPRWHEQ 341
I+ S S D+S +FSSARFS+I G +ED ++GK R PLVLKNKPPRWHEQ
Sbjct: 300 ITSSSSRKITYDYSNDFSSARFSDILGPLSEDQEVVLEEGKERNSPPLVLKNKPPRWHEQ 359
Query: 342 LQCWCLNFRGRVTVASVKNFQLIASTQP-PAGAPTP-----SQPAPPEH----DKIILQF 391
LQCWCLNFRGRVTVASVKNFQLIA+ QP P P P +QP P DKIILQF
Sbjct: 360 LQCWCLNFRGRVTVASVKNFQLIAANQPQPQPQPQPQPQPLTQPQPSGQTDGPDKIILQF 419
Query: 392 GKVGKDMFTMDYRYPLSAFQAFAICLSSFDTKLACE 427
GKVGKDMFTMD+RYPLSAFQAFAICLSSFDTKLACE
Sbjct: 420 GKVGKDMFTMDFRYPLSAFQAFAICLSSFDTKLACE 455
>At1g43640 unknown protein (At1g43640)
Length = 429
Score = 588 bits (1516), Expect = e-168
Identities = 302/442 (68%), Positives = 356/442 (80%), Gaps = 28/442 (6%)
Query: 1 MSFRSIVRDVRDSFGSLSRRSFDVKLTGH--HHRGKSHGSVQDLHDQPLVIQNSCWANLP 58
MSF SIVRDVRD+ GS SRRSFDV+++ H R KSHG + D +VI+N+ WANLP
Sbjct: 1 MSFLSIVRDVRDTVGSFSRRSFDVRVSNGTTHQRSKSHGVEAHIEDL-IVIKNTRWANLP 59
Query: 59 PELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKDIVRSPEFCGKLTFPVSLKQP 118
LL DV+++L+ESE+TWP+RK VVACA VC++WR MCKDIV+SPEF GKLTFPVSLKQP
Sbjct: 60 AALLRDVMKKLDESESTWPARKQVVACAGVCKTWRLMCKDIVKSPEFSGKLTFPVSLKQP 119
Query: 119 GPRDGIIQCFIKRDKSNLTYHLFLCLSPALLVENGKFLLSAKRTRRTTYTEYVISMDADN 178
GPRDGIIQC+IKRDKSN+TYHL+L LSPA+LVE+GKFLLSAKR+RR TYTEYVISMDADN
Sbjct: 120 GPRDGIIQCYIKRDKSNMTYHLYLSLSPAILVESGKFLLSAKRSRRATYTEYVISMDADN 179
Query: 179 ISRSSNTYIGKLRSNFLGTKFIIYDTQPPCTSAHICPPGTGKTSRRF-SKKVSPKVPSGS 237
ISRSS+TYIGKL+SNFLGTKFI+YDT P S+ I P SR F SKKVSPKVPSGS
Sbjct: 180 ISRSSSTYIGKLKSNFLGTKFIVYDTAPAYNSSQILSP--PNRSRSFNSKKVSPKVPSGS 237
Query: 238 YNIAQVTYELNVLGTRGPRKMHCIMHSIPNSALDAGGSVPGQPELIHRSLEDSFRSISFS 297
YNIAQVTYELN+LGTRGPR+M+CIMHSIP+ AL+ GG+VP QPE + RSL++SFRSI S
Sbjct: 238 YNIAQVTYELNLLGTRGPRRMNCIMHSIPSLALEPGGTVPSQPEFLQRSLDESFRSIGSS 297
Query: 298 KSLDHSIEFSSARFSEIGGSCNEDDDGKMRPLVLKNKPPRWHEQLQCWCLNFRGRVTVAS 357
K ++HS +F+ + +++GK+RPLVLK KPPRW + L+CWCLNF+GRVTVAS
Sbjct: 298 KIVNHSGDFTRPK----------EEEGKVRPLVLKTKPPRWLQPLRCWCLNFKGRVTVAS 347
Query: 358 VKNFQLI--ASTQPPAGAP----------TPSQPAPPEHDKIILQFGKVGKDMFTMDYRY 405
VKNFQL+ A+ QP +G+ +P QP HDKIIL FGKVGKDMFTMDYRY
Sbjct: 348 VKNFQLMSAATVQPGSGSDGGALATRPSLSPQQPEQSNHDKIILHFGKVGKDMFTMDYRY 407
Query: 406 PLSAFQAFAICLSSFDTKLACE 427
PLSAFQAFAI LS+FDTKLACE
Sbjct: 408 PLSAFQAFAISLSTFDTKLACE 429
>At2g47900 Tub family like protein
Length = 406
Score = 477 bits (1227), Expect = e-135
Identities = 245/432 (56%), Positives = 315/432 (72%), Gaps = 31/432 (7%)
Query: 1 MSFRSIVRDVRDSFGSLSRRSFDVKLTGHHHRGKSHGSVQDLHDQPLVIQNSCWANLPPE 60
MSF+S+++D+R GS+SR+ FDV+ + R +S VQD + SCWA++PPE
Sbjct: 1 MSFKSLIQDMRGELGSISRKGFDVRFG--YGRSRSQRVVQDTSVPVDAFKQSCWASMPPE 58
Query: 61 LLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKDIVRSPEFCGKLTFPVSLKQPGP 120
LL DV+ R+E+SE+TWPSRK+VV+CA VC++WR + K+IVR PE KLTFP+SLKQPGP
Sbjct: 59 LLRDVLMRIEQSEDTWPSRKNVVSCAGVCRNWREIVKEIVRVPELSSKLTFPISLKQPGP 118
Query: 121 RDGIIQCFIKRDKSNLTYHLFLCLSPALLVENGKFLLSAKRTRRTTYTEYVISMDADNIS 180
R ++QC+I R++SN TY+L+L L+ A ++GKFLL+AKR RR T T+Y+IS++ D++S
Sbjct: 119 RGSLVQCYIMRNRSNQTYYLYLGLNQAASNDDGKFLLAAKRFRRPTCTDYIISLNCDDVS 178
Query: 181 RSSNTYIGKLRSNFLGTKFIIYDTQPPCTSAHICPPGTGKTSRRFS-----KKVSPKVPS 235
R SNTYIGKLRSNFLGTKF +YD QP PGT T R S K+VSP++PS
Sbjct: 179 RGSNTYIGKLRSNFLGTKFTVYDAQP-------TNPGTQVTRTRSSRLLSLKQVSPRIPS 231
Query: 236 GSYNIAQVTYELNVLGTRGPRKMHCIMHSIPNSALDAGGSVPGQPELIHRSLEDSFRSIS 295
G+Y +A ++YELNVLG+RGPR+M C+M +IP SA++ GG+ P Q EL+H +L DSF S S
Sbjct: 232 GNYPVAHISYELNVLGSRGPRRMQCVMDAIPASAVEPGGTAPTQTELVHSNL-DSFPSFS 290
Query: 296 FSKSLDHSIEFSSARFSEIGGSCNEDDDGKMRPLVLKNKPPRWHEQLQCWCLNFRGRVTV 355
F +S SI S G S +G LVLKNK PRWHEQLQCWCLNF GRVTV
Sbjct: 291 FFRS--KSIRAESL---PSGPSSAAQKEGL---LVLKNKAPRWHEQLQCWCLNFNGRVTV 342
Query: 356 ASVKNFQLIASTQPPAGAPTPSQPAPPEHDKIILQFGKVGKDMFTMDYRYPLSAFQAFAI 415
ASVKNFQL+A+ + + PA PEH+ +ILQFGKVGKD+FTMDY+YP+SAFQAF I
Sbjct: 343 ASVKNFQLVAAPE--------NGPAGPEHENVILQFGKVGKDVFTMDYQYPISAFQAFTI 394
Query: 416 CLSSFDTKLACE 427
CLSSFDTK+ACE
Sbjct: 395 CLSSFDTKIACE 406
>At2g18280 tubby-like protein 2
Length = 394
Score = 415 bits (1066), Expect = e-116
Identities = 231/434 (53%), Positives = 283/434 (64%), Gaps = 48/434 (11%)
Query: 1 MSFRSIVRD---VRDSFGSLSRRSFDVKLTGHHHRGKSHGSVQDLHDQPLV-IQNSCWAN 56
MS +SI+RD VRD G +S+RS+ KS D PL I S WA+
Sbjct: 1 MSLKSILRDLKEVRDGLGGISKRSWS----------KSSHIAPDQTTPPLDNIPQSPWAS 50
Query: 57 LPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKDIVRSPEFCGKLTFPVSLK 116
LPPELL D+I R+EESE WP+R VV+CASVC+SWR + +IVR PE CGKLTFP+SLK
Sbjct: 51 LPPELLHDIIWRVEESETAWPARAAVVSCASVCKSWRGITMEIVRIPEQCGKLTFPISLK 110
Query: 117 QPGPRDGIIQCFIKRDKSNLTYHLFLCLSPALLVENGKFLLSAKRTRRTTYTEYVISMDA 176
QPGPRD IQCFIKR+++ TY L+ L P+ EN K LL+A+R RR T T+++IS+ A
Sbjct: 111 QPGPRDSPIQCFIKRNRATATYILYYGLMPS-ETENDKLLLAARRIRRATCTDFIISLSA 169
Query: 177 DNISRSSNTYIGKLRSNFLGTKFIIYDTQPPCTSAHICPPGTGKTSRRFSKKVSPKVPSG 236
N SRSS+TY+GKLRS FLGTKF IYD Q ++A P R K+ +PK+P+
Sbjct: 170 KNFSRSSSTYVGKLRSGFLGTKFTIYDNQTASSTAQAQP-----NRRLHPKQAAPKLPTN 224
Query: 237 SYNIAQVTYELNVLGTRGPRKMHCIMHSIPNSALDAGGS-VPGQPELIHRSLEDSFRSIS 295
S + +TYELNVL TRGPR+MHC M SIP S++ A S V G E + S +I+
Sbjct: 225 SSTVGNITYELNVLRTRGPRRMHCAMDSIPLSSVIAEPSVVQGIEEEVSSSPSPKGETIT 284
Query: 296 FSKSLDHSIEFSSARFSEIGGSCNEDDDGKMR--PLVLKNKPPRWHEQLQCWCLNFRGRV 353
K + D+ +R PLVLKNK PRWHEQLQCWCLNF+GRV
Sbjct: 285 TDKEI-------------------PDNSPSLRDQPLVLKNKSPRWHEQLQCWCLNFKGRV 325
Query: 354 TVASVKNFQLIASTQPPAGAPTPSQPAPPEHDKIILQFGKVGKDMFTMDYRYPLSAFQAF 413
TVASVKNFQL+A AP P EH+++ILQFGK+GKD+FTMDYRYPLSAFQAF
Sbjct: 326 TVASVKNFQLVAEIDASLDAP------PEEHERVILQFGKIGKDIFTMDYRYPLSAFQAF 379
Query: 414 AICLSSFDTKLACE 427
AIC+SSFDTK ACE
Sbjct: 380 AICISSFDTKPACE 393
>At1g47270 unknown protein
Length = 388
Score = 400 bits (1028), Expect = e-112
Identities = 217/429 (50%), Positives = 274/429 (63%), Gaps = 43/429 (10%)
Query: 1 MSFRSIVRDVRDSFGSLSRRSFDVKLTGHHHRGKSHGSVQDLHDQPLVIQNSCWANLPPE 60
MS ++IV++ + ++ RR G H SV + S W +LPPE
Sbjct: 1 MSLKNIVKN---KYKAIGRR-------GRSHIAPEGSSVSSSLSTNEGLNQSIWVDLPPE 50
Query: 61 LLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKDIVRSPEFCGKLTFPVSLKQPGP 120
LL D+I+R+E ++ WP R+ VVACASVC+SWR M K++V+ PE G +TFP+SL+QPGP
Sbjct: 51 LLLDIIQRIESEQSLWPGRRDVVACASVCKSWREMTKEVVKVPELSGLITFPISLRQPGP 110
Query: 121 RDGIIQCFIKRDKSNLTYHLFLCLSPALLVENGKFLLSAKRTRRTTYTEYVISMDADNIS 180
RD IQCFIKR+++ Y L+L LSPAL + K LLSAKR RR T E+V+S+ ++ S
Sbjct: 111 RDAPIQCFIKRERATGIYRLYLGLSPALSGDKSKLLLSAKRVRRATGAEFVVSLSGNDFS 170
Query: 181 RSSNTYIGKLRSNFLGTKFIIYDTQPPCTSAHICPPGTGKTSRRFSKKVSPKV--PSGSY 238
RSS+ YIGKLRSNFLGTKF +Y+ QPP + + P S +VSP V S SY
Sbjct: 171 RSSSNYIGKLRSNFLGTKFTVYENQPPPFNRKLPP----------SMQVSPWVSSSSSSY 220
Query: 239 NIAQVTYELNVLGTRGPRKMHCIMHSIPNSALDAGGSVPGQPELIHRSLEDSFRSISFSK 298
NIA + YELNVL TRGPR+M CIMHSIP SA+ GG + E ++ K
Sbjct: 221 NIASILYELNVLRTRGPRRMQCIMHSIPISAIQEGGKIQSPTEFTNQG----------KK 270
Query: 299 SLDHSIEFSSARFSEIGGSCNEDDDGKMRPLVLKNKPPRWHEQLQCWCLNFRGRVTVASV 358
++F S +GG + PL+LKNK PRWHEQLQCWCLNF+GRVTVASV
Sbjct: 271 KKKPLMDFCS---GNLGG-----ESVIKEPLILKNKSPRWHEQLQCWCLNFKGRVTVASV 322
Query: 359 KNFQLIASTQPPAGAPTPSQPAPPEHDKIILQFGKVGKDMFTMDYRYPLSAFQAFAICLS 418
KNFQL+A+ A A E D++ILQFGK+GKD+FTMDYRYP+SAFQAFAICLS
Sbjct: 323 KNFQLVAAA---AEAGKNMNIPEEEQDRVILQFGKIGKDIFTMDYRYPISAFQAFAICLS 379
Query: 419 SFDTKLACE 427
SFDTK CE
Sbjct: 380 SFDTKPVCE 388
>At3g06380 unknown protein
Length = 380
Score = 372 bits (955), Expect = e-103
Identities = 197/379 (51%), Positives = 260/379 (67%), Gaps = 36/379 (9%)
Query: 54 WANLPPELLFDVIRRLEESEN-TWPSRKHVVACASVCQSWRNMCKDIVRSPEFCGKLTFP 112
W+ LP ELL +++ R+E + WPSR++VVACA VC+SWR + K+IV PEF KLTFP
Sbjct: 33 WSELPEELLREILIRVETVDGGDWPSRRNVVACAGVCRSWRILTKEIVAVPEFSSKLTFP 92
Query: 113 VSLKQPGPRDGIIQCFIKRDKSNLTYHLFLCLSPALLVENGKFLLSAKRTRRTTYTEYVI 172
+SLKQ GPRD ++QCFIKR+++ +YHL+L L+ + L +NGKFLL+A + +R T T+Y+I
Sbjct: 93 ISLKQSGPRDSLVQCFIKRNRNTQSYHLYLGLTTS-LTDNGKFLLAASKLKRATCTDYII 151
Query: 173 SMDADNISRSSNTYIGKLRSNFLGTKFIIYDTQPPCTSAHICPPGTGKTSRRFSK---KV 229
S+ +D+IS+ SN Y+G++RSNFLGTKF ++D G K + S KV
Sbjct: 152 SLRSDDISKRSNAYLGRMRSNFLGTKFTVFDGS---------QTGAAKMQKSRSSNFIKV 202
Query: 230 SPKVPSGSYNIAQVTYELNVLGTRGPRKMHCIMHSIPNSALDAGGSVPGQPELIHRSLED 289
SP+VP GSY IA ++YELNVLG+RGPR+M CIM +IP S +++ G V S+
Sbjct: 203 SPRVPQGSYPIAHISYELNVLGSRGPRRMRCIMDTIPMSIVESRGVVAS------TSISS 256
Query: 290 -SFRSISFSKSLDHSIEFSSARFSEIGGSCNEDDDGKMRPLVLKNKPPRWHEQLQCWCLN 348
S RS +S + +SA S+ G + + PLVL NK PRWHEQL+CWCLN
Sbjct: 257 FSSRSSPVFRSHSKPLRSNSASCSDSGNNLGDP------PLVLSNKAPRWHEQLRCWCLN 310
Query: 349 FRGRVTVASVKNFQLIASTQPPAGAPTPSQPAPPEHDKIILQFGKVGKDMFTMDYRYPLS 408
F GRVTVASVKNFQL+A + AG + ++IILQFGKVGKDMFTMDY YP+S
Sbjct: 311 FHGRVTVASVKNFQLVAVSDCEAGQTS---------ERIILQFGKVGKDMFTMDYGYPIS 361
Query: 409 AFQAFAICLSSFDTKLACE 427
AFQAFAICLSSF+T++ACE
Sbjct: 362 AFQAFAICLSSFETRIACE 380
>At5g18680 tub family-like protein
Length = 389
Score = 360 bits (925), Expect = e-100
Identities = 202/411 (49%), Positives = 265/411 (64%), Gaps = 47/411 (11%)
Query: 32 RGKSHGSVQDL------HDQPLVIQNSCWANLPPELLFDVIRRLEESENT-WPSRKHVVA 84
R + H V DL + Q+ W+ +P ELL +++ R+E ++ WPSR+ VVA
Sbjct: 11 RSRPHRVVHDLAAAAAADSTSVSSQDYRWSEIPEELLREILIRVEAADGGGWPSRRSVVA 70
Query: 85 CASVCQSWRNMCKDIVRSPEFCGKLTFPVSLKQPGPRDGIIQCFIKRDKSNLTYHLFLCL 144
CA VC+ WR + + V PE KLTFP+SLKQPGPRD ++QCFIKR++ +YHL+L L
Sbjct: 71 CAGVCRGWRLLMNETVVVPEISSKLTFPISLKQPGPRDSLVQCFIKRNRITQSYHLYLGL 130
Query: 145 SPALLVENGKFLLSAKRTRRTTYTEYVISMDADNISRSSNTYIGKLRSNFLGTKFIIYDT 204
+ + L ++GKFLL+A + + TT T+Y+IS+ +D++SR S Y+GK+RSNFLGTKF ++D
Sbjct: 131 TNS-LTDDGKFLLAACKLKHTTCTDYIISLRSDDMSRRSQAYVGKVRSNFLGTKFTVFDG 189
Query: 205 QPPCTSAHICPPGTGKTSRRFSK-----KVSPKVPSGSYNIAQVTYELNVLGTRGPRKMH 259
P TG R S+ KVS KVP GSY +A +TYELNVLG+RGPRKM
Sbjct: 190 N--------LLPSTGAAKLRKSRSYNPAKVSAKVPLGSYPVAHITYELNVLGSRGPRKMQ 241
Query: 260 CIMHSIPNSALDAGG--SVPGQ-PELIHRSLEDSFRSISFSKSLDHSIEFSSARFSEIGG 316
C+M +IP S ++ G S P + P L RS + S+S + SS+ E
Sbjct: 242 CLMDTIPTSTMEPQGVASEPSEFPLLGTRS--------TLSRSQSKPLRSSSSHLKE--- 290
Query: 317 SCNEDDDGKMRPLVLKNKPPRWHEQLQCWCLNFRGRVTVASVKNFQLIASTQPPAGAPTP 376
PLVL NK PRWHEQL+CWCLNF GRVTVASVKNFQL+A+ G+ T
Sbjct: 291 ----------TPLVLSNKTPRWHEQLRCWCLNFHGRVTVASVKNFQLVAA-GASCGSGTG 339
Query: 377 SQPAPPEHDKIILQFGKVGKDMFTMDYRYPLSAFQAFAICLSSFDTKLACE 427
P + ++IILQFGKVGKDMFTMDY YP+SAFQAFAICLSSF+T++ACE
Sbjct: 340 MSP-ERQSERIILQFGKVGKDMFTMDYGYPISAFQAFAICLSSFETRIACE 389
>At1g53320 tubby like protein (Tub family protein)
Length = 379
Score = 338 bits (867), Expect = 3e-93
Identities = 185/379 (48%), Positives = 239/379 (62%), Gaps = 42/379 (11%)
Query: 51 NSCWANLPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKDIVRSPEFCGKLT 110
+S W+ + PELL ++IRR+EE+E+ WP R+ VV CA V + WR + D RS GK+T
Sbjct: 41 SSSWSAMLPELLGEIIRRVEETEDRWPQRRDVVTCACVSKKWREITHDFARSSLNSGKIT 100
Query: 111 FPVSLKQPGPRDGIIQCFIKRDKSNLTYHLFLCLSPALLVENGKFLLSAKRTRRTTYTEY 170
FP LK PGPRD QC IKR+K T++L+L L+P+ + GKFLL+A+R R YTEY
Sbjct: 101 FPSCLKLPGPRDFSNQCLIKRNKKTSTFYLYLALTPS-FTDKGKFLLAARRFRTGAYTEY 159
Query: 171 VISMDADNISRSSNTYIGKLRSNFLGTKFIIYDTQPPCTSAHICPPGTGKTSRRF-SKKV 229
+IS+DAD+ S+ SN Y+GKLRS+FLGT F +YD+QPP A P GK SRRF SK++
Sbjct: 160 IISLDADDFSQGSNAYVGKLRSDFLGTNFTVYDSQPPHNGA---KPSNGKASRRFASKQI 216
Query: 230 SPKVPSGSYNIAQVTYELNVLGTRGPRKMHCIMHSIPNSALDAGGSVPGQPELIHRSLED 289
SP+VP+G++ + V+Y+ N+L +RGPR+M S L P
Sbjct: 217 SPQVPAGNFEVGHVSYKFNLLKSRGPRRM--------VSTLRCPSPSPS----------- 257
Query: 290 SFRSISFSKSLDHSIEFSSARFSEIGGSCNEDDDGKMRPLVLKNKPPRWHEQLQCWCLNF 349
S S S + ++I N+D +LKNK PRWHE LQCWCLNF
Sbjct: 258 -------SSSAGLSSDQKPCDVTKIMKKPNKDGSSL---TILKNKAPRWHEHLQCWCLNF 307
Query: 350 RGRVTVASVKNFQLIASTQPPAGAPTPSQPA-PPEHDKIILQFGKVGKDMFTMDYRYPLS 408
GRVTVASVKNFQL+A+ SQP+ + + ++LQFGKVG D FTMDYR PLS
Sbjct: 308 HGRVTVASVKNFQLVATVD-------QSQPSGKGDEETVLLQFGKVGDDTFTMDYRQPLS 360
Query: 409 AFQAFAICLSSFDTKLACE 427
AFQAFAICL+SF TKLACE
Sbjct: 361 AFQAFAICLTSFGTKLACE 379
>At1g61940 hypothetical protein, 5' partial
Length = 265
Score = 118 bits (296), Expect = 5e-27
Identities = 49/86 (56%), Positives = 69/86 (79%)
Query: 57 LPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKDIVRSPEFCGKLTFPVSLK 116
+PPELL DV+ R+E SE+TWPSRK+VV+C VC++WR + K+IV PE K TFP+SLK
Sbjct: 1 MPPELLRDVLMRIERSEDTWPSRKNVVSCVGVCKNWRQIFKEIVNVPEVSSKFTFPISLK 60
Query: 117 QPGPRDGIIQCFIKRDKSNLTYHLFL 142
QPGP ++QC++KR++SN T++L+L
Sbjct: 61 QPGPGGSLVQCYVKRNRSNQTFYLYL 86
Score = 40.0 bits (92), Expect = 0.002
Identities = 39/140 (27%), Positives = 51/140 (35%), Gaps = 57/140 (40%)
Query: 256 RKMHCIMHSIPNSALDAGGSVPGQPELIHRSLEDSFRSISFSKSLDHSIEFSSARFSEIG 315
R+ HC+M +I SA+ GG+ Q EL D+ + F S + G
Sbjct: 109 RETHCVMDAISASAVKPGGTATTQTEL------------------DNFVSFRSPSGQKEG 150
Query: 316 GSCNEDDDGKMRPLVLKNKPPRWHEQLQCWCLNFRGRVTVASVKNFQLIASTQPPAGAPT 375
LVLK+K PR E Q WCL+F G
Sbjct: 151 ------------VLVLKSKVPRLEE--QSWCLDFNG------------------------ 172
Query: 376 PSQPAPPEHDKIILQFGKVG 395
S PE++ I QF KVG
Sbjct: 173 -SPENEPENENDIFQFAKVG 191
>At1g16070 unknown protein
Length = 397
Score = 58.9 bits (141), Expect = 5e-09
Identities = 38/99 (38%), Positives = 50/99 (50%), Gaps = 26/99 (26%)
Query: 333 NKPPRWHEQLQCWCLNFRGRVTV-----ASVKNFQLIASTQPPAGAPTPSQPAPPEHDKI 387
NK + HE L+FR R +SVKNFQL + P +
Sbjct: 318 NKISKQHE------LDFRDRGRTGLRIQSSVKNFQLTLTETPR---------------QT 356
Query: 388 ILQFGKVGKDMFTMDYRYPLSAFQAFAICLSSFDTKLAC 426
ILQ G+V K + +D+RYP S +QAF ICL+S D+KL C
Sbjct: 357 ILQMGRVDKARYVIDFRYPFSGYQAFCICLASIDSKLCC 395
Score = 28.1 bits (61), Expect = 9.7
Identities = 14/49 (28%), Positives = 29/49 (58%), Gaps = 1/49 (2%)
Query: 156 LLSAKRTRRTTYTEYVISMDADNISRSSN-TYIGKLRSNFLGTKFIIYD 203
L A +RR + + ++ + + SS+ +Y+G + +N LG+K+ I+D
Sbjct: 189 LAVAYHSRRNGKSIFRVAQNVKGLLCSSDESYVGSMTANLLGSKYYIWD 237
>At4g05490 N7 -like protein
Length = 307
Score = 37.4 bits (85), Expect = 0.016
Identities = 20/47 (42%), Positives = 26/47 (54%), Gaps = 7/47 (14%)
Query: 52 SCWANLPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKD 98
S WA LPP+LL ++ RL E +RK VC+SWR + KD
Sbjct: 25 SNWAELPPDLLSSILLRLSPLEILENARK-------VCRSWRRVSKD 64
>At4g05460 N7 like-protein
Length = 302
Score = 36.6 bits (83), Expect = 0.027
Identities = 18/49 (36%), Positives = 27/49 (54%), Gaps = 7/49 (14%)
Query: 50 QNSCWANLPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKD 98
+++ W LPPEL ++ RL E ++K VC+SWR +CKD
Sbjct: 15 ESTNWTELPPELTSAILHRLGAIEILENAQK-------VCRSWRRVCKD 56
>At4g05480 N7 -like protein
Length = 322
Score = 35.8 bits (81), Expect = 0.046
Identities = 16/45 (35%), Positives = 26/45 (57%), Gaps = 7/45 (15%)
Query: 54 WANLPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKD 98
W +LPPEL ++ RL ++ +RK +C++WR +CKD
Sbjct: 40 WVDLPPELTTSILLRLSVTDILDNARK-------LCRAWRRICKD 77
>At5g25350 leucine-rich repeats containing protein
Length = 623
Score = 34.7 bits (78), Expect = 0.10
Identities = 19/46 (41%), Positives = 25/46 (54%), Gaps = 7/46 (15%)
Query: 57 LPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKDIVRS 102
LP E LF+++RRL PS + ACA V + W N+ I RS
Sbjct: 58 LPEECLFEILRRL-------PSGQERSACACVSKHWLNLLSSISRS 96
>At4g05470 N7 -like protein
Length = 304
Score = 34.7 bits (78), Expect = 0.10
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 7/45 (15%)
Query: 54 WANLPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKD 98
W +LPPEL ++ RL ++ ++K VC+ WR +CKD
Sbjct: 46 WVDLPPELTTSILLRLSLTDILDNAQK-------VCKEWRRICKD 83
>At5g55150 putative protein
Length = 340
Score = 33.9 bits (76), Expect = 0.18
Identities = 16/59 (27%), Positives = 29/59 (49%), Gaps = 7/59 (11%)
Query: 47 LVIQNSCWANLPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKDIVRSPEF 105
+ + +S W+ PELL V L ++ + ++ CA+VC SW++ + S F
Sbjct: 1 MALPSSSWSEFLPELLNTVFHNLNDARD-------ILNCATVCSSWKDSSSAVYYSRTF 52
>At4g15475 unknown protein
Length = 610
Score = 30.4 bits (67), Expect = 1.9
Identities = 16/45 (35%), Positives = 24/45 (52%), Gaps = 10/45 (22%)
Query: 51 NSCWANLPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNM 95
N+C LP EL+ ++ RRLE N AC+ VC+ W ++
Sbjct: 8 NNC---LPEELILEIFRRLESKPNR-------DACSLVCKRWLSL 42
>At1g14000 putative protein kinase
Length = 438
Score = 30.0 bits (66), Expect = 2.5
Identities = 20/57 (35%), Positives = 30/57 (52%), Gaps = 8/57 (14%)
Query: 28 GHHHRGKSHGSVQDLHDQPLVIQNSCW---ANLPPELLFDVIRRLEESENTWPSRKH 81
GH +S G DL + L+++ CW N P L D+++RLE+ + T PS H
Sbjct: 381 GHRPTFRSKGCTPDLRE--LIVK--CWDADMNQRPSFL-DILKRLEKIKETLPSDHH 432
>At4g05500
Length = 449
Score = 29.6 bits (65), Expect = 3.3
Identities = 15/45 (33%), Positives = 22/45 (48%), Gaps = 7/45 (15%)
Query: 54 WANLPPELLFDVIRRLEESENTWPSRKHVVACASVCQSWRNMCKD 98
WA LP +L ++ RL E ++K VC+ W +CKD
Sbjct: 181 WAELPSKLTSSILLRLGAIEILQNAQK-------VCKPWHRVCKD 218
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,557,057
Number of Sequences: 26719
Number of extensions: 488232
Number of successful extensions: 1612
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 1543
Number of HSP's gapped (non-prelim): 37
length of query: 427
length of database: 11,318,596
effective HSP length: 102
effective length of query: 325
effective length of database: 8,593,258
effective search space: 2792808850
effective search space used: 2792808850
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136472.10


