

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136451.6 + phase: 0 /pseudo
(210 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
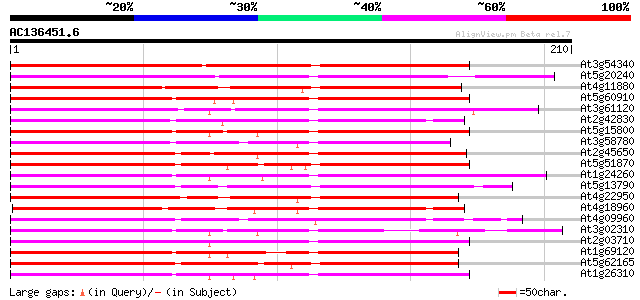
Score E
Sequences producing significant alignments: (bits) Value
At3g54340 floral homeotic protein APETALA3 (AP3) 191 3e-49
At5g20240 PI protein (PISTILLATA) 125 1e-29
At4g11880 MADS-box protein AGL14 122 2e-28
At5g60910 floral homeotic protein AGL8 116 7e-27
At3g61120 MADS-box protein AGL13 114 3e-26
At2g42830 MADS box transcription factor (AGL5) 114 4e-26
At5g15800 transcription factor (AGL2) 113 8e-26
At3g58780 shatterproof 1 (SHP1)/ agamous -like 1 (AGL1) 113 8e-26
At2g45650 MADS-box protein (AGL6) 112 1e-25
At5g51870 MADS box transcription factor-like 111 3e-25
At1g24260 floral homeotic protein, AGL9 111 3e-25
At5g13790 floral homeotic protein AGL15 (sp|Q38847) 110 4e-25
At4g22950 putative MADS Box / AGL protein 110 4e-25
At4g18960 floral homeotic protein agamous 110 7e-25
At4g09960 MADS-box protein AGL11 110 7e-25
At3g02310 floral homeotic protein AGL4 109 1e-24
At2g03710 MADS-box protein (AGL3) 109 1e-24
At1g69120 unknown protein 109 1e-24
At5g62165 unknown protein 108 3e-24
At1g26310 cauliflower (CAL) 107 3e-24
>At3g54340 floral homeotic protein APETALA3 (AP3)
Length = 232
Score = 191 bits (484), Expect = 3e-49
Identities = 94/172 (54%), Positives = 129/172 (74%), Gaps = 4/172 (2%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGKI+IK IEN TNRQVTYSKRRNG+FKKAHEL+VLCDA+VS+IMFS +NK+HEYI+P
Sbjct: 1 MARGKIQIKRIENQTNRQVTYSKRRNGLFKKAHELTVLCDARVSIIMFSSSNKLHEYISP 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
+TK+I+D YQ T+ D+D+W + YE+M E +KL + N LR QI+ R+GE LD+L
Sbjct: 61 NTTTKEIVDLYQ-TISDVDVWATQYERMQETKRKLLETNRNLRTQIKQRLGEC---LDEL 116
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
Q+LR LE++M ++ +RERKF + + +T +KK +S + + NL+ EL
Sbjct: 117 DIQELRRLEDEMENTFKLVRERKFKSLGNQIETTKKKNKSQQDIQKNLIHEL 168
>At5g20240 PI protein (PISTILLATA)
Length = 208
Score = 125 bits (315), Expect = 1e-29
Identities = 70/204 (34%), Positives = 112/204 (54%), Gaps = 14/204 (6%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIK IEN NR VT+SKRRNG+ KKA E++VLCDAKV+LI+F+ N KM +Y P
Sbjct: 1 MGRGKIEIKRIENANNRVVTFSKRRNGLVKKAKEITVLCDAKVALIIFASNGKMIDYCCP 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
+ ++DQYQK G LW + +E + + ++K N L+ ++RH GE ++ L
Sbjct: 61 SMDLGAMLDQYQKLSGK-KLWDAKHENLSNEIDRIKKENDSLQLELRHLKGE---DIQSL 116
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELVRTLFLSS 180
+ + L ++E + + K+R+ + ++ ++ + Q+ T L
Sbjct: 117 NLKNLMAVEHAIEHGLDKVRDHQMEILISKRRNEKMMAEEQRQL----------TFQLQQ 166
Query: 181 YLYAFCQHHSHLNLPHHHGEEGYK 204
A + + + H G+ GY+
Sbjct: 167 QEMAIASNARGMMMRDHDGQFGYR 190
>At4g11880 MADS-box protein AGL14
Length = 221
Score = 122 bits (305), Expect = 2e-28
Identities = 72/176 (40%), Positives = 110/176 (61%), Gaps = 15/176 (8%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGK E+K IEN T+RQVT+SKRRNG+ KKA ELSVLCDA+V+LI+FS K++E+ +
Sbjct: 1 MVRGKTEMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIIFSPRGKLYEF-SS 59
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRH-------RIGEG 113
S K +++YQK + D+ K +N ++ KD + L R+I H +GEG
Sbjct: 60 SSSIPKTVERYQKRIQDL----GSNHKRNDNSQQSKDETYGLARKIEHLEISTRKMMGEG 115
Query: 114 GMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLL 169
LD S ++L+ LE ++ S+ KIR +K+ +++ T+ ++K R+L N L+
Sbjct: 116 ---LDASSIEELQQLENQLDRSLMKIRAKKYQLLREETEKLKEKERNLIAENKMLM 168
>At5g60910 floral homeotic protein AGL8
Length = 242
Score = 116 bits (291), Expect = 7e-27
Identities = 71/176 (40%), Positives = 109/176 (61%), Gaps = 8/176 (4%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++++K IEN NRQVT+SKRR+G+ KKAHE+SVLCDA+V+LI+FS K+ EY T
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALIVFSSKGKLFEYSTD 60
Query: 61 GLSTKKIIDQYQKTL-GDIDLWR---SHYEKMLENLKKLKDINHKLRRQIRHRIGEGGME 116
++I+++Y + L D L S E + KLK L + R+ +GE +
Sbjct: 61 S-CMERILERYDRYLYSDKQLVGRDVSQSENWVLEHAKLKARVEVLEKNKRNFMGE---D 116
Query: 117 LDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
LD LS ++L+SLE ++++I IR RK + +KK ++L+ N +LL ++
Sbjct: 117 LDSLSLKELQSLEHQLDAAIKSIRSRKNQAMFESISALQKKDKALQDHNNSLLKKI 172
>At3g61120 MADS-box protein AGL13
Length = 244
Score = 114 bits (286), Expect = 3e-26
Identities = 74/208 (35%), Positives = 118/208 (56%), Gaps = 16/208 (7%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGK+E+K IEN RQVT+SKR++G+ KKA+ELSVLCDA+VSLI+FS K++E+
Sbjct: 1 MGRGKVEVKRIENKITRQVTFSKRKSGLLKKAYELSVLCDAEVSLIIFSTGGKLYEFSNV 60
Query: 61 GLSTKKIIDQYQK---TLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMEL 117
G+ + I++Y + L D D + + + + KLK L R R+ +GE +L
Sbjct: 61 GVG--RTIERYYRCKDNLLDNDTLED-TQGLRQEVTKLKCKYESLLRTHRNLVGE---DL 114
Query: 118 DDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL----- 172
+ +S ++L++LE + +++ R++K V+ + + R+K R L +N L LE
Sbjct: 115 EGMSIKELQTLERQLEGALSATRKQKTQVMMEQMEELRRKERELGDINNKLKLETEDHDF 174
Query: 173 --VRTLFLSSYLYAFCQHHSHLNLPHHH 198
+ L L+ L A C L H +
Sbjct: 175 KGFQDLLLNPVLTAGCSTDFSLQSTHQN 202
>At2g42830 MADS box transcription factor (AGL5)
Length = 246
Score = 114 bits (285), Expect = 4e-26
Identities = 65/174 (37%), Positives = 105/174 (59%), Gaps = 11/174 (6%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
+GRGKIEIK IEN TNRQVT+ KRRNG+ KKA+ELSVLCDA+V+L++FS +++EY
Sbjct: 16 IGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVIFSTRGRLYEYANN 75
Query: 61 GLSTKKIIDQYQKTLGDI----DLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGME 116
S + I++Y+K D + ++ + + KL+ ++ RH +GE
Sbjct: 76 --SVRGTIERYKKACSDAVNPPTITEANTQYYQQEASKLRRQIRDIQNLNRHILGE---S 130
Query: 117 LDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLL 170
L L+F++L++LE + I+++R +K ++ + +K R +E N N+ L
Sbjct: 131 LGSLNFKELKNLESRLEKGISRVRSKKHEMLVAEIEYMQK--REIELQNDNMYL 182
>At5g15800 transcription factor (AGL2)
Length = 251
Score = 113 bits (282), Expect = 8e-26
Identities = 70/177 (39%), Positives = 110/177 (61%), Gaps = 10/177 (5%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++E+K IEN NRQVT++KRRNG+ KKA+ELSVLCDA+V+LI+FS K++E+ +
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
Query: 61 GLSTKKIIDQYQK-TLGDIDLWRSHYEKMLEN----LKKLKDINHKLRRQIRHRIGEGGM 115
+ K +D+YQK + G I++ + K LEN KLK L+RQ R+ +GE
Sbjct: 61 S-NMLKTLDRYQKCSYGSIEV-NNKPAKELENSYREYLKLKGRYENLQRQQRNLLGE--- 115
Query: 116 ELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
+L L+ ++L LE ++ S+ ++R K + + + K + L + N L ++L
Sbjct: 116 DLGPLNSKELEQLERQLDGSLKQVRSIKTQYMLDQLSDLQNKEQMLLETNRALAMKL 172
>At3g58780 shatterproof 1 (SHP1)/ agamous -like 1 (AGL1)
Length = 248
Score = 113 bits (282), Expect = 8e-26
Identities = 69/172 (40%), Positives = 103/172 (59%), Gaps = 15/172 (8%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
+GRGKIEIK IEN TNRQVT+ KRRNG+ KKA+ELSVLCDA+V+L++FS +++EY
Sbjct: 16 LGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVIFSTRGRLYEYANN 75
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQI-------RHRIGEG 113
S + I++Y+K D S E N + + KLRRQI RH +GE
Sbjct: 76 --SVRGTIERYKKACSDAVNPPSVTE---ANTQYYQQEASKLRRQIRDIQNSNRHIVGE- 129
Query: 114 GMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMN 165
L L+F++L++LE + I+++R +K ++ + +K+ L+ N
Sbjct: 130 --SLGSLNFKELKNLEGRLEKGISRVRSKKNELLVAEIEYMQKREMELQHNN 179
>At2g45650 MADS-box protein (AGL6)
Length = 252
Score = 112 bits (281), Expect = 1e-25
Identities = 66/175 (37%), Positives = 109/175 (61%), Gaps = 10/175 (5%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++E+K IEN NRQVT+SKRRNG+ KKA+ELSVLCDA+V+LI+FS K++E+ +
Sbjct: 1 MGRGRVEMKRIENKINRQVTFSKRRNGLLKKAYELSVLCDAEVALIIFSSRGKLYEFGSV 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLEN----LKKLKDINHKLRRQIRHRIGEGGME 116
G+ + I++Y + + L + E+ ++ + KLK L R R+ +GE +
Sbjct: 61 GI--ESTIERYNRCY-NCSLSNNKPEETTQSWCQEVTKLKSKYESLVRTNRNLLGE---D 114
Query: 117 LDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLE 171
L ++ ++L++LE + +++ R+RK V+ + RKK R L +N L ++
Sbjct: 115 LGEMGVKELQALERQLEAALTATRQRKTQVMMEEMEDLRKKERQLGDINKQLKIK 169
>At5g51870 MADS box transcription factor-like
Length = 207
Score = 111 bits (277), Expect = 3e-25
Identities = 71/179 (39%), Positives = 110/179 (60%), Gaps = 14/179 (7%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGKIEIK IEN T+RQVT+SKRR+G+FKKAHELSVLCDA+V+ I+FS++ ++HEY +
Sbjct: 1 MVRGKIEIKKIENVTSRQVTFSKRRSGLFKKAHELSVLCDAQVAAIVFSQSGRLHEYSSS 60
Query: 61 GLSTKKIIDQYQKTLGDIDL-WRSHYEKMLENLKKLKDINHKLRR----QIRHR--IGEG 113
+ +KIID+Y K + R E+ L+ LK +I+ +++ ++ HR +G+G
Sbjct: 61 QM--EKIIDRYGKFSNAFYVAERPQVERYLQELK--MEIDRMVKKIDLLEVHHRKLLGQG 116
Query: 114 GMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
LD S +L+ ++ + S+ +R RK + + ++K R L LL E+
Sbjct: 117 ---LDSCSVTELQEIDTQIEKSLRIVRSRKAELYADQLKKLKEKERELLNERKRLLEEV 172
>At1g24260 floral homeotic protein, AGL9
Length = 251
Score = 111 bits (277), Expect = 3e-25
Identities = 74/208 (35%), Positives = 115/208 (54%), Gaps = 11/208 (5%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++E+K IEN NRQVT++KRRNG+ KKA+ELSVLCDA+V+LI+FS K++E+ +
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
Query: 61 GLSTKKIIDQYQK-TLGDIDLWRSHYEKMLENLK------KLKDINHKLRRQIRHRIGEG 113
S + +++YQK G + E + L KLK+ L+R R+ +GE
Sbjct: 61 S-SMLRTLERYQKCNYGAPEPNVPSREALAVELSSQQEYLKLKERYDALQRTQRNLLGE- 118
Query: 114 GMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELV 173
+L LS ++L SLE ++SS+ +IR + + + + + K R L + N L L L
Sbjct: 119 --DLGPLSTKELESLERQLDSSLKQIRALRTQFMLDQLNDLQSKERMLTETNKTLRLRLA 176
Query: 174 RTLFLSSYLYAFCQHHSHLNLPHHHGEE 201
+ L + H HH ++
Sbjct: 177 DGYQMPLQLNPNQEEVDHYGRHHHQQQQ 204
>At5g13790 floral homeotic protein AGL15 (sp|Q38847)
Length = 268
Score = 110 bits (276), Expect = 4e-25
Identities = 72/188 (38%), Positives = 112/188 (59%), Gaps = 11/188 (5%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIK IEN +RQVT+SKRR+G+ KKA ELSVLCDA+V++I+FSK+ K+ EY +
Sbjct: 1 MGRGKIEIKRIENANSRQVTFSKRRSGLLKKARELSVLCDAEVAVIVFSKSGKLFEYSST 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
G+ K+ + +Y S E+ + LKD KL+ + G+G L+ L
Sbjct: 61 GM--KQTLSRYGNHQSSS---ASKAEEDCAEVDILKDQLSKLQEKHLQLQGKG---LNPL 112
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELVRTLFLSS 180
+F++L+SLE+ + ++ +RERK ++ + + R K + E N L ++ L S
Sbjct: 113 TFKELQSLEQQLYHALITVRERKERLLTNQLEESRLKEQRAELENETLRRQVQE---LRS 169
Query: 181 YLYAFCQH 188
+L +F +
Sbjct: 170 FLPSFTHY 177
>At4g22950 putative MADS Box / AGL protein
Length = 219
Score = 110 bits (276), Expect = 4e-25
Identities = 63/175 (36%), Positives = 111/175 (63%), Gaps = 16/175 (9%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGK E+K IEN T+RQVT+SKRRNG+ KKA ELSVLCDA+V+L++FS +K++E+ +
Sbjct: 1 MVRGKTEMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALVIFSPRSKLYEFSSS 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQI-------RHRIGEG 113
++ I++YQ+ + +I + K +N ++ +D L ++I R +GEG
Sbjct: 61 SIAA--TIERYQRRIKEI----GNNHKRNDNSQQARDETSGLTKKIEQLEISKRKLLGEG 114
Query: 114 GMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNL 168
+D S ++L+ LE ++ S+++IR +K+ +++ + + + R+L + N +L
Sbjct: 115 ---IDACSIEELQQLENQLDRSLSRIRAKKYQLLREEIEKLKAEERNLVKENKDL 166
>At4g18960 floral homeotic protein agamous
Length = 284
Score = 110 bits (274), Expect = 7e-25
Identities = 73/177 (41%), Positives = 108/177 (60%), Gaps = 19/177 (10%)
Query: 2 GRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITPG 61
GRGKIEIK IEN TNRQVT+ KRRNG+ KKA+ELSVLCDA+V+LI+FS +++EY
Sbjct: 50 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEY--SN 107
Query: 62 LSTKKIIDQYQKTLGDIDLWRSHYEKMLE-NLKKLKDINHKLRRQI-------RHRIGEG 113
S K I++Y+K + D S+ + E N + + + KLR+QI R +GE
Sbjct: 108 NSVKGTIERYKKAISD----NSNTGSVAEINAQYYQQESAKLRQQIISIQNSNRQLMGE- 162
Query: 114 GMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLL 170
+ +S ++LR+LE + SI +IR +K ++ + D +K R ++ N N +L
Sbjct: 163 --TIGSMSPKELRNLEGRLERSITRIRSKKNELLFSEIDYMQK--REVDLHNDNQIL 215
>At4g09960 MADS-box protein AGL11
Length = 230
Score = 110 bits (274), Expect = 7e-25
Identities = 74/196 (37%), Positives = 109/196 (54%), Gaps = 16/196 (8%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIK IEN TNRQVT+ KRRNG+ KKA+ELSVLCDA+V+LI+FS +++EY
Sbjct: 1 MGRGKIEIKRIENSTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSTRGRLYEYANN 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEG----GME 116
+ + I++Y+K D + E N + + KLR+QI+ G
Sbjct: 61 NI--RSTIERYKKACSDSTNTSTVQE---INAAYYQQESAKLRQQIQTIQNSNRNLMGDS 115
Query: 117 LDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELVRTL 176
L LS ++L+ +E + +I++IR +K ++ + +K R +E N N+ L RT
Sbjct: 116 LSSLSVKELKQVENRLEKAISRIRSKKHELLLVEIENAQK--REIELDNENIYL---RTK 170
Query: 177 FLSSYLYAFCQHHSHL 192
Y QHH +
Sbjct: 171 VAEVERYQ--QHHHQM 184
>At3g02310 floral homeotic protein AGL4
Length = 250
Score = 109 bits (272), Expect = 1e-24
Identities = 77/217 (35%), Positives = 122/217 (55%), Gaps = 36/217 (16%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++E+K IEN NRQVT++KRRNG+ KKA+ELSVLCDA+VSLI+FS K++E+ +
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVSLIVFSNRGKLYEFCST 60
Query: 61 GLSTKKIIDQYQK-TLGDIDLWRSHYEKMLEN----LKKLKDINHKLRRQIRHRIGEGGM 115
+ K +++YQK + G I++ + K LEN KLK L+RQ R+ +GE
Sbjct: 61 S-NMLKTLERYQKCSYGSIEV-NNKPAKELENSYREYLKLKGRYENLQRQQRNLLGE--- 115
Query: 116 ELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNG-----NLLL 170
+L L+ ++L LE ++ S+ ++R C K L+Q++ ++LL
Sbjct: 116 DLGPLNSKELEQLERQLDGSLKQVR-------------CIKTQYMLDQLSDLQGKEHILL 162
Query: 171 ELVRTLFLSSYLYAFCQHHSHLNLPHHHGEEGYKNDD 207
+ R L + + + + HHH G++ D
Sbjct: 163 DANRALSM--------KLEDMIGVRHHHIGGGWEGGD 191
>At2g03710 MADS-box protein (AGL3)
Length = 257
Score = 109 bits (272), Expect = 1e-24
Identities = 62/175 (35%), Positives = 101/175 (57%), Gaps = 6/175 (3%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGK+E+K IEN NRQVT++KRRNG+ KKA+ELSVLCDA+++L++FS K++E+ +
Sbjct: 1 MGRGKVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEIALLIFSNRGKLYEFCSS 60
Query: 61 GLSTKKIIDQYQK---TLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMEL 117
+ +D+Y+K D + + ++ KLK L+ RH +GE EL
Sbjct: 61 PSGMARTVDKYRKHSYATMDPNQSAKDLQDKYQDYLKLKSRVEILQHSQRHLLGE---EL 117
Query: 118 DDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
++ +L LE +++S+ +IR K + + + K L + N +L +L
Sbjct: 118 SEMDVNELEHLERQVDASLRQIRSTKARSMLDQLSDLKTKEEMLLETNRDLRRKL 172
>At1g69120 unknown protein
Length = 256
Score = 109 bits (272), Expect = 1e-24
Identities = 62/179 (34%), Positives = 109/179 (60%), Gaps = 22/179 (12%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++++K IEN NRQVT+SKRR G+ KKAHE+SVLCDA+V+L++FS K+ EY T
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALVVFSHKGKLFEYSTD 60
Query: 61 GLSTKKIIDQYQK----------TLGDIDL-WRSHYEKMLENLKKLKDINHKLRRQIRHR 109
+KI+++Y++ D++ W Y ++ ++ L+ R RH
Sbjct: 61 S-CMEKILERYERYSYAERQLIAPESDVNTNWSMEYNRLKAKIELLE-------RNQRHY 112
Query: 110 IGEGGMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNL 168
+GE +L +S ++L++LE+ +++++ IR RK ++ + +KK +++++ N L
Sbjct: 113 LGE---DLQAMSPKELQNLEQQLDTALKHIRTRKNQLMYESINELQKKEKAIQEQNSML 168
>At5g62165 unknown protein
Length = 210
Score = 108 bits (269), Expect = 3e-24
Identities = 63/174 (36%), Positives = 107/174 (61%), Gaps = 13/174 (7%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGKIE+K IEN T+RQVT+SKRRNG+ KKA+ELSVLCDA++SLI+FS+ +++E+ +
Sbjct: 1 MVRGKIEMKKIENATSRQVTFSKRRNGLLKKAYELSVLCDAQLSLIIFSQRGRLYEFSSS 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRR------QIRHRIGEGG 114
+ +K I++Y+K D + + L+ LK ++ +H + + R +G+G
Sbjct: 61 DM--QKTIERYRKYTKDHETSNHDSQIHLQQLK--QEASHMITKIELLEFHKRKLLGQG- 115
Query: 115 MELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNL 168
+ S ++L+ ++ + S+ K+RERK + K + + + K + L + N L
Sbjct: 116 --IASCSLEELQEIDSQLQRSLGKVRERKAQLFKEQLEKLKAKEKQLLEENVKL 167
>At1g26310 cauliflower (CAL)
Length = 255
Score = 107 bits (268), Expect = 3e-24
Identities = 64/178 (35%), Positives = 106/178 (58%), Gaps = 10/178 (5%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++E+K IEN NRQVT+SKRR G+ KKA E+SVLCDA+VSLI+FS K+ EY +
Sbjct: 1 MGRGRVELKRIENKINRQVTFSKRRTGLLKKAQEISVLCDAEVSLIVFSHKGKLFEYSSE 60
Query: 61 GLSTKKIIDQYQK-TLGDIDLWR--SHYEKMLE---NLKKLKDINHKLRRQIRHRIGEGG 114
+K++++Y++ + + L SH +LK L R RH +GE
Sbjct: 61 S-CMEKVLERYERYSYAERQLIAPDSHVNAQTNWSMEYSRLKAKIELLERNQRHYLGE-- 117
Query: 115 MELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
EL+ +S + L++LE+ + +++ IR RK ++ + ++K + +++ N L ++
Sbjct: 118 -ELEPMSLKDLQNLEQQLETALKHIRSRKNQLMNESLNHLQRKEKEIQEENSMLTKQI 174
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,710,933
Number of Sequences: 26719
Number of extensions: 188952
Number of successful extensions: 793
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 108
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 652
Number of HSP's gapped (non-prelim): 138
length of query: 210
length of database: 11,318,596
effective HSP length: 95
effective length of query: 115
effective length of database: 8,780,291
effective search space: 1009733465
effective search space used: 1009733465
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC136451.6


