

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136449.6 - phase: 0
(478 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
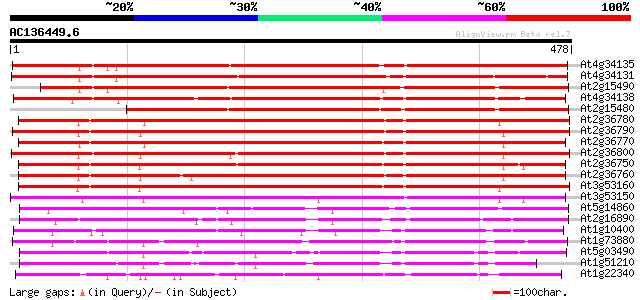
Score E
Sequences producing significant alignments: (bits) Value
At4g34135 glucosyltransferase like protein 503 e-142
At4g34131 glucosyltransferase -like protein 498 e-141
At2g15490 putative glucosyltransferase 464 e-131
At4g34138 glucosyltransferase -like protein 457 e-129
At2g15480 glucosyltransferase like protein 421 e-118
At2g36780 putative glucosyl transferase 413 e-116
At2g36790 putative glucosyl transferase 405 e-113
At2g36770 putative glucosyl transferase 400 e-112
At2g36800 putative glucosyl transferase 398 e-111
At2g36750 putative glucosyl transferase 388 e-108
At2g36760 putative glucosyl transferase 387 e-108
At3g53160 glucosyltransferase - like protein 376 e-104
At3g53150 glucosyltransferase - like protein 358 3e-99
At5g14860 glucosyltransferase -like protein 259 2e-69
At2g16890 glucosyltransferase like protein 258 7e-69
At1g10400 glucosyl transferase - like protein 238 4e-63
At1g73880 putative glucosyltransferase (At1g73880) 237 9e-63
At5g03490 UDPG glucosyltransferase - like protein 224 1e-58
At1g51210 putative glucosyl transferase 221 7e-58
At1g22340 hypothetical protein 204 7e-53
>At4g34135 glucosyltransferase like protein
Length = 483
Score = 503 bits (1294), Expect = e-142
Identities = 253/486 (52%), Positives = 344/486 (70%), Gaps = 20/486 (4%)
Query: 3 NENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAK---- 58
+ +++LH++FFPF+A GH+IP +D+A++FSSRG K TI+TT LN ++ + I K
Sbjct: 5 HHHRKLHVMFFPFMAYGHMIPTLDMAKLFSSRGAKSTILTTSLNSKILQKPIDTFKNLNP 64
Query: 59 ---INIKTIKFPSPEETGLPEGCENSE---SALAPDK---FIKFMKSTLLLREPLEHVLE 109
I+I+ FP E GLPEGCEN + S DK +KF ST ++ LE +L
Sbjct: 65 GLEIDIQIFNFPCVE-LGLPEGCENVDFFTSNNNDDKNEMIVKFFFSTRFFKDQLEKLLG 123
Query: 110 QEKPDCLVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPF 169
+PDCL+ADMFFPW+T++A KFN+PR+VFHG G+F LC C +KPQ +V+S +EPF
Sbjct: 124 TTRPDCLIADMFFPWATEAAGKFNVPRLVFHGTGYFSLCAGYCIGVHKPQKRVASSSEPF 183
Query: 170 VVPNLPGEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYR 229
V+P LPG I +T+ Q+ + K + E ESEVKS GV+ N+FYELE YAD Y+
Sbjct: 184 VIPELPGNIVITEEQIID-GDGESDMGKFMTEVRESEVKSSGVVLNSFYELEHDYADFYK 242
Query: 230 NELGRKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVF 289
+ + ++AWH+GP+S+ NR EEKA RG++A+IDE ECLKWL SK+PNSVIYV FGS+ F
Sbjct: 243 SCVQKRAWHIGPLSVYNRGFEEKAERGKKANIDEAECLKWLDSKKPNSVIYVSFGSVAFF 302
Query: 290 SDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQ 349
+ QL EIA GLEAS FIWVVRK+ + + EWLPEGFEER++G KG+IIRGWAPQ
Sbjct: 303 KNEQLFEIAAGLEASGTSFIWVVRKT---KDDREEWLPEGFEERVKG--KGMIIRGWAPQ 357
Query: 350 VMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQ 409
V+ILDH++ GGFVTHCGWNS LEGV+AGLPMVTWP+ EQFYN K ++ +++ GV VG
Sbjct: 358 VLILDHQATGGFVTHCGWNSLLEGVAAGLPMVTWPVGAEQFYNEKLVTQVLRTGVSVGAS 417
Query: 410 TWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNL 469
+ + G+ + ++ ++KAVR ++ G+ AEE R RAK+ MA+ AVE GGSS+ND ++
Sbjct: 418 KHMKVMMGDFISREKVDKAVREVLAGEAAEERRRRAKKLAAMAKAAVEEGGSSFNDLNSF 477
Query: 470 IEDLKS 475
+E+ S
Sbjct: 478 MEEFSS 483
>At4g34131 glucosyltransferase -like protein
Length = 481
Score = 498 bits (1283), Expect = e-141
Identities = 252/487 (51%), Positives = 349/487 (70%), Gaps = 21/487 (4%)
Query: 2 TNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAK--- 58
++ +++LH++FFPF+A GH+IP +D+A++FSSRG K TI+TT LN + + I + K
Sbjct: 3 SDPHRKLHVVFFPFMAYGHMIPTLDMAKLFSSRGAKSTILTTPLNSKIFQKPIERFKNLN 62
Query: 59 ----INIKTIKFPSPEETGLPEGCENSESALAPDK------FIKFMKSTLLLREPLEHVL 108
I+I+ FP + GLPEGCEN + + + +KF KST ++ LE +L
Sbjct: 63 PSFEIDIQIFDFPCVD-LGLPEGCENVDFFTSNNNDDRQYLTLKFFKSTRFFKDQLEKLL 121
Query: 109 EQEKPDCLVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEP 168
E +PDCL+ADMFFPW+T++A KFN+PR+VFHG G+F LC C R + PQ+ V+S EP
Sbjct: 122 ETTRPDCLIADMFFPWATEAAEKFNVPRLVFHGTGYFSLCSEYCIRVHNPQNIVASRYEP 181
Query: 169 FVVPNLPGEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHY 228
FV+P+LPG I +T+ Q+ + ++ K + E ES+VKS GVI N+FYELEP YAD Y
Sbjct: 182 FVIPDLPGNIVITQEQIADRDEESEM-GKFMIEVKESDVKSSGVIVNSFYELEPDYADFY 240
Query: 229 RNELGRKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTV 288
++ + ++AWH+GP+S+ NR EEKA RG++ASI+E ECLKWL SK+P+SVIY+ FGS+
Sbjct: 241 KSVVLKRAWHIGPLSVYNRGFEEKAERGKKASINEVECLKWLDSKKPDSVIYISFGSVAC 300
Query: 289 FSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAP 348
F + QL EIA GLE S FIWVVRK+ E E EWLPEGFEER++ GKG+IIRGWAP
Sbjct: 301 FKNEQLFEIAAGLETSGANFIWVVRKNIGIEKE--EWLPEGFEERVK--GKGMIIRGWAP 356
Query: 349 QVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGV 408
QV+ILDH++ GFVTHCGWNS LEGV+AGLPMVTWP+ EQFYN K ++ +++ GV VG
Sbjct: 357 QVLILDHQATCGFVTHCGWNSLLEGVAAGLPMVTWPVAAEQFYNEKLVTQVLRTGVSVGA 416
Query: 409 QTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSN 468
+ + G+ + ++ + KAVR ++VG+EA+E R RAK+ +MA+ AVE GGSS+ND ++
Sbjct: 417 KKNV-RTTGDFISREKVVKAVREVLVGEEADERRERAKKLAEMAKAAVE-GGSSFNDLNS 474
Query: 469 LIEDLKS 475
IE+ S
Sbjct: 475 FIEEFTS 481
>At2g15490 putative glucosyltransferase
Length = 460
Score = 464 bits (1195), Expect = e-131
Identities = 241/465 (51%), Positives = 315/465 (66%), Gaps = 21/465 (4%)
Query: 27 LARVFSSRGLKVTIVTTHLNVPLISRTIGKAK-------INIKTIKFPSPEETGLPEGCE 79
+A++F+ RG K T++TT +N ++ + I K I IK + FP E GLPEGCE
Sbjct: 1 MAKLFARRGAKSTLLTTPINAKILEKPIEAFKVQNPDLEIGIKILNFPCVE-LGLPEGCE 59
Query: 80 NSE------SALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADMFFPWSTDSAAKFN 133
N + + + D F+KF+ ST +++ LE +E KP LVADMFFPW+T+SA K
Sbjct: 60 NRDFINSYQKSDSFDLFLKFLFSTKYMKQQLESFIETTKPSALVADMFFPWATESAEKIG 119
Query: 134 IPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITLTKMQLPQLPQHDK 193
+PR+VFHG F LC R +KP KV+S + PFV+P LPG+I +T+ Q + +
Sbjct: 120 VPRLVFHGTSSFALCCSYNMRIHKPHKKVASSSTPFVIPGLPGDIVITEDQA-NVTNEET 178
Query: 194 VFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLGPVSLCNRDTEEKA 253
F K +E ESE SFGV+ N+FYELE YAD YR+ + +KAWH+GP+SL NR EKA
Sbjct: 179 PFGKFWKEVRESETSSFGVLVNSFYELESSYADFYRSFVAKKAWHIGPLSLSNRGIAEKA 238
Query: 254 CRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVR 313
RG++A+IDE ECLKWL SK P SV+Y+ FGS T + QL EIA GLE S FIWVV
Sbjct: 239 GRGKKANIDEQECLKWLDSKTPGSVVYLSFGSGTGLPNEQLLEIAFGLEGSGQNFIWVVS 298
Query: 314 KSAK--SEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTL 371
K+ GEN +WLP+GFEER GKGLIIRGWAPQV+ILDH+++GGFVTHCGWNSTL
Sbjct: 299 KNENQVGTGENEDWLPKGFEER--NKGKGLIIRGWAPQVLILDHKAIGGFVTHCGWNSTL 356
Query: 372 EGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPVKKDVIEKAVRR 431
EG++AGLPMVTWPM EQFYN K L+ +++IGV VG + G+ + + +EKAVR
Sbjct: 357 EGIAAGLPMVTWPMGAEQFYNEKLLTKVLRIGVNVGATELVKK--GKLISRAQVEKAVRE 414
Query: 432 IMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLKSR 476
++ G++AEE R RAKE G+MA+ AVE GGSSYND + +E+L R
Sbjct: 415 VIGGEKAEERRLRAKELGEMAKAAVEEGGSSYNDVNKFMEELNGR 459
>At4g34138 glucosyltransferase -like protein
Length = 488
Score = 457 bits (1175), Expect = e-129
Identities = 236/485 (48%), Positives = 334/485 (68%), Gaps = 27/485 (5%)
Query: 4 ENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISR---------TI 54
E +LH + FPF+A+GH+IP +D+A++F+++G K TI+TT LN L
Sbjct: 6 EVSKLHFLLFPFMAHGHMIPTLDMAKLFATKGAKSTILTTPLNAKLFFEKPIKSFNQDNP 65
Query: 55 GKAKINIKTIKFPSPEETGLPEGCENSESALA-PDKFI-----KFMKSTLLLREPLEHVL 108
G I I+ + FP E GLP+GCEN++ + PD + KF+ + EPLE +L
Sbjct: 66 GLEDITIQILNFPCTE-LGLPDGCENTDFIFSTPDLNVGDLSQKFLLAMKYFEEPLEELL 124
Query: 109 EQEKPDCLVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEP 168
+PDCLV +MFFPWST A KF +PR+VFHG G+F LC C R K V++ +EP
Sbjct: 125 VTMRPDCLVGNMFFPWSTKVAEKFGVPRLVFHGTGYFSLCASHCIRLPK---NVATSSEP 181
Query: 169 FVVPNLPGEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHY 228
FV+P+LPG+I +T+ Q+ + + + V + ++ +SE SFGV+ N+FYELE Y+D++
Sbjct: 182 FVIPDLPGDILITEEQVMET-EEESVMGRFMKAIRDSERDSFGVLVNSFYELEQAYSDYF 240
Query: 229 RNELGRKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTV 288
++ + ++AWH+GP+SL NR EEKA RG++ASIDEHECLKWL SK+ +SVIY+ FG+M+
Sbjct: 241 KSFVAKRAWHIGPLSLGNRKFEEKAERGKKASIDEHECLKWLDSKKCDSVIYMAFGTMSS 300
Query: 289 FSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAP 348
F + QL EIA GL+ S F+WVV + S+ E +WLPEGFEE+ + GKGLIIRGWAP
Sbjct: 301 FKNEQLIEIAAGLDMSGHDFVWVVNRKG-SQVEKEDWLPEGFEEKTK--GKGLIIRGWAP 357
Query: 349 QVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGV 408
QV+IL+H+++GGF+THCGWNS LEGV+AGLPMVTWP+ EQFYN K ++ ++K GV VGV
Sbjct: 358 QVLILEHKAIGGFLTHCGWNSLLEGVAAGLPMVTWPVGAEQFYNEKLVTQVLKTGVSVGV 417
Query: 409 QTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSN 468
+ + + G+ + ++ +E AVR +MVG EE R RAKE +MA+ AV+ GGSS +
Sbjct: 418 KKMMQV-VGDFISREKVEGAVREVMVG---EERRKRAKELAEMAKNAVKEGGSSDLEVDR 473
Query: 469 LIEDL 473
L+E+L
Sbjct: 474 LMEEL 478
>At2g15480 glucosyltransferase like protein
Length = 372
Score = 421 bits (1083), Expect = e-118
Identities = 205/377 (54%), Positives = 275/377 (72%), Gaps = 6/377 (1%)
Query: 100 LREPLEHVLEQEKPDCLVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQ 159
+++ LE +E KP LVADMFFPW+T+SA K +PR+VFHG FF LC R +KP
Sbjct: 1 MKQQLESFIETTKPSALVADMFFPWATESAEKLGVPRLVFHGTSFFSLCCSYNMRIHKPH 60
Query: 160 DKVSSYTEPFVVPNLPGEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYE 219
KV++ + PFV+P LPG+I +T+ Q + + + K ++E ESE SFGV+ N+FYE
Sbjct: 61 KKVATSSTPFVIPGLPGDIVITEDQA-NVAKEETPMGKFMKEVRESETNSFGVLVNSFYE 119
Query: 220 LEPVYADHYRNELGRKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVI 279
LE YAD YR+ + ++AWH+GP+SL NR+ EKA RG++A+IDE ECLKWL SK P SV+
Sbjct: 120 LESAYADFYRSFVAKRAWHIGPLSLSNRELGEKARRGKKANIDEQECLKWLDSKTPGSVV 179
Query: 280 YVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGK 339
Y+ FGS T F++ QL EIA GLE S FIWVVRK+ +++G+N EWLPEGF+ER +GK
Sbjct: 180 YLSFGSGTNFTNDQLLEIAFGLEGSGQSFIWVVRKN-ENQGDNEEWLPEGFKERT--TGK 236
Query: 340 GLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDI 399
GLII GWAPQV+ILDH+++GGFVTHCGWNS +EG++AGLPMVTWPM EQFYN K L+ +
Sbjct: 237 GLIIPGWAPQVLILDHKAIGGFVTHCGWNSAIEGIAAGLPMVTWPMGAEQFYNEKLLTKV 296
Query: 400 VKIGVGVGVQTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVG 459
++IGV VG + G+ + + +EKAVR ++ G++AEE R AK+ G+MA+ AVE G
Sbjct: 297 LRIGVNVGATELVKK--GKLISRAQVEKAVREVIGGEKAEERRLWAKKLGEMAKAAVEEG 354
Query: 460 GSSYNDFSNLIEDLKSR 476
GSSYND + +E+L R
Sbjct: 355 GSSYNDVNKFMEELNGR 371
>At2g36780 putative glucosyl transferase
Length = 496
Score = 413 bits (1062), Expect = e-116
Identities = 217/484 (44%), Positives = 315/484 (64%), Gaps = 18/484 (3%)
Query: 8 LHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKA-----KINIK 62
LH + FPF+A GH+IP +D+AR+ + RG+ +TIVTT N + +A INI
Sbjct: 13 LHFVLFPFMAQGHMIPMIDIARLLAQRGVTITIVTTPHNAARFKNVLNRAIESGLAINIL 72
Query: 63 TIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKP--DCLVADM 120
+KFP +E GLPEG EN +S + + + F K+ LL +P+ ++E+ KP CL++D
Sbjct: 73 HVKFPY-QEFGLPEGKENIDSLDSTELMVPFFKAVNLLEDPVMKLMEEMKPRPSCLISDW 131
Query: 121 FFPWSTDSAAKFNIPRIVFHGLGFFPL-CVLACTRQYKPQDKVSSYTEPFVVPNLPGEIT 179
P+++ A FNIP+IVFHG+G F L C+ R + + V S E F+VP+ P +
Sbjct: 132 CLPYTSIIAKNFNIPKIVFHGMGCFNLLCMHVLRRNLEILENVKSDEEYFLVPSFPDRVE 191
Query: 180 LTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHL 239
TK+QLP + ++++E ++E S+GVI NTF ELEP Y Y+ + K W +
Sbjct: 192 FTKLQLPVKANASGDWKEIMDEMVKAEYTSYGVIVNTFQELEPPYVKDYKEAMDGKVWSI 251
Query: 240 GPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAM 299
GPVSLCN+ +KA RG +A+ID+ ECL+WL SKE SV+YVC GS+ +QLKE+ +
Sbjct: 252 GPVSLCNKAGADKAERGSKAAIDQDECLQWLDSKEEGSVLYVCLGSICNLPLSQLKELGL 311
Query: 300 GLEASEVPFIWVVRKSAKSEGENLEWLPE-GFEERIEGSGKGLIIRGWAPQVMILDHESV 358
GLE S FIWV+R S K + E EW+ E GFEERI+ +GL+I+GWAPQV+IL H SV
Sbjct: 312 GLEESRRSFIWVIRGSEKYK-ELFEWMLESGFEERIK--ERGLLIKGWAPQVLILSHPSV 368
Query: 359 GGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGG-- 416
GGF+THCGWNSTLEG+++G+P++TWP++G+QF N K + ++K GV GV+ + G
Sbjct: 369 GGFLTHCGWNSTLEGITSGIPLITWPLFGDQFCNQKLVVQVLKAGVSAGVEEVMKWGEED 428
Query: 417 --GEPVKKDVIEKAVRRIM-VGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDL 473
G V K+ ++KAV +M D+A+E R R KE G++A +AVE GGSS+++ + L++D+
Sbjct: 429 KIGVLVDKEGVKKAVEELMGDSDDAKERRRRVKELGELAHKAVEKGGSSHSNITLLLQDI 488
Query: 474 KSRA 477
A
Sbjct: 489 MQLA 492
>At2g36790 putative glucosyl transferase
Length = 495
Score = 405 bits (1041), Expect = e-113
Identities = 214/489 (43%), Positives = 316/489 (63%), Gaps = 18/489 (3%)
Query: 3 NENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKA----- 57
NE LH + FPF+A GH+IP VD+AR+ + RG+ +TIVTT N + +A
Sbjct: 7 NEPFPLHFVLFPFMAQGHMIPMVDIARLLAQRGVLITIVTTPHNAARFKNVLNRAIESGL 66
Query: 58 KINIKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQ--EKPDC 115
IN+ +KFP +E GL EG EN + ++ F K+ LL+EP+++++E+ +P C
Sbjct: 67 PINLVQVKFPY-QEAGLQEGQENMDLLTTMEQITSFFKAVNLLKEPVQNLIEEMSPRPSC 125
Query: 116 LVADMFFPWSTDSAAKFNIPRIVFHGLGFFPL-CVLACTRQYKPQDKVSSYTEPFVVPNL 174
L++DM ++++ A KF IP+I+FHG+G F L CV + + D + S E F+VP
Sbjct: 126 LISDMCLSYTSEIAKKFKIPKILFHGMGCFCLLCVNVLRKNREILDNLKSDKEYFIVPYF 185
Query: 175 PGEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGR 234
P + T+ Q+P + ++LE+ E++ S+GVI N+F ELEP YA ++
Sbjct: 186 PDRVEFTRPQVPVETYVPAGWKEILEDMVEADKTSYGVIVNSFQELEPAYAKDFKEARSG 245
Query: 235 KAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQL 294
KAW +GPVSLCN+ +KA RG ++ ID+ ECL+WL SKEP SV+YVC GS+ +QL
Sbjct: 246 KAWTIGPVSLCNKVGVDKAERGNKSDIDQDECLEWLDSKEPGSVLYVCLGSICNLPLSQL 305
Query: 295 KEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPE-GFEERIEGSGKGLIIRGWAPQVMIL 353
E+ +GLE S+ PFIWV+R K + E +EW E GFE+RI+ +GL+I+GW+PQ++IL
Sbjct: 306 LELGLGLEESQRPFIWVIRGWEKYK-ELVEWFSESGFEDRIQ--DRGLLIKGWSPQMLIL 362
Query: 354 DHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIG 413
H SVGGF+THCGWNSTLEG++AGLPM+TWP++ +QF N K + I+K+GV V+ +
Sbjct: 363 SHPSVGGFLTHCGWNSTLEGITAGLPMLTWPLFADQFCNEKLVVQILKVGVSAEVKEVMK 422
Query: 414 MGGGEP----VKKDVIEKAVRRIM-VGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSN 468
G E V K+ ++KAV +M D+A+E R RAKE G+ A +AVE GGSS+++ +
Sbjct: 423 WGEEEKIGVLVDKEGVKKAVEELMGESDDAKERRRRAKELGESAHKAVEEGGSSHSNITF 482
Query: 469 LIEDLKSRA 477
L++D+ A
Sbjct: 483 LLQDIMQLA 491
>At2g36770 putative glucosyl transferase
Length = 496
Score = 400 bits (1028), Expect = e-112
Identities = 211/480 (43%), Positives = 305/480 (62%), Gaps = 18/480 (3%)
Query: 8 LHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAK-----INIK 62
LH I FPF+A GH+IP +D+AR+ + RG VTIVTT N + +A INI
Sbjct: 13 LHFILFPFMAQGHMIPMIDIARLLAQRGATVTIVTTRYNAGRFENVLSRAMESGLPINIV 72
Query: 63 TIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKP--DCLVADM 120
+ FP +E GLPEG EN +S + + + F ++ +L +P+ ++E+ KP C+++D+
Sbjct: 73 HVNFPY-QEFGLPEGKENIDSYDSMELMVPFFQAVNMLEDPVMKLMEEMKPRPSCIISDL 131
Query: 121 FFPWSTDSAAKFNIPRIVFHGLGFFPL-CVLACTRQYKPQDKVSSYTEPFVVPNLPGEIT 179
P+++ A KF+IP+IVFHG G F L C+ R + + S + F+VP+ P +
Sbjct: 132 LLPYTSKIARKFSIPKIVFHGTGCFNLLCMHVLRRNLEILKNLKSDKDYFLVPSFPDRVE 191
Query: 180 LTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHL 239
TK Q+P + L+E E+E S+GVI NTF ELEP Y Y K W +
Sbjct: 192 FTKPQVPVETTASGDWKAFLDEMVEAEYTSYGVIVNTFQELEPAYVKDYTKARAGKVWSI 251
Query: 240 GPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAM 299
GPVSLCN+ +KA RG +A+ID+ ECL+WL SKE SV+YVC GS+ +QLKE+ +
Sbjct: 252 GPVSLCNKAGADKAERGNQAAIDQDECLQWLDSKEDGSVLYVCLGSICNLPLSQLKELGL 311
Query: 300 GLEASEVPFIWVVRKSAKSEGENLEWLPE-GFEERIEGSGKGLIIRGWAPQVMILDHESV 358
GLE S+ FIWV+R K E EW+ E GFEERI+ +GL+I+GW+PQV+IL H SV
Sbjct: 312 GLEKSQRSFIWVIRGWEK-YNELYEWMMESGFEERIK--ERGLLIKGWSPQVLILSHPSV 368
Query: 359 GGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGE 418
GGF+THCGWNSTLEG+++G+P++TWP++G+QF N K + ++K GV GV+ + G E
Sbjct: 369 GGFLTHCGWNSTLEGITSGIPLITWPLFGDQFCNQKLVVQVLKAGVSAGVEEVMKWGEEE 428
Query: 419 P----VKKDVIEKAVRRIM-VGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDL 473
V K+ ++KAV +M D+A+E R R KE G+ A +AVE GGSS+++ + L++D+
Sbjct: 429 KIGVLVDKEGVKKAVEELMGASDDAKERRRRVKELGESAHKAVEEGGSSHSNITYLLQDI 488
>At2g36800 putative glucosyl transferase
Length = 495
Score = 398 bits (1023), Expect = e-111
Identities = 215/493 (43%), Positives = 314/493 (63%), Gaps = 23/493 (4%)
Query: 2 TNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKA---- 57
T ++ LH + FPF+A GH+IP VD+AR+ + RG+ +TIVTT N + +A
Sbjct: 5 TTKSSPLHFVLFPFMAQGHMIPMVDIARLLAQRGVIITIVTTPHNAARFKNVLNRAIESG 64
Query: 58 -KINIKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQ--EKPD 114
IN+ +KFP E GL EG EN +S ++ I F K+ L EP++ ++E+ +P
Sbjct: 65 LPINLVQVKFPYLE-AGLQEGQENIDSLDTMERMIPFFKAVNFLEEPVQKLIEEMNPRPS 123
Query: 115 CLVADMFFPWSTDSAAKFNIPRIVFHGLGFFPL-CVLACTRQYKPQDKVSSYTEPFVVPN 173
CL++D P+++ A KFNIP+I+FHG+G F L C+ + + D + S E F VP+
Sbjct: 124 CLISDFCLPYTSKIAKKFNIPKILFHGMGCFCLLCMHVLRKNREILDNLKSDKELFTVPD 183
Query: 174 LPGEITLTKMQLP---QLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRN 230
P + T+ Q+P +P D + + + E+ S+GVI N+F ELEP YA Y+
Sbjct: 184 FPDRVEFTRTQVPVETYVPAGD--WKDIFDGMVEANETSYGVIVNSFQELEPAYAKDYKE 241
Query: 231 ELGRKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFS 290
KAW +GPVSLCN+ +KA RG ++ ID+ ECLKWL SK+ SV+YVC GS+
Sbjct: 242 VRSGKAWTIGPVSLCNKVGADKAERGNKSDIDQDECLKWLDSKKHGSVLYVCLGSICNLP 301
Query: 291 DAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPE-GFEERIEGSGKGLIIRGWAPQ 349
+QLKE+ +GLE S+ PFIWV+R K + E +EW E GFE+RI+ +GL+I+GW+PQ
Sbjct: 302 LSQLKELGLGLEESQRPFIWVIRGWEKYK-ELVEWFSESGFEDRIQ--DRGLLIKGWSPQ 358
Query: 350 VMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQ 409
++IL H SVGGF+THCGWNSTLEG++AGLP++TWP++ +QF N K + +++K GV GV+
Sbjct: 359 MLILSHPSVGGFLTHCGWNSTLEGITAGLPLLTWPLFADQFCNEKLVVEVLKAGVRSGVE 418
Query: 410 TWIGMGGGEP----VKKDVIEKAVRRIM-VGDEAEEMRSRAKEFGKMARRAVEVGGSSYN 464
+ G E V K+ ++KAV +M D+A+E R RAKE G A +AVE GGSS++
Sbjct: 419 QPMKWGEEEKIGVLVDKEGVKKAVEELMGESDDAKERRRRAKELGDSAHKAVEEGGSSHS 478
Query: 465 DFSNLIEDLKSRA 477
+ S L++D+ A
Sbjct: 479 NISFLLQDIMELA 491
>At2g36750 putative glucosyl transferase
Length = 491
Score = 388 bits (997), Expect = e-108
Identities = 210/481 (43%), Positives = 306/481 (62%), Gaps = 21/481 (4%)
Query: 8 LHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKA-----KINIK 62
LH + FPF+A GH+IP VD+AR+ + RG+ +TIVTT N + +A IN+
Sbjct: 9 LHFVLFPFMAQGHMIPMVDIARLLAQRGVTITIVTTPQNAGRFKNVLSRAIQSGLPINLV 68
Query: 63 TIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLE--QEKPDCLVADM 120
+KFPS +E+G PEG EN + + + F K+ LL EP+E +L+ Q +P+C++ADM
Sbjct: 69 QVKFPS-QESGSPEGQENLDLLDSLGASLTFFKAFSLLEEPVEKLLKEIQPRPNCIIADM 127
Query: 121 FFPWSTDSAAKFNIPRIVFHGLGFFPL-CVLACTRQYKPQDKVSSYTEPFVVPNLPGEIT 179
P++ A IP+I+FHG+ F L C + ++ + + S E F +PN P +
Sbjct: 128 CLPYTNRIAKNLGIPKIIFHGMCCFNLLCTHIMHQNHEFLETIESDKEYFPIPNFPDRVE 187
Query: 180 LTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHL 239
TK QLP + + L+ E + S+GVI NTF ELEP Y Y+ K W +
Sbjct: 188 FTKSQLPMVLVAGD-WKDFLDGMTEGDNTSYGVIVNTFEELEPAYVRDYKKVKAGKIWSI 246
Query: 240 GPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAM 299
GPVSLCN+ E++A RG +A ID+ EC+KWL SKE SV+YVC GS+ +QLKE+ +
Sbjct: 247 GPVSLCNKLGEDQAERGNKADIDQDECIKWLDSKEEGSVLYVCLGSICNLPLSQLKELGL 306
Query: 300 GLEASEVPFIWVVRKSAKSEGENLEWLPE-GFEERIEGSGKGLIIRGWAPQVMILDHESV 358
GLE S+ PFIWV+R K E LEW+ E G++ERI+ +GL+I GW+PQ++IL H +V
Sbjct: 307 GLEESQRPFIWVIRGWEK-YNELLEWISESGYKERIK--ERGLLITGWSPQMLILTHPAV 363
Query: 359 GGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGE 418
GGF+THCGWNSTLEG+++G+P++TWP++G+QF N K I+K GV GV+ + G E
Sbjct: 364 GGFLTHCGWNSTLEGITSGVPLLTWPLFGDQFCNEKLAVQILKAGVRAGVEESMRWGEEE 423
Query: 419 P----VKKDVIEKAVRRIMVGD--EAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIED 472
V K+ ++KAV +M GD +A+E R R KE G++A +AVE GGSS+++ + L++D
Sbjct: 424 KIGVLVDKEGVKKAVEELM-GDSNDAKERRKRVKELGELAHKAVEEGGSSHSNITFLLQD 482
Query: 473 L 473
+
Sbjct: 483 I 483
>At2g36760 putative glucosyl transferase
Length = 496
Score = 387 bits (993), Expect = e-108
Identities = 202/482 (41%), Positives = 308/482 (62%), Gaps = 22/482 (4%)
Query: 8 LHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKA-----KINIK 62
LH + FPF+A GH+IP VD+AR+ + RG+ +TIVTT N + +A I ++
Sbjct: 13 LHFVLFPFMAQGHMIPMVDIARILAQRGVTITIVTTPHNAARFKDVLNRAIQSGLHIRVE 72
Query: 63 TIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQ--EKPDCLVADM 120
+KFP +E GL EG EN + + + + F K+ +L P+ ++E+ KP CL++D
Sbjct: 73 HVKFPF-QEAGLQEGQENVDFLDSMELMVHFFKAVNMLENPVMKLMEEMKPKPSCLISDF 131
Query: 121 FFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACT---RQYKPQDKVSSYTEPFVVPNLPGE 177
P+++ A +FNIP+IVFHG+ F C+L+ R + + S E F+VP+ P
Sbjct: 132 CLPYTSKIAKRFNIPKIVFHGVSCF--CLLSMHILHRNHNILHALKSDKEYFLVPSFPDR 189
Query: 178 ITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAW 237
+ TK+Q+ + ++++E +++ S+GVI NTF +LE Y +Y K W
Sbjct: 190 VEFTKLQVTVKTNFSGDWKEIMDEQVDADDTSYGVIVNTFQDLESAYVKNYTEARAGKVW 249
Query: 238 HLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEI 297
+GPVSLCN+ E+KA RG +A+ID+ EC+KWL SK+ SV+YVC GS+ AQL+E+
Sbjct: 250 SIGPVSLCNKVGEDKAERGNKAAIDQDECIKWLDSKDVESVLYVCLGSICNLPLAQLREL 309
Query: 298 AMGLEASEVPFIWVVRKSAKSEGENLEWLPE-GFEERIEGSGKGLIIRGWAPQVMILDHE 356
+GLEA++ PFIWV+R K E EW+ E GFEER + + L+I+GW+PQ++IL H
Sbjct: 310 GLGLEATKRPFIWVIRGGGKYH-ELAEWILESGFEERTK--ERSLLIKGWSPQMLILSHP 366
Query: 357 SVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGG 416
+VGGF+THCGWNSTLEG+++G+P++TWP++G+QF N K + ++K GV VGV+ + G
Sbjct: 367 AVGGFLTHCGWNSTLEGITSGVPLITWPLFGDQFCNQKLIVQVLKAGVSVGVEEVMKWGE 426
Query: 417 GEP----VKKDVIEKAVRRIM-VGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIE 471
E V K+ ++KAV IM DEA+E R R +E G++A +AVE GGSS+++ L++
Sbjct: 427 EESIGVLVDKEGVKKAVDEIMGESDEAKERRKRVRELGELAHKAVEEGGSSHSNIIFLLQ 486
Query: 472 DL 473
D+
Sbjct: 487 DI 488
>At3g53160 glucosyltransferase - like protein
Length = 490
Score = 376 bits (966), Expect = e-104
Identities = 206/484 (42%), Positives = 299/484 (61%), Gaps = 18/484 (3%)
Query: 8 LHIIFFPFLANGHIIPCVDLARVFSSR-GLKVTIVTTHLNVPLISRTIGK----AKINIK 62
LH + PF+A GH+IP VD++R+ S R G+ V I+TT NV I ++ A INI
Sbjct: 7 LHFVVIPFMAQGHMIPLVDISRLLSQRQGVTVCIITTTQNVAKIKTSLSFSSLFATINIV 66
Query: 63 TIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLE---QEKPDCLVAD 119
+KF S ++TGLPEGCE+ + + +KF + L E +E +E Q +P C++ D
Sbjct: 67 EVKFLS-QQTGLPEGCESLDMLASMGDMVKFFDAANSLEEQVEKAMEEMVQPRPSCIIGD 125
Query: 120 MFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEIT 179
M P+++ A KF IP+++FHG F L + R+ + S E F +P LP ++
Sbjct: 126 MSLPFTSRLAKKFKIPKLIFHGFSCFSLMSIQVVRESGILKMIESNDEYFDLPGLPDKVE 185
Query: 180 LTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHL 239
TK Q+ L + + + E++ S+GVI NTF ELE YA YR K W +
Sbjct: 186 FTKPQVSVLQPVEGNMKESTAKIIEADNDSYGVIVNTFEELEVDYAREYRKARAGKVWCV 245
Query: 240 GPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAM 299
GPVSLCNR +KA RG +ASI + +CL+WL S+E SV+YVC GS+ AQLKE+ +
Sbjct: 246 GPVSLCNRLGLDKAKRGDKASIGQDQCLQWLDSQETGSVLYVCLGSLCNLPLAQLKELGL 305
Query: 300 GLEASEVPFIWVVRKSAKSEGENLEWLPE-GFEERIEGSGKGLIIRGWAPQVMILDHESV 358
GLEAS PFIWV+R+ K G+ W+ + GFEERI+ +GL+I+GWAPQV IL H S+
Sbjct: 306 GLEASNKPFIWVIREWGK-YGDLANWMQQSGFEERIK--DRGLVIKGWAPQVFILSHASI 362
Query: 359 GGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGG-- 416
GGF+THCGWNSTLEG++AG+P++TWP++ EQF N K + I+K G+ +GV+ + G
Sbjct: 363 GGFLTHCGWNSTLEGITAGVPLLTWPLFAEQFLNEKLVVQILKAGLKIGVEKLMKYGKEE 422
Query: 417 --GEPVKKDVIEKAVRRIM-VGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDL 473
G V ++ + KAV +M +EAEE R + E +A +A+E GGSS ++ + LI+D+
Sbjct: 423 EIGAMVSRECVRKAVDELMGDSEEAEERRRKVTELSDLANKALEKGGSSDSNITLLIQDI 482
Query: 474 KSRA 477
++
Sbjct: 483 MEQS 486
>At3g53150 glucosyltransferase - like protein
Length = 507
Score = 358 bits (920), Expect = 3e-99
Identities = 199/498 (39%), Positives = 291/498 (57%), Gaps = 27/498 (5%)
Query: 1 MTNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKIN 60
+ ++ + LH + P +A GH+IP VD++++ + +G VTIVTT N ++T+ +A++
Sbjct: 5 IVSKAKRLHFVLIPLMAQGHLIPMVDISKILARQGNIVTIVTTPQNASRFAKTVDRARLE 64
Query: 61 ----IKTIKFPSP-EETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEK--P 113
I +KFP P +E GLP+ CE ++ + D +F + L+EP+E LEQ+ P
Sbjct: 65 SGLEINVVKFPIPYKEFGLPKDCETLDTLPSKDLLRRFYDAVDKLQEPMERFLEQQDIPP 124
Query: 114 DCLVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPN 173
C+++D W++ +A +F IPRIVFHG+ F L + P VSS EPF +P
Sbjct: 125 SCIISDKCLFWTSRTAKRFKIPRIVFHGMCCFSLLSSHNIHLHSPHLSVSSAVEPFPIPG 184
Query: 174 LPGEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELG 233
+P I + + QLP + + E+ ESE ++FGVI N+F ELEP YA+ Y +
Sbjct: 185 MPHRIEIARAQLPGAFEKLANMDDVREKMRESESEAFGVIVNSFQELEPGYAEAYAEAIN 244
Query: 234 RKAWHLGPVSLCNRDTEEKACRGREASI--DEHECLKWLQSKEPNSVIYVCFGSMTVFSD 291
+K W +GPVSLCN + RG +I E ECL++L S P SV+YV GS+
Sbjct: 245 KKVWFVGPVSLCNDRMADLFDRGSNGNIAISETECLQFLDSMRPRSVLYVSLGSLCRLIP 304
Query: 292 AQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLP-EGFEERIEGSGKGLIIRGWAPQV 350
QL E+ +GLE S PFIWV++ K E EWL E FEER+ G +G++I+GW+PQ
Sbjct: 305 NQLIELGLGLEESGKPFIWVIKTEEKHMIELDEWLKRENFEERVRG--RGIVIKGWSPQA 362
Query: 351 MILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQT 410
MIL H S GGF+THCGWNST+E + G+PM+TWP++ EQF N K + +++ IGV VGV+
Sbjct: 363 MILSHGSTGGFLTHCGWNSTIEAICFGVPMITWPLFAEQFLNEKLIVEVLNIGVRVGVEI 422
Query: 411 WIGMGG----GEPVKKDVIEKAVRRIMVGD-----------EAEEMRSRAKEFGKMARRA 455
+ G G VKK + KA++ +M D E R R +E MA++A
Sbjct: 423 PVRWGDEERLGVLVKKPSVVKAIKLLMDQDCQRVDENDDDNEFVRRRRRIQELAVMAKKA 482
Query: 456 VEVGGSSYNDFSNLIEDL 473
VE GSS + S LI+D+
Sbjct: 483 VEEKGSSSINVSILIQDV 500
>At5g14860 glucosyltransferase -like protein
Length = 492
Score = 259 bits (663), Expect = 2e-69
Identities = 170/493 (34%), Positives = 264/493 (53%), Gaps = 46/493 (9%)
Query: 9 HIIFFPFLANGHIIPCVDLARVF-----------SSRGLKVTIVTTHLNVPLISRTIGKA 57
H + FP+++ GH IP + AR+ + VT+ TT N P +S +
Sbjct: 8 HAVLFPYMSKGHTIPLLQFARLLLRHRRIVSVDDEEPTISVTVFTTPKNQPFVSNFLSDV 67
Query: 58 KINIKTIKFPSPEE-TGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQ-EKPDC 115
+IK I P PE G+P G E+++ + ++ F ++T L+ E L+ EK
Sbjct: 68 ASSIKVISLPFPENIAGIPPGVESTDMLPSISLYVPFTRATKSLQPFFEAELKNLEKVSF 127
Query: 116 LVADMFFPWSTDSAAKFNIPRIVFHGLGFFP--LC-VLACTRQYKPQDKVSSYTEPFVVP 172
+V+D F W+++SAAKF IPR+ F+G+ + +C ++ + + V S TEP VP
Sbjct: 128 MVSDGFLWWTSESAAKFEIPRLAFYGMNSYASAMCSAISVHELFTKPESVKSDTEPVTVP 187
Query: 173 NLPGEITLTKMQ----LPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADH- 227
+ P I + K + L + Q D F L++ ++ KS GVI N+FYELE + D+
Sbjct: 188 DFPW-ICVKKCEFDPVLTEPDQSDPAFELLIDHLMSTK-KSRGVIVNSFYELESTFVDYR 245
Query: 228 YRNELGRKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSK--EPNSVIYVCFGS 285
R+ K W +GP+ L N E D+ + + WL K E V+YV FG+
Sbjct: 246 LRDNDEPKPWCVGPLCLVNPPKPES---------DKPDWIHWLDRKLEERCPVMYVAFGT 296
Query: 286 MTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRG 345
S+ QLKEIA+GLE S+V F+WV RK + L GFE+R++ G +I+R
Sbjct: 297 QAEISNEQLKEIALGLEDSKVNFLWVTRKDLEEVTGGL-----GFEKRVKEHG--MIVRD 349
Query: 346 WAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVG 405
W Q IL H+SV GF++HCGWNS E + AG+P++ WPM EQ NAK + + +KIGV
Sbjct: 350 WVDQWEILSHKSVKGFLSHCGWNSAQESICAGVPLLAWPMMAEQPLNAKLVVEELKIGVR 409
Query: 406 VGVQTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVG-GSSYN 464
+ + G V ++ + + V+++M G+ + KE+ KMA++A+ G GSS+
Sbjct: 410 IETEDVSVKGF---VTREELSRKVKQLMEGEMGKTTMKNVKEYAKMAKKAMAQGTGSSWK 466
Query: 465 DFSNLIEDL-KSR 476
+L+E+L KSR
Sbjct: 467 SLDSLLEELCKSR 479
>At2g16890 glucosyltransferase like protein
Length = 478
Score = 258 bits (658), Expect = 7e-69
Identities = 174/487 (35%), Positives = 258/487 (52%), Gaps = 43/487 (8%)
Query: 9 HIIFFPFLANGHIIPCVDLARVFSSRGLK-----VTIVTTHLNVPLISRTIGKAKINIKT 63
H++ FPF++ GHIIP + R+ K VT+ TT N P IS + IK
Sbjct: 9 HVVLFPFMSKGHIIPLLQFGRLLLRHHRKEPTITVTVFTTPKNQPFISDFLSDTP-EIKV 67
Query: 64 IKFPSPEE-TGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQ-EKPDCLVADMF 121
I P PE TG+P G EN+E + F+ F ++T LL+ E L+ K +V+D F
Sbjct: 68 ISLPFPENITGIPPGVENTEKLPSMSLFVPFTRATKLLQPFFEETLKTLPKVSFMVSDGF 127
Query: 122 FPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYK----PQDKVSSYTEPFVVPNLPGE 177
W+++SAAKFNIPR V +G+ + V +++ P+ K S TEP VP+ P
Sbjct: 128 LWWTSESAAKFNIPRFVSYGMNSYSAAVSISVFKHELFTEPESK--SDTEPVTVPDFPW- 184
Query: 178 ITLTKMQLPQ---LPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGR 234
I + K P+ +L + +S S G + N+FYELE + D+ N +
Sbjct: 185 IKVKKCDFDHGTTEPEESGAALELSMDQIKSTTTSHGFLVNSFYELESAFVDYNNNSGDK 244
Query: 235 -KAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSK--EPNSVIYVCFGSMTVFSD 291
K+W +GP+ L + + A + + WL K E V+YV FG+ S+
Sbjct: 245 PKSWCVGPLCLTDPPKQGSA---------KPAWIHWLDQKREEGRPVLYVAFGTQAEISN 295
Query: 292 AQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVM 351
QL E+A GLE S+V F+WV RK + E + EGF +RI SG +I+R W Q
Sbjct: 296 KQLMELAFGLEDSKVNFLWVTRKDVE------EIIGEGFNDRIRESG--MIVRDWVDQWE 347
Query: 352 ILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTW 411
IL HESV GF++HCGWNS E + G+P++ WPM EQ NAK + + +K VGV V+T
Sbjct: 348 ILSHESVKGFLSHCGWNSAQESICVGVPLLAWPMMAEQPLNAKMVVEEIK--VGVRVETE 405
Query: 412 IGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRA-VEVGGSSYNDFSNLI 470
G G ++++ K ++ +M G+ + R KE+ KMA+ A VE GSS+ + ++
Sbjct: 406 DGSVKGFVTREELSGK-IKELMEGETGKTARKNVKEYSKMAKAALVEGTGSSWKNLDMIL 464
Query: 471 EDL-KSR 476
++L KSR
Sbjct: 465 KELCKSR 471
>At1g10400 glucosyl transferase - like protein
Length = 467
Score = 238 bits (608), Expect = 4e-63
Identities = 161/490 (32%), Positives = 258/490 (51%), Gaps = 51/490 (10%)
Query: 4 ENQELHIIFFPFLANGHIIPCVDLARVFSSRG----LKVTIVTTHLNVPLISRTIGKAKI 59
E +++H++ FP+L+ GH+IP + LAR+ S + VT+ TT LN P I ++ K
Sbjct: 2 ELEKVHVVLFPYLSKGHMIPMLQLARLLLSHSFAGDISVTVFTTPLNRPFIVDSLSGTKA 61
Query: 60 NIKTIKFPS--PEETGLPEG--CENSESALAPDKFIKFMKSTLLLREPLEH-VLEQEKPD 114
I + FP PE +P G C + AL+ F+ F ++T ++ E ++ +
Sbjct: 62 TIVDVPFPDNVPE---IPPGVECTDKLPALSSSLFVPFTRATKSMQADFERELMSLPRVS 118
Query: 115 CLVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNL 174
+V+D F W+ +SA K PR+VF G+ + Q + V S TEP VP
Sbjct: 119 FMVSDGFLWWTQESARKLGFPRLVFFGMNCASTVICDSVFQNQLLSNVKSETEPVSVPEF 178
Query: 175 PGEITLTKMQLPQLPQHDKVFT----KLLEESNESEVKSFGVIANTFYELEPVYADHYRN 230
P I + K + K T KL+ + S +S G+I NTF +LEPV+ D Y+
Sbjct: 179 PW-IKVRKCDFVKDMFDPKTTTDPGFKLILDQVTSMNQSQGIIFNTFDDLEPVFIDFYKR 237
Query: 231 ELGRKAWHLGPVSLCNR----DTEEKACRGREASIDEHECLKWLQSKEPN--SVIYVCFG 284
+ K W +GP+ N + EEK + +KWL K +V+YV FG
Sbjct: 238 KRKLKLWAVGPLCYVNNFLDDEVEEKV---------KPSWMKWLDEKRDKGCNVLYVAFG 288
Query: 285 SMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIR 344
S S QL+EIA+GLE S+V F+WVV+ + + +GFEER+ +G+++R
Sbjct: 289 SQAEISREQLEEIALGLEESKVNFLWVVKGNE---------IGKGFEERV--GERGMMVR 337
Query: 345 G-WAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIG 403
W Q IL+HESV GF++HCGWNS E + + +P++ +P+ EQ NA + + +++
Sbjct: 338 DEWVDQRKILEHESVRGFLSHCGWNSLTESICSEVPILAFPLAAEQPLNAILVVEELRVA 397
Query: 404 VGVGVQTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVG-GSS 462
V + G ++++ EK V+ +M G++ +E+R + +GKMA++A+E G GSS
Sbjct: 398 ERV-----VAASEGVVRREEIAEK-VKELMEGEKGKELRRNVEAYGKMAKKALEEGIGSS 451
Query: 463 YNDFSNLIED 472
+ NLI +
Sbjct: 452 RKNLDNLINE 461
>At1g73880 putative glucosyltransferase (At1g73880)
Length = 473
Score = 237 bits (605), Expect = 9e-63
Identities = 154/483 (31%), Positives = 239/483 (48%), Gaps = 32/483 (6%)
Query: 3 NENQELHIIFFPFLANGHIIPCVDLARVFSSRG---LKVTIVTTHLNVPLISRTIGKAKI 59
N+ + H++ FPF A GH+IP +D + RG LK+T++ T N+P +S + A +
Sbjct: 8 NKPTKTHVLIFPFPAQGHMIPLLDFTHRLALRGGAALKITVLVTPKNLPFLSPLLS-AVV 66
Query: 60 NIKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEK--PDCLV 117
NI+ + P P +P G EN + L P F + + L PL + P +V
Sbjct: 67 NIEPLILPFPSHPSIPSGVENVQD-LPPSGFPLMIHALGNLHAPLISWITSHPSPPVAIV 125
Query: 118 ADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKP----QDKVSSYTEPFVVPN 173
+D F W+ + IPR F C+L P +D + +PN
Sbjct: 126 SDFFLGWTKN----LGIPRFDFSPSAAITCCILNTLWIEMPTKINEDDDNEILHFPKIPN 181
Query: 174 LPGEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELG 233
P L + H + + +S V S+G++ N+F +E VY +H + E+G
Sbjct: 182 CPKYRFDQISSLYRSYVHGDPAWEFIRDSFRDNVASWGLVVNSFTAMEGVYLEHLKREMG 241
Query: 234 R-KAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDA 292
+ W +GP+ + D RG S+ + WL ++E N V+YVCFGS V +
Sbjct: 242 HDRVWAVGPIIPLSGDN-----RGGPTSVSVDHVMSWLDAREDNHVVYVCFGSQVVLTKE 296
Query: 293 QLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMI 352
Q +A GLE S V FIW V++ + + L +GF++R+ +G+GL+IRGWAPQV +
Sbjct: 297 QTLALASGLEKSGVHFIWAVKEPVEKDSTRGNIL-DGFDDRV--AGRGLVIRGWAPQVAV 353
Query: 353 LDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWI 412
L H +VG F+THCGWNS +E V AG+ M+TWPM +Q+ +A + D +K+GV
Sbjct: 354 LRHRAVGAFLTHCGWNSVVEAVVAGVLMLTWPMRADQYTDASLVVDELKVGVRA------ 407
Query: 413 GMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIED 472
G D + + + G++ E R +A E K A A++ GSS ND I+
Sbjct: 408 CEGPDTVPDPDELARVFADSVTGNQTE--RIKAVELRKAALDAIQERGSSVNDLDGFIQH 465
Query: 473 LKS 475
+ S
Sbjct: 466 VVS 468
>At5g03490 UDPG glucosyltransferase - like protein
Length = 465
Score = 224 bits (570), Expect = 1e-58
Identities = 150/474 (31%), Positives = 242/474 (50%), Gaps = 37/474 (7%)
Query: 9 HIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKINIKTIKFPS 68
HI+ FPF A GH++P +DL RG V+++ T N+ +S + ++ ++ FP
Sbjct: 19 HIVVFPFPAQGHLLPLLDLTHQLCLRGFNVSVIVTPGNLTYLSPLLSAHPSSVTSVVFPF 78
Query: 69 PEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEK--PDCLVADMFFPWST 126
P L G EN + + + M S LREP+ + + P L++D F W+
Sbjct: 79 PPHPSLSPGVENVKD-VGNSGNLPIMASLRQLREPIINWFQSHPNPPIALISDFFLGWTH 137
Query: 127 DSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITLTKMQLP 186
D + IPR F + FF + VL ++ D + S T+P + +LP + LP
Sbjct: 138 DLCNQIGIPRFAFFSISFFLVSVLQFC--FENIDLIKS-TDPIHLLDLPRAPIFKEEHLP 194
Query: 187 QLPQHD-KVFTKLLEESNESEVK--SFGVIANTFYELEPVYADHYRNELGR-KAWHLGPV 242
+ + + + LE + + S+G + N+ LE Y + + +G + + +GP
Sbjct: 195 SIVRRSLQTPSPDLESIKDFSMNLLSYGSVFNSSEILEDDYLQYVKQRMGHDRVYVIGP- 253
Query: 243 SLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLE 302
LC+ + K+ G S+D L WL SV+YVCFGS + Q +A+GLE
Sbjct: 254 -LCSIGSGLKSNSG---SVDP-SLLSWLDGSPNGSVLYVCFGSQKALTKDQCDALALGLE 308
Query: 303 ASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFV 362
S F+WVV+K +P+GFE+R+ SG+GL++RGW Q+ +L H +VGGF+
Sbjct: 309 KSMTRFVWVVKKDP---------IPDGFEDRV--SGRGLVVRGWVSQLAVLRHVAVGGFL 357
Query: 363 THCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPV-K 421
+HCGWNS LEG+++G ++ WPM +QF NA+ L + +GV V V GGE V
Sbjct: 358 SHCGWNSVLEGITSGAVILGWPMEADQFVNARLL--VEHLGVAVRV-----CEGGETVPD 410
Query: 422 KDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAV-EVGGSSYNDFSNLIEDLK 474
D + + + M G+ E+ +RA+E + AV E GSS + L+++ +
Sbjct: 411 SDELGRVIAETM-GEGGREVAARAEEIRRKTEAAVTEANGSSVENVQRLVKEFE 463
>At1g51210 putative glucosyl transferase
Length = 433
Score = 221 bits (563), Expect = 7e-58
Identities = 146/447 (32%), Positives = 224/447 (49%), Gaps = 39/447 (8%)
Query: 9 HIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKINIKTIKFPS 68
HI+ FP+ A GH++P +DL RGL V+I+ T N+P +S + + + P
Sbjct: 20 HIMVFPYPAQGHLLPLLDLTHQLCLRGLTVSIIVTPKNLPYLSPLLSAHPSAVSVVTLPF 79
Query: 69 PEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEK--PDCLVADMFFPWST 126
P +P G EN + + M S LREP+ + L P L++D F W+
Sbjct: 80 PHHPLIPSGVENVKDLGGYGNPL-IMASLRQLREPIVNWLSSHPNPPVALISDFFLGWTK 138
Query: 127 DSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITLTKMQLP 186
D IPR F G F +L KP + TEP + +LP LP
Sbjct: 139 D----LGIPRFAFFSSGAFLASILHFVSD-KPH--LFESTEPVCLSDLPRSPVFKTEHLP 191
Query: 187 QL-PQHDKVFTKLLEESNESEVK--SFGVIANTFYELEPVYADHYRNELGR-KAWHLGPV 242
L PQ ++ LE +S + S+G I NT LE Y ++ + ++ + + +GP+
Sbjct: 192 SLIPQSP--LSQDLESVKDSTMNFSSYGCIFNTCECLEEDYMEYVKQKVSENRVFGVGPL 249
Query: 243 SLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLE 302
S E+ +++D L WL +SV+Y+CFGS V + Q ++A+GLE
Sbjct: 250 SSVGLSKEDSV-----SNVDAKALLSWLDGCPDDSVLYICFGSQKVLTKEQCDDLALGLE 304
Query: 303 ASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFV 362
S F+WVV+K +P+GFE+R+ +G+G+I+RGWAPQV +L H +VGGF+
Sbjct: 305 KSMTRFVWVVKKDP---------IPDGFEDRV--AGRGMIVRGWAPQVAMLSHVAVGGFL 353
Query: 363 THCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPVKK 422
HCGWNS LE +++G ++ WPM +QF +A+ + + +GV V V GG+ V
Sbjct: 354 IHCGWNSVLEAMASGTMILAWPMEADQFVDARLV--VEHMGVAVSV-----CEGGKTVPD 406
Query: 423 DVIEKAVRRIMVGDEAEEMRSRAKEFG 449
+ +G+ E R+RAKE G
Sbjct: 407 PYEMGRIIADTMGESGGEARARAKEMG 433
>At1g22340 hypothetical protein
Length = 487
Score = 204 bits (520), Expect = 7e-53
Identities = 147/495 (29%), Positives = 249/495 (49%), Gaps = 56/495 (11%)
Query: 6 QELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKINIKTIK 65
Q+ H++ P+ A GHI P + +A++ ++G VT V T N + R+ G ++
Sbjct: 10 QKPHVVCVPYPAQGHINPMLKVAKLLYAKGFHVTFVNTLYNHNRLLRSRGPNALD----G 65
Query: 66 FPSPEETGLPEGCENSE---SALAPDKFIKFMKSTLLLREPLEHVLEQ--EKPD-----C 115
FPS +P+G ++ + P + K+ L P + +L + +K D C
Sbjct: 66 FPSFRFESIPDGLPETDGDRTQHTPTVCMSIEKNCLA---PFKEILRRINDKDDVPPVSC 122
Query: 116 LVADMFFPWSTDSAAKFNIPRIVF---HGLGF-----FPLCVLACTRQYKPQDKVSSYTE 167
+V+D ++ D+A + +P ++F GF F L + +K + +S
Sbjct: 123 IVSDGVMSFTLDAAEELGVPEVIFWTNSACGFMTILHFYLFIEKGLSPFKDESYMSKEHL 182
Query: 168 PFVVPNLPGEITLTKMQLPQLPQH------DKVFTKLLEESNESEVKSFGVIANTFYELE 221
V+ +P ++ ++L +P + D + L E ++ +I NTF ELE
Sbjct: 183 DTVIDWIP---SMKNLRLKDIPSYIRTTNPDNIMLNFLIREVERSKRASAIILNTFDELE 239
Query: 222 PVYADHYRNELGRKAWHLGPVSLCNRDTEEKACRGREASI----DEHECLKWLQSKEPNS 277
++ L + +GP+ L ++ +A + + +E ECL WL +K PNS
Sbjct: 240 HDVIQSMQSILP-PVYSIGPLHLLVKEEINEASEIGQMGLNLWREEMECLDWLDTKTPNS 298
Query: 278 VIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGF-EERIEG 336
V++V FG +TV S QL+E A GL AS F+WV+R + GE + LP+ F E I+
Sbjct: 299 VLFVNFGCITVMSAKQLEEFAWGLAASRKEFLWVIRPNLVV-GEAMVVLPQEFLAETIDR 357
Query: 337 SGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFL 396
++ W PQ +L H ++GGF+THCGWNSTLE ++ G+PM+ WP + EQ N KF
Sbjct: 358 R----MLASWCPQEKVLSHPAIGGFLTHCGWNSTLESLAGGVPMICWPCFSEQPTNCKFC 413
Query: 397 SDIVKIGVGVGVQTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAV 456
D + GVG+ + G+ VK++ +E VR +M G++ +++R +A+E+ ++A A
Sbjct: 414 CD--EWGVGIEI--------GKDVKREEVETVVRELMDGEKGKKLREKAEEWRRLAEEAT 463
Query: 457 EV-GGSSYNDFSNLI 470
GSS + LI
Sbjct: 464 RYKHGSSVMNLETLI 478
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,065,196
Number of Sequences: 26719
Number of extensions: 489344
Number of successful extensions: 1735
Number of sequences better than 10.0: 122
Number of HSP's better than 10.0 without gapping: 117
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1174
Number of HSP's gapped (non-prelim): 155
length of query: 478
length of database: 11,318,596
effective HSP length: 103
effective length of query: 375
effective length of database: 8,566,539
effective search space: 3212452125
effective search space used: 3212452125
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC136449.6


